Team:Tokyo Tech/Experiment/Inducible Plaque Forming Assay
From 2013.igem.org
Shinya0825 (Talk | contribs) |
|||
| (67 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{tokyotechmenudark}} | {{tokyotechmenudark}} | ||
<div id="text-area"><br> | <div id="text-area"><br> | ||
| - | + | <div class="box" id="title"> | |
| - | + | <p style="line-height:0em; text-indent:0em;" name="top">Inducible Plaque Forming Assay</p> | |
| - | < | + | </div> |
| - | </ | + | <div class="box"> |
| + | <h1>1. Introduction</h1> | ||
<h2> | <h2> | ||
| - | <p>Our previous assay revealed that the M13 phage genes and the M13 origin worked together with the pSB origin. Next, we confirmed whether the release of the M13 phage particle depends on the induction of <i>g2p</i> expression. We inserted <i>lux</i> promoter (which is activated by | + | <p>Our previous assay revealed that the M13 phage genes and the M13 origin worked together with the pSB origin. Next, we confirmed whether the release of the M13 phage particle depends on the induction of <i>g2p</i> expression. We inserted <i>lux</i> promoter (which is activated by 3OC6HSL-LuxR complex) upstream of <i>g2p</i>, as an inducible promoter (Fig. 3-4-1)([https://2013.igem.org/Team:Tokyo_Tech/Project/M13_supplement#1._M13_plasmid_construction See more about our plasmid construction]). The method for our assay is briefly shown in Fig. 3-4-2. First, we cultured <i>E. coli</i> to which was introduced our new part ([http://parts.igem.org/Part:BBa_K1139021 BBa_K1139021]), and added 3OC6HSL, which induces the phage release. Second, we centrifuged the culture and obtained phage particles. Finally, we added the phage particles and lawn culture (<i>E. coli</i> containing pSB6A1-Ptet-<i>luxR</i>) to the soft agar, and poured it on a YT plate. Moreover, we confirmed that the inducible release of M13 phage was also realized with BAD promoter. |
</p> | </p> | ||
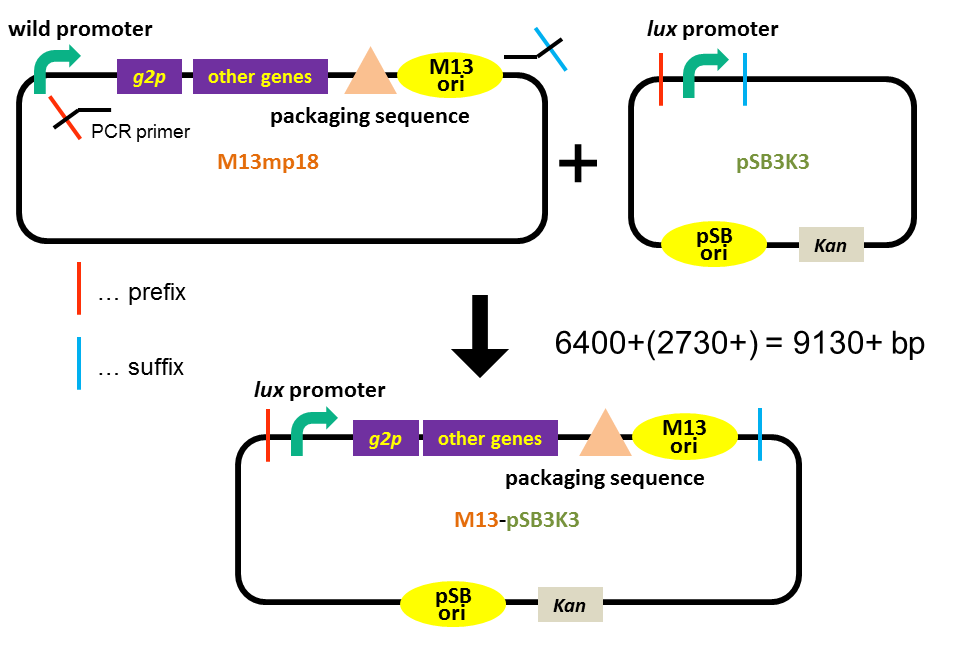
| - | [[Image:Titech2013_pSB-M13_Phage_replication_Fig.1.PNG|600px|thumb|center|Fig. 1. Our plasmid construction]] | + | [[Image:Titech2013_pSB-M13_Phage_replication_Fig.1.PNG|600px|thumb|center|Fig. 3-4-1. Our plasmid construction]] |
| - | [[Image: | + | [[Image:Titech2013_inducible_Fig1.png|600px|thumb|center|Fig. 3-4-2. How to perform our inducible phage release assay]] |
</h2> | </h2> | ||
| - | < | + | <h1>2. Materials and Methods</h1> |
| - | </h3> | + | <h3>2-1. Plasmid construction</h3> |
<h2> | <h2> | ||
| - | + | <p>pSB3K3-Plux-M13-Plac-<i>GFP</i> ([http://parts.igem.org/Part:BBa_K1139021 BBa_K1139021]) | |
| - | <p>pSB3K3-Plux-M13-Plac-<i>GFP</i> ( | + | |
</p> | </p> | ||
| - | <p>pSB6A1-Ptet-<i>luxR</i> | + | <p>pSB6A1-Ptet-<i>luxR</i> ([http://parts.igem.org/Part:BBa_S03119 BBa_S03119]) |
| - | + | ||
| - | + | ||
| - | + | ||
</p> | </p> | ||
| - | <p> | + | <p>pSB3K3-Pbad-M13-Plac-<i>GFP</i> ([http://parts.igem.org/Part:BBa_K1139026 BBa_K1139026]) |
| - | + | ||
| - | + | ||
| - | + | ||
</p> | </p> | ||
| - | <p> | + | <p>pSB6A1-Ptet-<i>araC</i> ([http://parts.igem.org/Part:BBa_I13458 BBa_I13458]) |
</p> | </p> | ||
| - | <p> | + | </h2> |
| + | <h3>2-2. Strains</h3> | ||
| + | <h2> | ||
| + | <p>DH5α (<i>E. coli</i> of high competence) | ||
</p> | </p> | ||
| - | <p> | + | <p>JM109 (F+ strain <i>E. coli</i>) |
| - | </p> | + | </p><br> |
| - | <br>< | + | </h2> |
| - | </ | + | <h3>2-3. Media</h3> |
| - | < | + | <h2> |
| - | + | <div style="margin-left:80px;"> | |
| - | + | LB | |
| - | + | {| class="wikitable" cellpadding="6" | |
| - | + | |Bacto tryptone||10 g/L | |
| - | + | |- | |
| - | + | |Yeast Extract||5 g/L | |
| - | + | |- | |
| - | + | |NaCl||10 g/L | |
| - | + | |} | |
| - | + | ||
| - | + | YT soft agar | |
| - | + | {| class="wikitable" cellpadding="6" | |
| - | + | |Bacto tryptone||8 g/L | |
| - | + | |- | |
| - | + | |Yeast Extract||5 g/L | |
| - | + | |- | |
| - | </ | + | |NaCl||5 g/L |
| - | <br>2-4 Others | + | |- |
| - | <p>3OC6HSL dissolved in DMSO (>100 | + | |Agarose||6 g/L |
| + | |} | ||
| + | |||
| + | YT plate | ||
| + | {| class="wikitable" cellpadding="6" | ||
| + | |Bacto tryptone||8 g/L | ||
| + | |- | ||
| + | |Yeast Extract||5 g/L | ||
| + | |- | ||
| + | |NaCl||5 g/L | ||
| + | |- | ||
| + | |Agarose||15 g/L | ||
| + | |} | ||
| + | </div> | ||
| + | <br> | ||
| + | </h2> | ||
| + | <h3>2-4. Others</h3> | ||
| + | <h2> | ||
| + | <p>3OC6HSL dissolved in DMSO (>100 µM) | ||
</p> | </p> | ||
<p>Autoclaved pieces of filter paper (about 1.5 cm in diameter) | <p>Autoclaved pieces of filter paper (about 1.5 cm in diameter) | ||
</p><br> | </p><br> | ||
| - | 2-5 Protocol | + | </h2> |
| - | < | + | <h3>2-5. Protocol</h3> |
| - | < | + | <h2> |
| + | <ol> | ||
| + | <em align="center">Preparation</em> | ||
| + | <li> Transform DH5α with pSB3K3-Plux-M13-Plac-<i>GFP</i> and pSB6A1-Ptet-<i>luxR</i>([http://parts.igem.org/Part:BBa_S03119 BBa_S03119]). | ||
| - | < | + | <li> Prepare overnight cultures of the transformed DH5α and JM109, DH5α in LB containing ampicillin (50 µg/mL) and kanamycin (30 µg/mL). |
| - | < | + | <li> Take 30 µL of the overnight cultures of the transformed DH5α into 3 mL LB medium containing ampicillin (50 µg/mL) and kanamycin (30 µg/mL) . (->fresh cultures) |
| - | < | + | <li> Incubate the fresh cultures at 37°C for 2 h. |
| - | < | + | <li> Add 3 µL of 5 µM 3OC6HSL in DMSO (final concentration of 5 nM) to the fresh cultures and incubate them at 37°C for 4 h. |
| - | < | + | <li> Centrifuge the overnight cultures of the transformed DH5α at 9,000g for 1 min. |
| - | < | + | <li> Pipette the supernatant into a 1.5 mL tube. |
| - | < | + | <li> Dilute it 100 times with water. (-> phage-particle-solution) |
| - | <br> | + | <br><br> |
| - | < | + | <em align="center">Plaque formation</em> |
| - | + | ||
| - | < | + | <li> Transform JM109 with pSB6A1-Ptet-<i>luxR</i>([http://parts.igem.org/Part:BBa_S03119 BBa_S03119]). |
| - | < | + | <li> Prepare overnight cultures of the transformed JM109 at 37°C. |
| - | < | + | <li> Melt the YT soft agar by using a microwave. |
| - | < | + | <li> Add ampicillin to the YT soft agar. |
| - | < | + | <li> Dispense 3.5 mL of the melted soft agar to a new round tube, and keep them at 50°C in an incubator. |
| - | < | + | <li> Dispense 400 µL of the overnight cultures of the transformed JM109 to a 1.5 mL tube. |
| - | < | + | <li> Into the 1.5 mL tube, add 100 µL of the phage-particle-solution. |
| - | < | + | <li> Transfer all the content of the 1.5 mL tube to the dispensed soft agar, mix them, and spread all of it on a YT plate. |
| - | < | + | <li> Wait for the YT soft agar to solidify at room temperature (for about 5 min.). |
| - | </h2> | + | |
| - | < | + | <li> Put an autoclaved piece of filter paper on the plate, and drip 20 mL of 3OC6HSL in DMSO (100 µM, 5 µM or DMSO only) into the piece of filter paper. |
| - | </ | + | </ol></h2> |
| + | <h1>3. Results</h1> | ||
<h2> | <h2> | ||
| - | <p>The result of the plaque forming assay is shown in Fig 3. On the plate, the plaques were formed at some distance from the piece of filter paper. The result shows that the phage release is regulated by the induction of 3OC6HSL. | + | <p>The result of the plaque forming assay is shown in Fig 3-4-3, 3-4-4. On the plate, the plaques were formed at some distance from the piece of filter paper. The result shows that the phage release is regulated by the induction of 3OC6HSL. |
| - | We also analyzed the distribution of the plaques (Fig. 4). The histogram result implicates that the plaques are formed only at the appropriate concentration of 3OC6HSL in the medium (Fig | + | We also analyzed the distribution of the plaques (Fig. 3-4-5, 3-4-6). The histogram result implicates that the plaques are formed only at the appropriate concentration of 3OC6HSL in the medium (Fig. 3-4-7). In the neighborhood of the piece of filter paper, the expression of <i>g2p</i> is so activated that the phage particles cannot be produced efficiently, because the production of the single stranded DNA largely exceeds that of the coat proteins. In contrast, far from the piece of filter paper, the expression of <i>g2p</i> is so activated that the phage particles cannot be produced efficiently, because the production of the single stranded DNA isn't enough to produce phage particles. |
</p></h2> | </p></h2> | ||
| - | + | <gallery widths="300px" heights="250px" style="margin-left: auto; margin-right: auto;"> | |
| - | + | Image:Titech2013_M13_Plux_Fig_3-4-3-A.jpg|Fig. 3-4-3. | |
| - | < | + | Image:Titech2013_M13_Plux_Fig_3-4-3-B.jpg|Fig. 3-4-4. |
| - | <h4><p>Fig. 3. The plaques formed with 3OC6HSL induction (plaque plotted)</p> | + | </gallery> |
| - | <p>The plaques were formed using the JM109 (with pSB6A1-Ptet-<i>luxR</i>) overnight cultures and the phage-particle-solution. The phage particles were obtained from the supernatant of the fresh cultures (+3OC6HSL) of transformed | + | <div style="text-align:center;"> |
| + | <h4><p>Fig. 3-4-3, 3-4-4. The plaques formed with 3OC6HSL induction (plaque plotted)</p> | ||
| + | </h4> | ||
| + | </div><h4> | ||
| + | <p>The plaques were formed using the JM109 (with pSB6A1-Ptet-<i>luxR</i>) overnight cultures and the phage-particle-solution. The phage particles were obtained from the supernatant of the fresh cultures (+3OC6HSL) of transformed DH5α (with pSB3K3-Plux-M13). A mixture of 3.5 mL YT soft agar, 100 µL of 0.01X supernatant and 400 µL of the overnight cultures of JM109 (with pSB6A1-Ptet-<i>luxR</i>) was poured on a YT plate. After solidifying the soft agar, we put a piece of filter paper on the plate and dripped 3OC6HSL in DMSO (or DMSO only) on the piece of filter paper. | ||
</p></h4> | </p></h4> | ||
| - | + | <gallery widths="300px" heights="250px" style="margin-left: auto; margin-right: auto;"> | |
| - | + | Image:Titech2013_M13_Plux_Fig_3-4-4-A.jpg|Fig. 3-4-5. | |
| - | + | Image:Titech2013_M13_Plux_Fig_3-4-4-B.jpg|Fig. 3-4-6. | |
| - | < | + | </gallery> |
| - | <h4><p>Fig. 4. The analysis of distribution of the plaques</p> | + | <div style="text-align:center;"> |
| + | <h4><p>Fig. 3-4-5, 3-4-6. The analysis of distribution of the plaques</p> | ||
<p>We totalize the distances from the center of the piece of filter paper to each plaque, by using ImageJ. | <p>We totalize the distances from the center of the piece of filter paper to each plaque, by using ImageJ. | ||
| - | </p></h4> | + | </p></h4></div> |
| - | [[Image:Titech2013_M13_plaque_hist.png|584px|thumb|center|Fig. | + | [[Image:Titech2013_M13_plaque_hist.png|584px|thumb|center|Fig. 3-4-7. The distribution histogram of the plaques(Fig. 3-4-6)<br> |
| - | < | + | This histogram shows distance from the piece of paper in a horizontal axis, and shows number of plaques over distance squared, which means the rate of plaque formation, in a vertical axis. The bin range is 0.2 cm.]] |
| + | <h2><p> | ||
| + | We hypothesize that in the vicinity of the piece of filter paper, the expression of g2p was too activated to produce the phage particles efficiently, because the production of the single stranded DNA largely exceeded that of the coat proteins. In contrast, too far from the piece of filter paper, the expression of g2p was not so activated that the phage particles could not be produced efficiently, because the production of the single stranded DNA wasn’t enough to produce phage particles. The plaques were formed only when the inducer existed in the medium at appropriate concentrations. | ||
</p> | </p> | ||
| - | </h2> | + | <p> |
| - | + | To demonstrate the hypothesis is true, we changed the waiting time before dropping AHL from 5 min. to 2h. In the meantime, phage coat proteins would be sufficiently produced in the lawn cells. | |
| + | Fig. 3-4-8 shows that the plaques were formed on the whole surface of the plate. From this assay, we can conclude that the balance of the production of coat proteins and single stranded DNA is very concerned with phage release. | ||
| + | </p></h2> | ||
| + | |||
| + | [[Image:Fig_3-4-8.png|400px|thumb|center|Fig. 3-4-8. Formed plaques after we changed the waiting time prior to dropping AHL]] | ||
| + | <h2><p> | ||
| + | We also confirmed that inducible phage release would be realized with another promoter. | ||
| + | We used BAD promoter(arabinose-inducible promorter) for g2p. | ||
| + | The result of the arabinose-inducible plaque forming assay is shown in Fig 3-4-9, 3-4-10 and 3-4-11. The result shows that the phage release is regulated by the induction of arabinose. | ||
| + | We confirmed that we can use two kinds of promoters with our part ([http://parts.igem.org/Part:BBa_K1139022 BBa_K1139022]). By replacement of the promoter upstream of g2p with various promoters, we have constructed a part collection of M13 phage. We have not only constructed the part collection, but also assayed the function of parts. | ||
| + | </p></h2> | ||
| + | |||
| + | [[Image:Fig_3-4-9.png|400px|thumb|center|Fig. 3-4-9. Formed plaques with arabinose and<i> araC</i><sup>+</sup> receiver]] | ||
| + | |||
| + | [[Image:Fig_3-4-10.png|400px|thumb|center|Fig. 3-4-10. Formed plaques without arabinose and with <i>araC</i><sup>+</sup> receiver]] | ||
| + | |||
| + | [[Image:Fig_3-4-11.png|400px|thumb|center|Fig. 3-4-11. No plaques with arabinose and without <i>araC</i><sup>+</sup> receiver]] | ||
| + | |||
| + | <div style="text-align:center;"> | ||
| + | <h4><p>The plaques were formed using the JM109 (with pSB6A1-Pcon-<i>araC</i>) overnight cultures and the transformed JM109 (with pSB3K3-Pbad-M13-Plac-<i>GFP</i>) overnight cultures. A mixture of 3.5 mL YT soft agar, 100 µL of the transformed JM109 (with pSB3K3-Pbad-M13-Plac-<i>GFP</i>) and 400 µL of the overnight cultures of JM109 (with pSB6A1-Ptet-luxR) was poured on a YT plate. After solidifying the soft agar, we put a piece of filter paper on the plate and dripped arabinose in sterile distilled water (or sterile distilled water only) on the piece of filter paper. | ||
| + | </p></h4></div> | ||
| + | <h2><p> | ||
| + | Plaques were formed with arabinose and <i>araC</i><sup>+</sup> receiver (Fig. 3-4-9). Even though we did not add arabinose, plaques were formed <i>araC</i><sup>+</sup> receiver (Fig. 3-4-10). However, no plaques were formed with arabinose and without <i>araC</i><sup>+</sup> receiver (Fig. 3-4-11). | ||
| + | The reason we thought why plaques were formed with no arabinose and <i>araC</i><sup>+</sup> receiver is that too much <i>araC</i> expressed in receiver led to leaky expression of g2p that is enough to form plaques (Fig. 3-4-12). | ||
| + | </p></h2> | ||
| + | |||
| + | [[Image:Fig_3-4-12.png|400px|thumb|center|Fig. 3-4-12. Summary of the result of the arabinose-inducible plaque forming assay]] | ||
| + | |||
| + | </div><br> | ||
| + | |||
| + | <html><div align="center"><a href="https://2013.igem.org/Team:Tokyo_Tech/Experiment/Inducible_Plaque_Forming_Assay#top"><img src="https://static.igem.org/mediawiki/2013/f/f0/Titeh2013_backtotop.png" width="200px"></a></div></html> | ||
</div> | </div> | ||
Latest revision as of 03:15, 29 October 2013
Inducible Plaque Forming Assay
Contents |
1. Introduction
Our previous assay revealed that the M13 phage genes and the M13 origin worked together with the pSB origin. Next, we confirmed whether the release of the M13 phage particle depends on the induction of g2p expression. We inserted lux promoter (which is activated by 3OC6HSL-LuxR complex) upstream of g2p, as an inducible promoter (Fig. 3-4-1)(See more about our plasmid construction). The method for our assay is briefly shown in Fig. 3-4-2. First, we cultured E. coli to which was introduced our new part ([http://parts.igem.org/Part:BBa_K1139021 BBa_K1139021]), and added 3OC6HSL, which induces the phage release. Second, we centrifuged the culture and obtained phage particles. Finally, we added the phage particles and lawn culture (E. coli containing pSB6A1-Ptet-luxR) to the soft agar, and poured it on a YT plate. Moreover, we confirmed that the inducible release of M13 phage was also realized with BAD promoter.
2. Materials and Methods
2-1. Plasmid construction
pSB3K3-Plux-M13-Plac-GFP ([http://parts.igem.org/Part:BBa_K1139021 BBa_K1139021])
pSB6A1-Ptet-luxR ([http://parts.igem.org/Part:BBa_S03119 BBa_S03119])
pSB3K3-Pbad-M13-Plac-GFP ([http://parts.igem.org/Part:BBa_K1139026 BBa_K1139026])
pSB6A1-Ptet-araC ([http://parts.igem.org/Part:BBa_I13458 BBa_I13458])
2-2. Strains
DH5α (E. coli of high competence)
JM109 (F+ strain E. coli)
2-3. Media
LB
Bacto tryptone 10 g/L
Yeast Extract 5 g/L
NaCl 10 g/L
YT soft agar
Bacto tryptone 8 g/L
Yeast Extract 5 g/L
NaCl 5 g/L
Agarose 6 g/L
YT plate
Bacto tryptone 8 g/L
Yeast Extract 5 g/L
NaCl 5 g/L
Agarose 15 g/L
LB
| Bacto tryptone | 10 g/L |
| Yeast Extract | 5 g/L |
| NaCl | 10 g/L |
YT soft agar
| Bacto tryptone | 8 g/L |
| Yeast Extract | 5 g/L |
| NaCl | 5 g/L |
| Agarose | 6 g/L |
YT plate
| Bacto tryptone | 8 g/L |
| Yeast Extract | 5 g/L |
| NaCl | 5 g/L |
| Agarose | 15 g/L |
2-4. Others
3OC6HSL dissolved in DMSO (>100 µM)
Autoclaved pieces of filter paper (about 1.5 cm in diameter)
2-5. Protocol
Preparation
- Transform DH5α with pSB3K3-Plux-M13-Plac-GFP and pSB6A1-Ptet-luxR([http://parts.igem.org/Part:BBa_S03119 BBa_S03119]).
- Prepare overnight cultures of the transformed DH5α and JM109, DH5α in LB containing ampicillin (50 µg/mL) and kanamycin (30 µg/mL).
- Take 30 µL of the overnight cultures of the transformed DH5α into 3 mL LB medium containing ampicillin (50 µg/mL) and kanamycin (30 µg/mL) . (->fresh cultures)
- Incubate the fresh cultures at 37°C for 2 h.
- Add 3 µL of 5 µM 3OC6HSL in DMSO (final concentration of 5 nM) to the fresh cultures and incubate them at 37°C for 4 h.
- Centrifuge the overnight cultures of the transformed DH5α at 9,000g for 1 min.
- Pipette the supernatant into a 1.5 mL tube.
- Dilute it 100 times with water. (-> phage-particle-solution)
Plaque formation
- Transform JM109 with pSB6A1-Ptet-luxR([http://parts.igem.org/Part:BBa_S03119 BBa_S03119]).
- Prepare overnight cultures of the transformed JM109 at 37°C.
- Melt the YT soft agar by using a microwave.
- Add ampicillin to the YT soft agar.
- Dispense 3.5 mL of the melted soft agar to a new round tube, and keep them at 50°C in an incubator.
- Dispense 400 µL of the overnight cultures of the transformed JM109 to a 1.5 mL tube.
- Into the 1.5 mL tube, add 100 µL of the phage-particle-solution.
- Transfer all the content of the 1.5 mL tube to the dispensed soft agar, mix them, and spread all of it on a YT plate.
- Wait for the YT soft agar to solidify at room temperature (for about 5 min.).
- Put an autoclaved piece of filter paper on the plate, and drip 20 mL of 3OC6HSL in DMSO (100 µM, 5 µM or DMSO only) into the piece of filter paper.
Plaque formation
3. Results
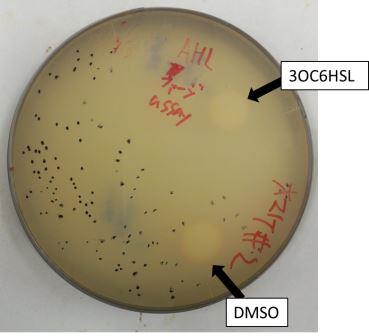
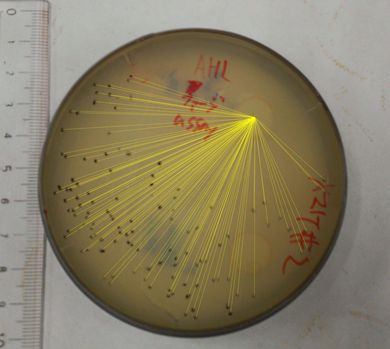
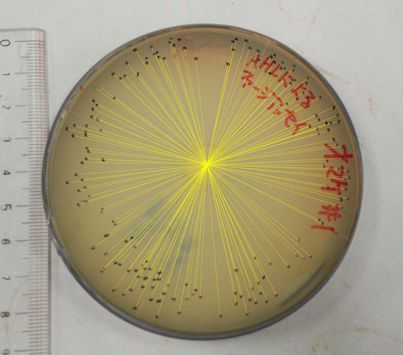
The result of the plaque forming assay is shown in Fig 3-4-3, 3-4-4. On the plate, the plaques were formed at some distance from the piece of filter paper. The result shows that the phage release is regulated by the induction of 3OC6HSL. We also analyzed the distribution of the plaques (Fig. 3-4-5, 3-4-6). The histogram result implicates that the plaques are formed only at the appropriate concentration of 3OC6HSL in the medium (Fig. 3-4-7). In the neighborhood of the piece of filter paper, the expression of g2p is so activated that the phage particles cannot be produced efficiently, because the production of the single stranded DNA largely exceeds that of the coat proteins. In contrast, far from the piece of filter paper, the expression of g2p is so activated that the phage particles cannot be produced efficiently, because the production of the single stranded DNA isn't enough to produce phage particles.
Fig. 3-4-3, 3-4-4. The plaques formed with 3OC6HSL induction (plaque plotted)
The plaques were formed using the JM109 (with pSB6A1-Ptet-luxR) overnight cultures and the phage-particle-solution. The phage particles were obtained from the supernatant of the fresh cultures (+3OC6HSL) of transformed DH5α (with pSB3K3-Plux-M13). A mixture of 3.5 mL YT soft agar, 100 µL of 0.01X supernatant and 400 µL of the overnight cultures of JM109 (with pSB6A1-Ptet-luxR) was poured on a YT plate. After solidifying the soft agar, we put a piece of filter paper on the plate and dripped 3OC6HSL in DMSO (or DMSO only) on the piece of filter paper.
Fig. 3-4-5, 3-4-6. The analysis of distribution of the plaques
We totalize the distances from the center of the piece of filter paper to each plaque, by using ImageJ.
We hypothesize that in the vicinity of the piece of filter paper, the expression of g2p was too activated to produce the phage particles efficiently, because the production of the single stranded DNA largely exceeded that of the coat proteins. In contrast, too far from the piece of filter paper, the expression of g2p was not so activated that the phage particles could not be produced efficiently, because the production of the single stranded DNA wasn’t enough to produce phage particles. The plaques were formed only when the inducer existed in the medium at appropriate concentrations.
To demonstrate the hypothesis is true, we changed the waiting time before dropping AHL from 5 min. to 2h. In the meantime, phage coat proteins would be sufficiently produced in the lawn cells. Fig. 3-4-8 shows that the plaques were formed on the whole surface of the plate. From this assay, we can conclude that the balance of the production of coat proteins and single stranded DNA is very concerned with phage release.
We also confirmed that inducible phage release would be realized with another promoter. We used BAD promoter(arabinose-inducible promorter) for g2p. The result of the arabinose-inducible plaque forming assay is shown in Fig 3-4-9, 3-4-10 and 3-4-11. The result shows that the phage release is regulated by the induction of arabinose. We confirmed that we can use two kinds of promoters with our part ([http://parts.igem.org/Part:BBa_K1139022 BBa_K1139022]). By replacement of the promoter upstream of g2p with various promoters, we have constructed a part collection of M13 phage. We have not only constructed the part collection, but also assayed the function of parts.
The plaques were formed using the JM109 (with pSB6A1-Pcon-araC) overnight cultures and the transformed JM109 (with pSB3K3-Pbad-M13-Plac-GFP) overnight cultures. A mixture of 3.5 mL YT soft agar, 100 µL of the transformed JM109 (with pSB3K3-Pbad-M13-Plac-GFP) and 400 µL of the overnight cultures of JM109 (with pSB6A1-Ptet-luxR) was poured on a YT plate. After solidifying the soft agar, we put a piece of filter paper on the plate and dripped arabinose in sterile distilled water (or sterile distilled water only) on the piece of filter paper.
Plaques were formed with arabinose and araC+ receiver (Fig. 3-4-9). Even though we did not add arabinose, plaques were formed araC+ receiver (Fig. 3-4-10). However, no plaques were formed with arabinose and without araC+ receiver (Fig. 3-4-11). The reason we thought why plaques were formed with no arabinose and araC+ receiver is that too much araC expressed in receiver led to leaky expression of g2p that is enough to form plaques (Fig. 3-4-12).
 "
"