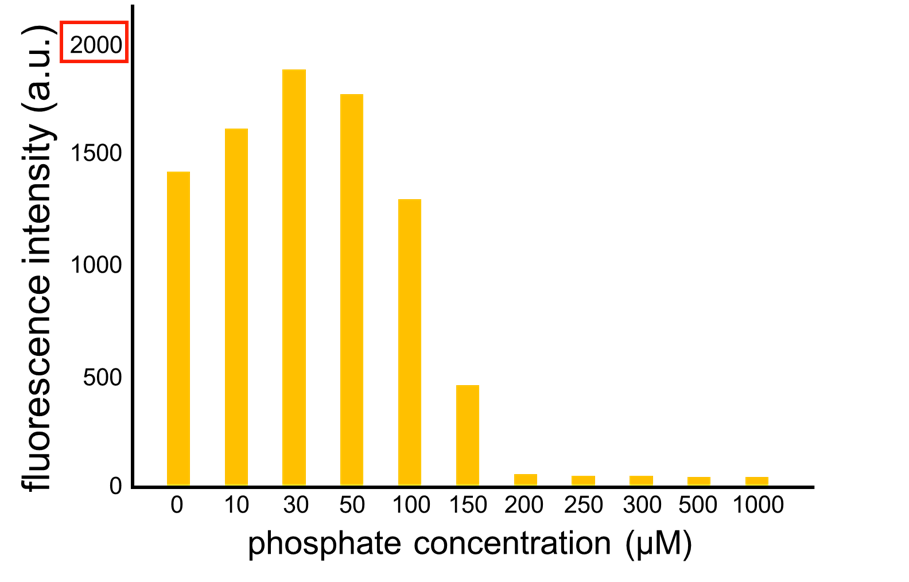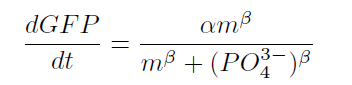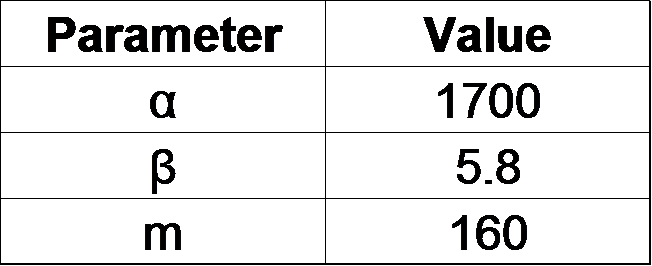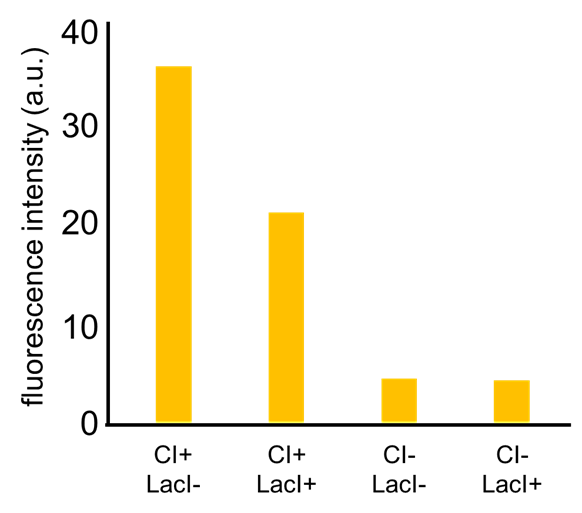Team:Tokyo Tech/Project/Farming
From 2013.igem.org
| (101 intermediate revisions not shown) | |||
| Line 2: | Line 2: | ||
<div id="text-area"><br> | <div id="text-area"><br> | ||
| - | <div class="box" id=" | + | <div class="box" id="title"> |
| - | <h1> | + | <p style="line-height:0em; text-indent:0em;" name="top">Farming</p> |
| - | [[Image:Titech2013_farming_Fig_2-3-1_farmer_ecoli.png| | + | </div> |
| - | + | <!-- | |
| + | <div class="box" id="silver"> | ||
| + | |||
| + | </div> | ||
| + | --> | ||
| + | <div class="box"> | ||
| + | <h1>1. Introduction</h1><h2> | ||
| + | [[Image:Titech2013_farming_Fig_2-3-1_farmer_ecoli.png|250px|thumb|right|Fig. 2-3-1. <i>E. ninja</i> turns to farming.]] | ||
<p> | <p> | ||
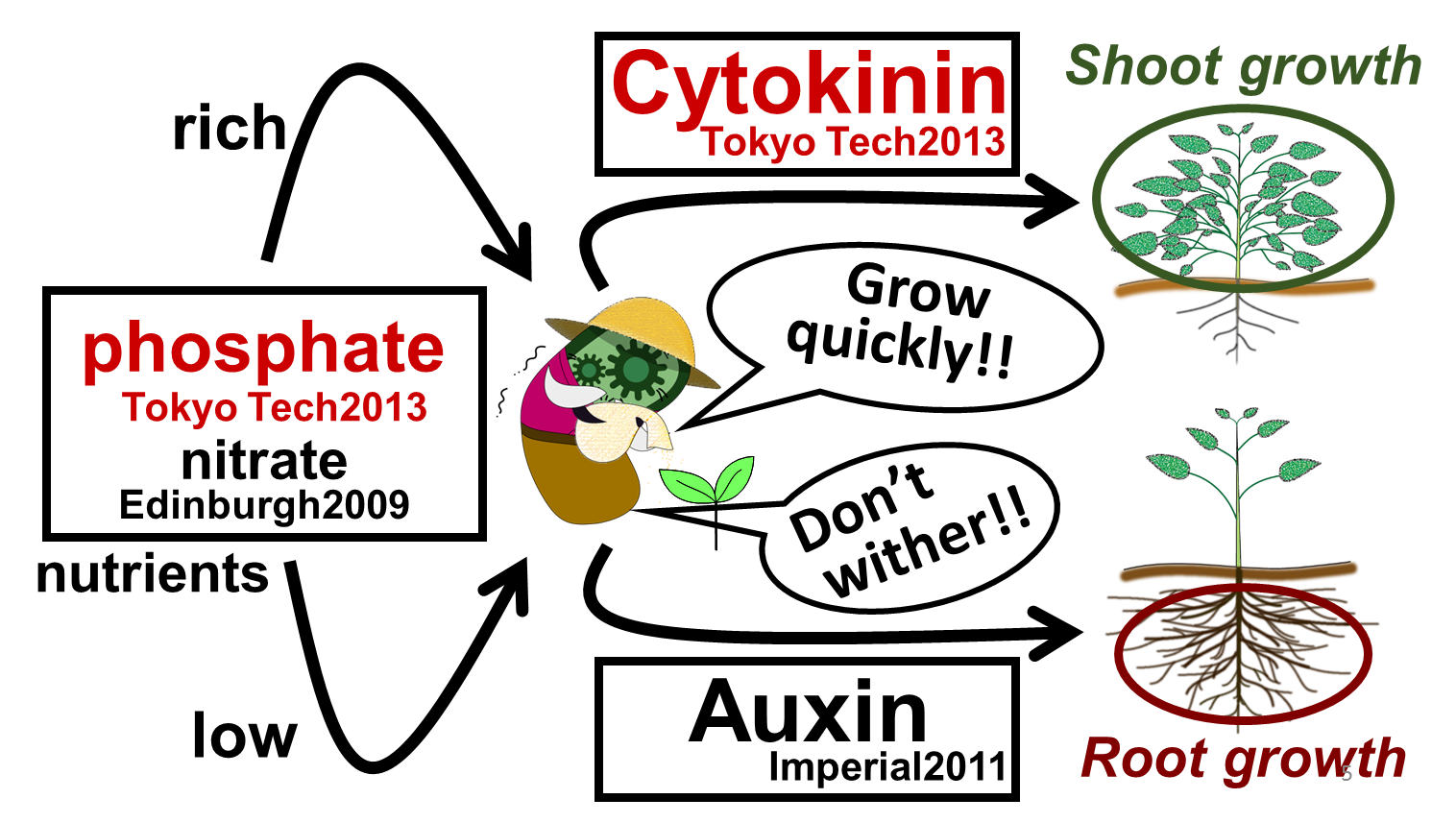
| - | In our story, <i>E. ninja</i> retires from being an undercover warrior, and turns to farming (Fig. 2-3-1). Our improved part for phosphate | + | In our story, <i>E. ninja</i> retires from being an undercover warrior, and turns to farming (Fig. 2-3-1). Our improved part for phosphate sensor and a new part for cytokinin synthesis will lead us to our goal, which is to create "Farmer <i>E. coli</i>" that could increase plant growth by synthesizing several plant hormones depending on the soil environment. As for sensing soil nutrients, we focused on phosphate and nitrate because these are known to be especially important for plants. When nutrient levels are low, plants are in danger of withering. Therefore, we are proposing <i>E. coli</i> to produce the plant hormone "auxin," which promotes the growth of plants' roots. Also, when the soil is rich in nutrients, plants can grow quickly. Therefore, we want to make <i>E. coli</i> produce the plant hormone "cytokinin," which promotes the growth of plants' shoots (Fig. 2-3-2). |
</p> | </p> | ||
[[Image:Titech2013_farming_Fig_2-3-2_whole.png|600px|thumb|center|Fig. 2-3-2. Our goal to create "Farmer <i>E. coli</i>"]] | [[Image:Titech2013_farming_Fig_2-3-2_whole.png|600px|thumb|center|Fig. 2-3-2. Our goal to create "Farmer <i>E. coli</i>"]] | ||
<p> | <p> | ||
| - | Our | + | Our "Farmer <i>E. coli</i>" can be implemented by combining our improved phosphate sensor part ([http://parts.igem.org/Part:BBa_K1139201 BBa_ K1139201]), our newly constructed cytokinin production part and two pre-existing parts which are the nitrate sensor part (Edinburgh 2009, [http://parts.igem.org/Part:BBa_K216005 BBa_K216005]) and the auxin production part (Imperial College 2011, [http://parts.igem.org/Part:BBa_K515100 BBa_K515100]) (Fig. 2-3-2). Since the existing phosphate sensor part (OUC-China 2012, [http://parts.igem.org/Part:BBa_K737024 BBa_K737024]) does not have sufficient data, we improved the part by using a different promoter. We also learned methods for quantitative analysis for cytokinin through bioassay of cucumber seed sprouts (Fig. 2-3-3) and through using ultra-performance liquid chromatography (UPLC). |
</p> | </p> | ||
[[Image:Titech2013_farming_Fig_2-3-3_cotyledons.png|300px|thumb|center|Fig. 2-3-3. Cucumber seed sprouts respond to cytokinin samples]] | [[Image:Titech2013_farming_Fig_2-3-3_cotyledons.png|300px|thumb|center|Fig. 2-3-3. Cucumber seed sprouts respond to cytokinin samples]] | ||
<p> | <p> | ||
| - | In addition, to achieve the natural | + | In addition, to achieve the natural plants' temporal pattern for producing plant hormones in <i>E. coli</i>, we introduced an incoherent feed forward loop to our circuit, including a new hybrid promoter part ([http://parts.igem.org/Part:BBa_K1139150 BBa_K1139150]). Our mathematical model for this system predicted that we can create temporal patterns for the plant hormone production. |
</p> | </p> | ||
</h2> | </h2> | ||
</div><br> | </div><br> | ||
<div class="box"> | <div class="box"> | ||
| - | <h1>Phosphate | + | <h1>2. Phosphate sensor</h1> |
| - | + | <h2> | |
| - | [[Image:Titech2013_farming_Fig_2-3-4_phosphate.png| | + | [[Image:Titech2013_farming_Fig_2-3-4_phosphate.png|350px|thumb|right|Fig. 2-3-4. Our phosphate sensor part including the <i>phoA</i> promoter]] |
| + | </h2> | ||
<h2> | <h2> | ||
<p> | <p> | ||
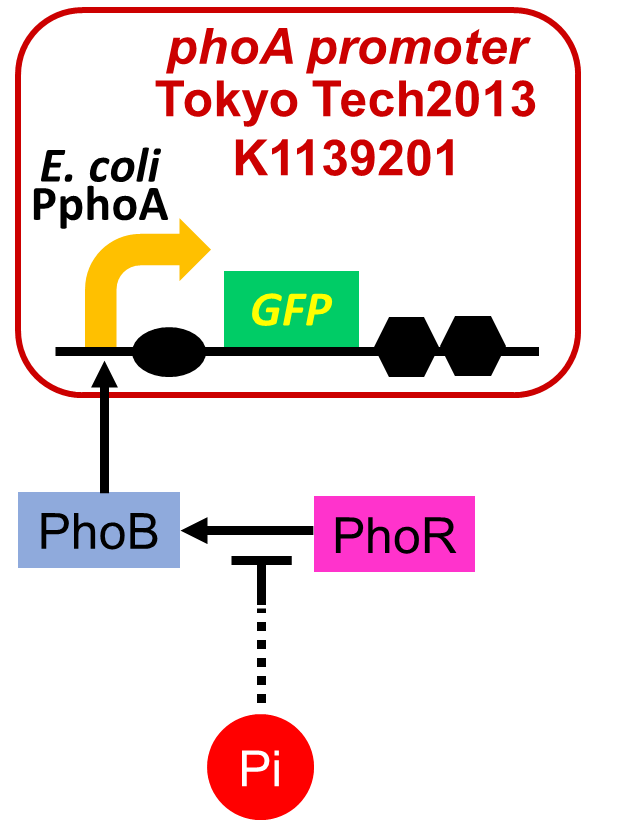
| - | We | + | We improved a phosphate sensor part by using the inducible promoter of the alkaline phosphatase gene (<i>phoA</i>) from <i>E. coli</i> (Dollard et al., 2003). This promoter is repressed by high phosphate concentrations (Fig. 2-3-4). To know more about the regulation of this promoter, please click <font size ="5">[https://2013.igem.org/Team:Tokyo_Tech/Experiment/phoA_Promoter_Assay#1._Introduction here]</font size>. We amplified the <i>phoA</i> promoter from <i>E. coli</i> (MG1655) and ligated this promoter into <i>GFP</i> part to construct the new part ([http://parts.igem.org/Part:BBa_K1139201 BBa_K1139201]). We then introduced this new part into <i>E. coli</i> (MG1655). Fig. 2-3-5 shows the result of our induction assay. It shows that the increase in phosphate concentration repressed the <i>phoA</i> promoter. Especially, we see that the <i>phoA</i> promoter is drastically repressed at phosphate concentrations of 100 to 200 µM. |
| - | + | ||
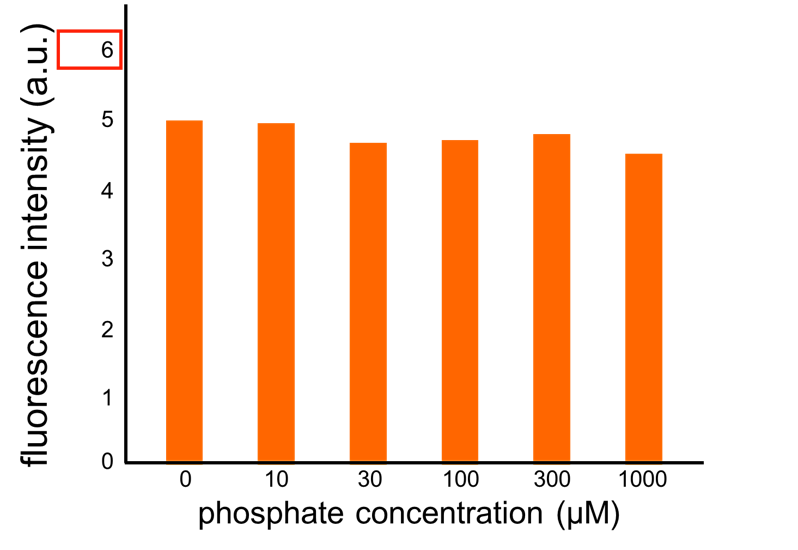
| - | [[Image:Titech2013_farming_Fig_2-3-5_phoa.png | + | Though we also assayed OUC-China's phosphate sensor part including the <i>phoB</i> promoter ([http://parts.igem.org/Part:BBa_K737024 BBa_K737024]) by the same method as that for our <i>phoA</i> promoter assay, their part did not respond to the increase in phosphate concentration (Fig. 2-3-6). Thus, we concluded that we improved the phosphate sensor part. (Note that the scales of the vertical axis are different between the two results.) |
| - | + | ||
| - | < | + | |
| - | + | ||
| - | From our results | + | To know more about this assay, please see <font size ="5">[https://2013.igem.org/Team:Tokyo_Tech/Experiment/phoA_Promoter_Assay#1._Introduction here]</font size>. |
| + | |||
| + | <gallery widths="350px" heights="250px" style="margin-left:auto; margin-right:auto; text-align: center;"> | ||
| + | Image:Titech2013_farming_Fig_2-3-5_phoa.png|Fig. 2-3-5. Our induction assay result for our <i>phoA</i> promoter | ||
| + | Image:Titech2013_farming_Fig_2-3-6_phob.png|Fig. 2-3-6. Our induction assay result for OUC-China 2012's <i>phoB</i> promoter | ||
| + | </gallery> | ||
| + | |||
| + | From our results above, we determined parameters for the induction mechanism by fitting the results to the following Hill equation (Fig. 2-3-7). α denotes the maximum GFP expression rate in this construct. m denotes the phosphate concentration at which the GFP expression rate is half of α. β denotes the hill coefficient. Those parameters (Fig. 2-3-8) can be used in future modeling. Plants are reported to be in phosphate starvation when its concentration is below 1 mM (Hoagland et al., 1950). Our part can sense the concentration below 1 mM, too (Fig. 2-3-9). Therefore, our improved part is useful for our farming circuit. Moreover, a sensor for phosphate concentration is valuable for various studies in synthetic biology. | ||
</p> | </p> | ||
| + | |||
| + | |||
| + | [[Image:Titech2013_farming_Fig_2-3-7.png|240px|thumb|left|Fig. 2-3-7. Equation for the induction mechanism]] | ||
| + | [[Image:Titech2013_farming_Fig_2-3-9.png|500px|thumb|right|Fig. 2-3-9. A model with fitting the results of our assay]] | ||
| + | <br><br><br> | ||
| + | [[Image:Titech2013_farming_Fig_2-3-8.png|240px|thumb|left|Fig. 2-3-8. Determined parameters <br><font size ="1">α denotes the maximum GFP expression rate in this construct. <br>m denotes the phosphate concentration at which the GFP expression rate is half of α. <br>β denotes the hill coefficient.</font size>]] | ||
| + | |||
| + | <br><br><br><br><br><br><br><br><br><br><br><br><br> | ||
</h2> | </h2> | ||
</div><br> | </div><br> | ||
<div class="box"> | <div class="box"> | ||
| - | <h1>Cytokinin synthesis</h1><h2> | + | <h1>3. Cytokinin synthesis</h1><h2> |
<p> | <p> | ||
| - | In order to construct cytokinin production part, we focused on | + | In order to construct the cytokinin production part, we focused on <i>At</i>IPT, a plant enzyme which catalyzes the synthesis of cytokinin. The mechanisms of cytokinin synthesis are shown in Fig. 2-3-10 (Takei et al., 2001). We ordered two DNA sequences of <i>At</i>IPT (<i>At</i>IPT4 and <i>At</i>IPT7) derived from <i>A. thaliana</i> and constructed the part including one of them. |
</p> | </p> | ||
| - | [[Image:Titech2013_farming_Fig_2-3-10.png|600px|thumb|center|Fig. 2-3-10. Mechanisms of cytokinin synthesis]] | + | [[Image:Titech2013_farming_Fig_2-3-10.png|600px|thumb|center|Fig. 2-3-10. Mechanisms of the cytokinin synthesis]] |
</h2><h5> | </h2><h5> | ||
<p> | <p> | ||
| - | + | <div style="margin-left:80px;"> | |
| - | </p></h5> | + | <i>At</i>IPT, a plant enzyme, catalyzes the synthesis of iPRMP from DMAPP and AMP. According to the previous research, DMAPP and AMP are provided by the authentic metabolism in <i>E. coli</i> . It is also expected in the report that <i>E. coli</i> also has enzymes which catalyze the synthesis of iP or tZ (both sorts of cytokinin) from iPRMP. |
| + | </div></p></h5> | ||
</div><br> | </div><br> | ||
<div class="box"> | <div class="box"> | ||
| - | <h1>Quantitative analysis for cytokinin by bioassay</h1><h2> | + | <h1>4. Quantitative analysis for cytokinin by bioassay</h1><h2> |
<p> | <p> | ||
| - | Before constructing genetic parts for cytokinin synthesis, we learned methods for quantitative analysis for cytokinin through a bioassay of cucumber seed sprouts ( | + | Before constructing genetic parts for cytokinin synthesis, we learned methods for quantitative analysis for cytokinin through a bioassay of cucumber seed sprouts (Fletcher et al., 1971; Porra et al., 1989) and through using ultra-performance liquid chromatography (UPLC). |
</p> | </p> | ||
<p> | <p> | ||
| - | In our bioassay of cucumber seed sprouts, we | + | In our bioassay of cucumber seed sprouts, we learned how to detect cytokinin and found which concentration of cytokinin can act on plants. We planted the seeds and germinated for 5 days. Then, we cultivated the sprouts in standard cytokinin sample solutions. After 24 hours in the dark and 24 hours in the light, we measured the weight of the sprouts and also homogenized them to measure the concentration of chlorophyll (Fig. 2-3-12). Fig. 2-3-11 shows the result that the sprouts in the cytokinin solution are larger and greener than the negative controls, which are the sprouts in the solution without cytokinin. The quantitative results (Fig. 2-3-13 and Fig. 2-3-14) also show that cytokinin increases the weight of the sprouts and the concentration of chlorophyll (iP, tZ are both sorts of cytokinin). To know more about this assay, please see <font size ="5">[https://2013.igem.org/Team:Tokyo_Tech/Experiment/Quantitative_Analysis_of_Cytokinin#1._Quantitative_analysis_of_cytokinins_using_cucumber_cotyledons here]</font size>. |
</p> | </p> | ||
| - | + | <gallery widths="350px" heights="160px" style="margin-left:auto; margin-right:auto;"> | |
| - | + | Image:Titech2013_farming_Fig_2-3-11_cotyledons.png|Fig. 2-3-11. Cucumber seed sprouts respond to cytokinin samples | |
| - | + | Image:Titech2013_farming_Fig_2-3-14.png|Fig. 2-3-12. Chlorophyll extraction | |
| - | + | </gallery> | |
| + | <gallery widths="350px" heights="350px" style="margin-left:auto; margin-right:auto;"> | ||
| + | Image:Titech2013_farming_Fig_2-3-12.png|Fig. 2-3-13. Quantitative results of bioassay | ||
| + | Image:Titech2013_farming_Fig_2-3-13.png|Fig. 2-3-14. Quantitative results of bioassay | ||
| + | </gallery> | ||
</h2><h5> | </h2><h5> | ||
<p> | <p> | ||
| - | iP : 6-(γ, γ-Dimethylallylamino) purine, tZ : trans-zeatin<br> | + | <div style="margin-left:80px;"> |
| - | The | + | iP : 6-(γ, γ-Dimethylallylamino) purine, tZ : trans-zeatin (both sorts of cytokinin)<br> |
| - | </p> | + | The absorbances of the supernatant were read at 663.6 and 646.6 nm. Calculations of concentrations of the chlorophyll were carried out as described by Porra et al.,1989. |
| + | </div></p> | ||
</h5><h2> | </h5><h2> | ||
<p> | <p> | ||
| - | Through ultra-performance liquid chromatography (UPLC), we | + | Through ultra-performance liquid chromatography (UPLC), we planned to confirm that our "Farmer <i>E. coli</i>" actually produces cytokinin. Before completing the system to produce cytokinin in <i>E. coli</i>, we determined the retention times of the authentic samples of iP and tZ by using UPLC. In addition, we confirmed that iP and tZ were able to be detected from the mixture of the <i>E. coli</i> culture medium and the cytokinin solution (Fig. 2-3-15). To see the detail method of this experiment, please click [https://2013.igem.org/Team:Tokyo_Tech/Experiment/Quantitative_Analysis_of_Cytokinin#2._Identification_of_cytokinins_by_ultra-performance_liquid_chromatography_.28UPLC.29 <font size ="5">here</font size>]. |
</p> | </p> | ||
| - | + | [[Image:Titech2013_farming_Fig_2-3-15.png|700px|thumb|center|Fig. 2-3-15. The UPLC chromatograms<br> The black line stands for the chromatogram of the diluted supernatant. The red line stands for the chromatogram of the diluted supernatant containing the cytokinin standards.]] | |
| - | [[Image:Titech2013_farming_Fig_2-3-15.png|700px|thumb|center|Fig. 2-3-15. The | + | </h2></div><br> |
| - | </div><br> | + | |
<div class="box"> | <div class="box"> | ||
| - | <h1>Hybrid promoter for temporal pattern generation</h1><h2> | + | <h1>5. Hybrid promoter for temporal pattern generation</h1><h2> |
<p> | <p> | ||
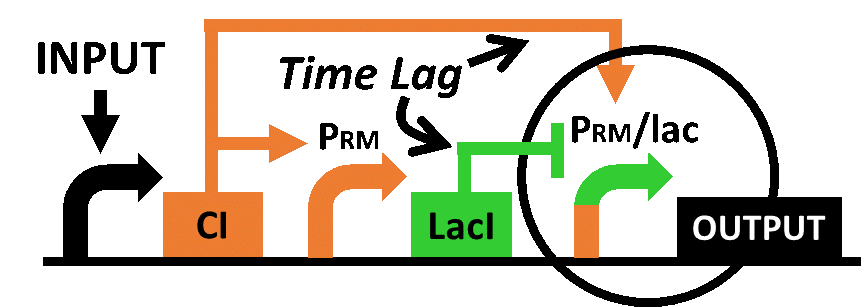
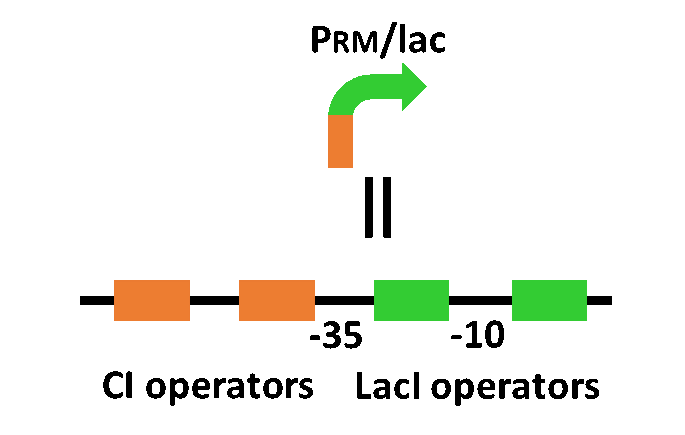
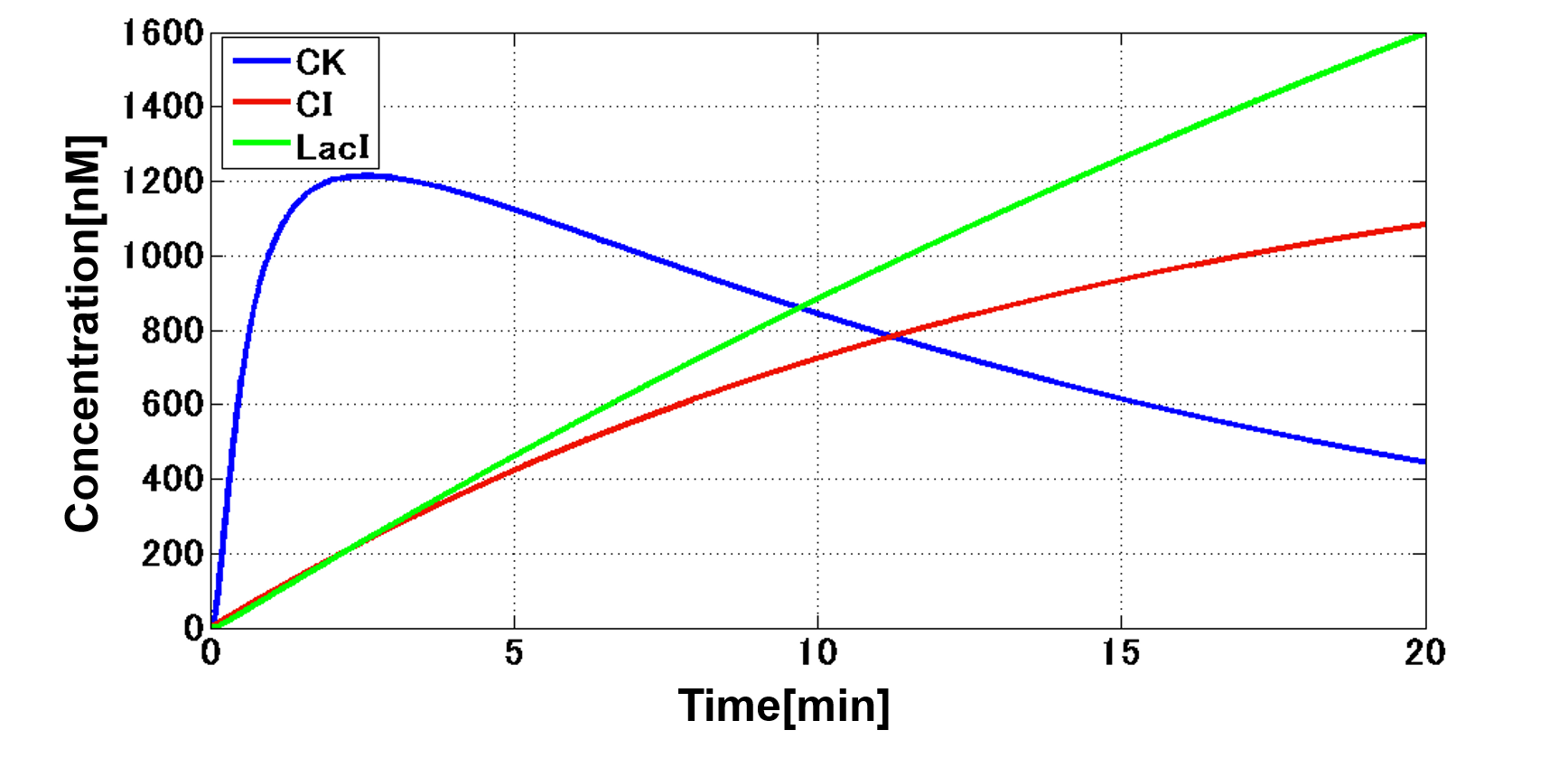
| - | In order to achieve the natural | + | In order to achieve the natural plants' temporal pattern for producing plant hormones in <i>E. coli</i>, we introduced an incoherent feed forward loop (Mangan et al., 2006) to our circuit, including a new hybrid promoter part ([http://parts.igem.org/Part:BBa_K1139150 BBa_K1139150]). Plants produce their hormones transiently rather than steadily (Takei et al., 2001). Moreover, continuous overexpression of hormones is harmful to plants (Thiman, 1937). Thus, we thought that it should be important to achieve this transient temporal pattern for producing plant hormones in <i>E. coli</i>, too. Our designed system with an incoherent feed forward loop is shown in Fig. 2-3-16. We newly developed the <i>RM/lac</i> hybrid promoter, which is activated by CI and repressed by LacI (Fig. 2-3-17). We planned to ligate a hormone synthase part downstream of this hybrid promoter. Our mathematical model (Fig. 2-3-18) shows the pulse wave which the temporal pattern for plant hormone production should achieve. While the <i>RM/lac</i> hybrid promoter activation by CI is a single-step reaction, the repression by LacI is a two-step reaction. Thus, the activation of the <i>RM/lac</i> hybrid promoter is faster than the repression. This time lag between the activation and the repression is important for generating a temporal pattern of plant hormone production (details about our mathematical model can be found <font size ="5">[https://2013.igem.org/Team:Tokyo_Tech/Modeling/Incoherent_Feed_Forward_Loop#1._Introduction here]</font size>). |
</p> | </p> | ||
| - | + | <gallery widths="350px" heights="200px" style="margin-left:auto; margin-right:auto;"> | |
| - | + | Image:Titech2013_farming_Fig_2-3-16.png|Fig. 2-3-16. Our designed system with an incoherent feed forward loop | |
| - | < | + | Image:Titech2013_farming_Fig_2-3-17.png|Fig. 2-3-17. Our newly designed hybrid promoter |
| - | [[Image:Titech2013_farming_Fig_2-3-18.png|500px|thumb|center|Fig. | + | </gallery> |
| - | + | [[Image:Titech2013_farming_Fig_2-3-18.png|500px|thumb|center|Fig. 2-3-18. Our mathematical model for temporal pattern generation]] | |
| + | <br> | ||
<p> | <p> | ||
| - | As a first step to achieve this incoherent feed forward loop, we constructed | + | As a first step to achieve this system with an incoherent feed forward loop, we constructed a circuit shown in Fig. 2-3-19 to confirm that our new <i>RM/lac</i> hybrid promoter actually works. We set GFP as an output of the <i>RM/lac</i> hybrid promoter and introduced the circuit into <i>E. coli</i> . Through an induction assay (Fig. 2-3-20), we saw that our new <i>RM/lac</i> hybrid promoter was actually activated by CI and repressed by LacI. Details about this assay can be found <font size ="5">[https://2013.igem.org/Team:Tokyo_Tech/Experiment/RM-lac_Hybrid_Promoter_Assay#1._Introduction here]</font size>. |
</p> | </p> | ||
| - | < | + | <gallery widths="350px" heights="250px" style="margin-left:auto; margin-right:auto; text-align:center;"> |
| - | + | Image:Titech2013_farming_Fig_2-3-19.png|Fig. 2-3-19. The circuit we constructed including the new part | |
| - | + | Image:Titech2013_farming_Fig_2-3-20.png|Fig. 2-3-20. Our <i>RM/lac</i> hybrid promoter was activated by CI and repressed by LacI | |
| - | </div><br> | + | </gallery> |
| + | </h2></div><br> | ||
<div class="box"> | <div class="box"> | ||
| - | <h1>Applications</h1><h2> | + | <h1>6. Applications</h1><h2> |
<p> | <p> | ||
| - | Our project | + | Our project can be applied to studying the plants' response to external plant hormones. By applying the incoherent feed forward loop to our circuit, we can achieve pulse-generation of the output (Basu et al., 2003). Moreover, the pulse wave can be customized with changing the parameters, for example, intensities of promoters or various interactions. Various artificial genetic circuits which can achieve the temporal pattern of the output, such as incoherent feed forward loop, have been reported (Elowitz et al., 2000; Fung et al., 2005). By combining plant hormone synthesis genes to these artificial genetic circuits, we will be able to make <i>E. coli</i> produce plant hormones in various temporal patterns. Analysis of the plants' response to plant hormones given in temporal change leads to elucidate the abilities of plants in signal transduction. Findings in plant science using this strategy will be important for contributing to new approaches in farming. We believe that such basic research can help to solve worldwide food shortages. We hope to meet public expectations by realizing agriculture aids by bacteria, though there may be some difficulties. |
| + | |||
| + | |||
</p> | </p> | ||
</h2> | </h2> | ||
</div><br> | </div><br> | ||
<div class="box"> | <div class="box"> | ||
| - | <h1> | + | <h1>7. References</h1><h2><OL> |
| - | Eileen Fung, Wilson W. Wong, Jason K. Suen, Thomas Bulter, Sun-gu Lee | + | <LI>Eileen Fung, Wilson W. Wong, Jason K. Suen, Thomas Bulter, Sun-gu Lee and James C. Liao. (2005) A synthetic gene–metabolic oscillator. Nature, Vol 435, 118-122 |
| - | < | + | <LI>Michael B. Elowitz and Stanislas Leibler. (2000) A synthetic oscillatory network of transcriptional regulators. Nature, Vol 403, 335-338 |
| - | Michael B. Elowitz | + | <LI>R. A. Fletcher, V. Kallidumbil and P. Steele. (1981) An Improved Bioassay for Cytokinins Using Cucumber Cotyledons. Plant Physiol, 69, 675-677 |
| - | < | + | <LI>R. A. Fletcher and Dianne McCullagh. (1971) Cytokinin-Induced Chlorophyll Formation in Cucumber Cotyledons. Planta (Berl.), 101, 88-90 |
| - | R. A. | + | <LI>Frederick C. Neidhardt, Philip L. Bloch and David F. Smith. (1974) Culture Medium for Enterobacteria. Journal of bacteriology, Vol 119, 736-747 |
| - | < | + | <LI>R. J. Porra, W.A. Thompson and P.E. Kriedemann. (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochimica et Biophysica Acta, 975, 384-394 |
| - | R. A. | + | <LI>Yi-Ju Hsieh and Barry L Wanner. (2010) Global regulation by the seven-component Pi signaling system. Current Opinion in Microbiology, 13, 198-203 |
| - | < | + | <LI>Marie-Andre´e Dollard, Patrick Billard. (2003) Whole-cell bacterial sensors for the monitoring of phosphate bioavailability. Journal of Microbiological Methods, 55, 221– 229. |
| - | + | <LI>Subhayu Basu, Rishabh Mehreja, Stephan Thiberge, Ming-Tang Chen, and Ron Weiss. (2003) Spatiotemporal control of gene expression with pulse-generating networks. Proc. Natl. Acad. Sci. USA, 101, 6355-6360. | |
| - | < | + | <LI>Hideo Shinagawa, Kozo Makino and Atsuo Nakata. (1983) Regulation of the pho Regulon in <i>Escherichia coli</i> K-12 Genetic and Physiological Regulation of the Positive |
| - | R.J. Porra, W.A. Thompson and P.E. Kriedemann. (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochimica et Biophysica Acta, 975, 384-394 | + | |
| - | < | + | |
| - | Yi-Ju Hsieh and Barry L Wanner. (2010) Global regulation by the seven-component Pi signaling system. Current Opinion in Microbiology, 13, 198-203 | + | |
| - | < | + | |
| - | Marie-Andre´e Dollard, Patrick Billard. (2003) Whole-cell bacterial sensors for the monitoring of phosphate bioavailability. Journal of Microbiological Methods, 55, 221– 229. | + | |
| - | < | + | |
| - | Subhayu Basu, Rishabh Mehreja, Stephan Thiberge, Ming-Tang Chen, and Ron Weiss. (2003) Spatiotemporal control of gene expression with pulse-generating networks. Proc. Natl. Acad. Sci. USA, 101, 6355-6360. | + | |
| - | < | + | |
| - | + | ||
Regulatory Gene <i>phoB</i>. J. Mol. Biol, 168, 477-488 | Regulatory Gene <i>phoB</i>. J. Mol. Biol, 168, 477-488 | ||
| - | < | + | <LI> |
Kenneth V. Thimann. (1938) On the Nature of Inhibitions Caused by Auxin. American Journal of Botany, 24, 407-412 | Kenneth V. Thimann. (1938) On the Nature of Inhibitions Caused by Auxin. American Journal of Botany, 24, 407-412 | ||
| - | < | + | <LI>Kentaro Takei, Hitoshi Sakakibara1, Mitsutaka Taniguchi and Tatsuo Sugiyama. (2001) Nitrogen-Dependent Accumulation of Cytokinins in Root and the Translocation to Leaf: Implication of Cytokinin Species that Induces Gene Expression of Maize Response Regulator. Plant Cell Physiol, 42, 85–93 |
| - | Kentaro Takei, Hitoshi Sakakibara1, Mitsutaka Taniguchi and Tatsuo Sugiyama. (2001) Nitrogen-Dependent Accumulation of Cytokinins in Root and the Translocation to Leaf: Implication of Cytokinin Species that Induces Gene Expression of Maize Response Regulator. Plant Cell Physiol, 42, 85–93 | + | <LI>Kentaro Takei, Hitoshi Sakakibara, and Tatsuo Sugiyama. (2001) Identification of Genes Encoding Adenylate Isopentenyltransferase, a Cytokinin Biosynthesis Enzyme, in Arabidopsis thaliana. The Journal of Biological Chemistry, 276, 26405-26410 |
| - | < | + | <LI>S. Mangan, S. Itzkovitz, A. Zaslaver, U. Alon. (2006) The Incoherent Feed-forward Loop Accelerates the Response-time of the <i>gal</i> System of <i>Escherichia coli</i>. J Mol Biol, 356(5):1073-81 |
| - | Kentaro Takei, Hitoshi Sakakibara, and Tatsuo Sugiyama. (2001) Identification of Genes Encoding Adenylate Isopentenyltransferase, a Cytokinin Biosynthesis Enzyme, in Arabidopsis thaliana. | + | </OL> |
| - | < | + | |
| - | Mangan S | + | |
| - | < | + | |
</h2> | </h2> | ||
</div><br> | </div><br> | ||
| + | <html><div align="center"><a href="https://2013.igem.org/Team:Tokyo_Tech/Project/Farming#top"><img src="https://static.igem.org/mediawiki/2013/f/f0/Titeh2013_backtotop.png" width="200px"></a></div></html> | ||
</div> | </div> | ||
Latest revision as of 19:20, 28 October 2013
Farming
Contents |
1. Introduction
In our story, E. ninja retires from being an undercover warrior, and turns to farming (Fig. 2-3-1). Our improved part for phosphate sensor and a new part for cytokinin synthesis will lead us to our goal, which is to create "Farmer E. coli" that could increase plant growth by synthesizing several plant hormones depending on the soil environment. As for sensing soil nutrients, we focused on phosphate and nitrate because these are known to be especially important for plants. When nutrient levels are low, plants are in danger of withering. Therefore, we are proposing E. coli to produce the plant hormone "auxin," which promotes the growth of plants' roots. Also, when the soil is rich in nutrients, plants can grow quickly. Therefore, we want to make E. coli produce the plant hormone "cytokinin," which promotes the growth of plants' shoots (Fig. 2-3-2).
Our "Farmer E. coli" can be implemented by combining our improved phosphate sensor part ([http://parts.igem.org/Part:BBa_K1139201 BBa_ K1139201]), our newly constructed cytokinin production part and two pre-existing parts which are the nitrate sensor part (Edinburgh 2009, [http://parts.igem.org/Part:BBa_K216005 BBa_K216005]) and the auxin production part (Imperial College 2011, [http://parts.igem.org/Part:BBa_K515100 BBa_K515100]) (Fig. 2-3-2). Since the existing phosphate sensor part (OUC-China 2012, [http://parts.igem.org/Part:BBa_K737024 BBa_K737024]) does not have sufficient data, we improved the part by using a different promoter. We also learned methods for quantitative analysis for cytokinin through bioassay of cucumber seed sprouts (Fig. 2-3-3) and through using ultra-performance liquid chromatography (UPLC).
In addition, to achieve the natural plants' temporal pattern for producing plant hormones in E. coli, we introduced an incoherent feed forward loop to our circuit, including a new hybrid promoter part ([http://parts.igem.org/Part:BBa_K1139150 BBa_K1139150]). Our mathematical model for this system predicted that we can create temporal patterns for the plant hormone production.
2. Phosphate sensor
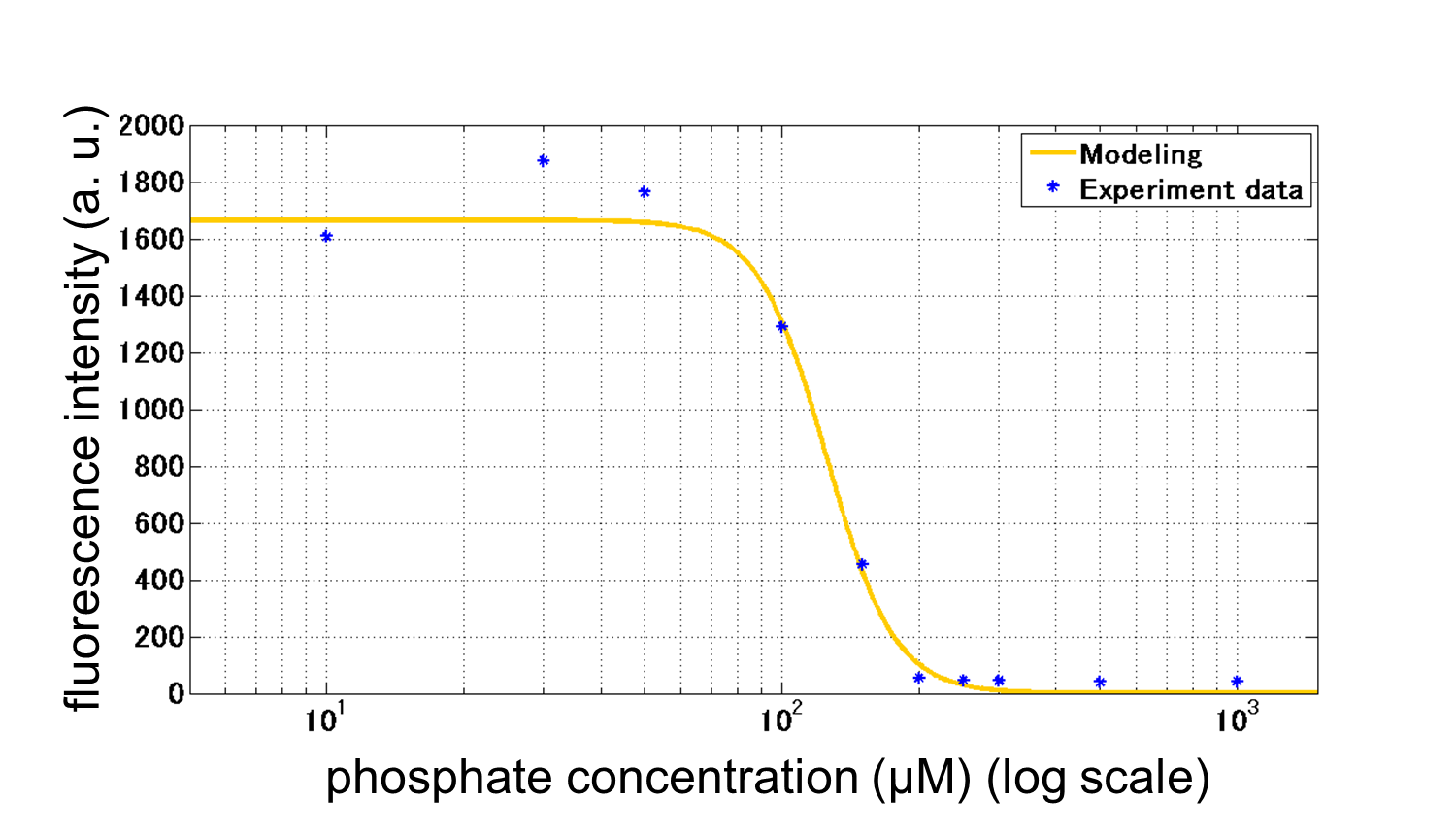
We improved a phosphate sensor part by using the inducible promoter of the alkaline phosphatase gene (phoA) from E. coli (Dollard et al., 2003). This promoter is repressed by high phosphate concentrations (Fig. 2-3-4). To know more about the regulation of this promoter, please click here. We amplified the phoA promoter from E. coli (MG1655) and ligated this promoter into GFP part to construct the new part ([http://parts.igem.org/Part:BBa_K1139201 BBa_K1139201]). We then introduced this new part into E. coli (MG1655). Fig. 2-3-5 shows the result of our induction assay. It shows that the increase in phosphate concentration repressed the phoA promoter. Especially, we see that the phoA promoter is drastically repressed at phosphate concentrations of 100 to 200 µM. Though we also assayed OUC-China's phosphate sensor part including the phoB promoter ([http://parts.igem.org/Part:BBa_K737024 BBa_K737024]) by the same method as that for our phoA promoter assay, their part did not respond to the increase in phosphate concentration (Fig. 2-3-6). Thus, we concluded that we improved the phosphate sensor part. (Note that the scales of the vertical axis are different between the two results.) To know more about this assay, please see here.
From our results above, we determined parameters for the induction mechanism by fitting the results to the following Hill equation (Fig. 2-3-7). α denotes the maximum GFP expression rate in this construct. m denotes the phosphate concentration at which the GFP expression rate is half of α. β denotes the hill coefficient. Those parameters (Fig. 2-3-8) can be used in future modeling. Plants are reported to be in phosphate starvation when its concentration is below 1 mM (Hoagland et al., 1950). Our part can sense the concentration below 1 mM, too (Fig. 2-3-9). Therefore, our improved part is useful for our farming circuit. Moreover, a sensor for phosphate concentration is valuable for various studies in synthetic biology.
3. Cytokinin synthesis
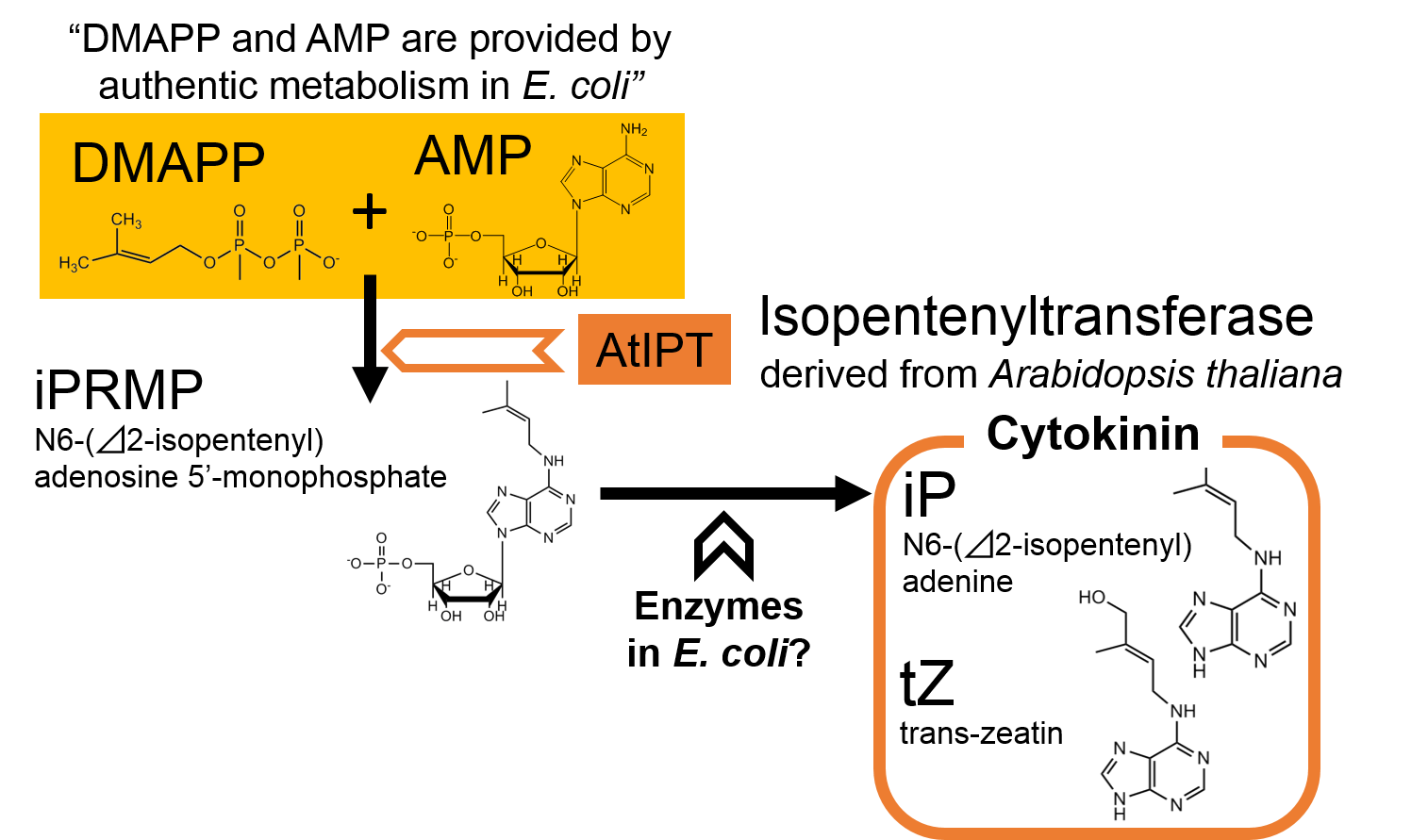
In order to construct the cytokinin production part, we focused on AtIPT, a plant enzyme which catalyzes the synthesis of cytokinin. The mechanisms of cytokinin synthesis are shown in Fig. 2-3-10 (Takei et al., 2001). We ordered two DNA sequences of AtIPT (AtIPT4 and AtIPT7) derived from A. thaliana and constructed the part including one of them.
AtIPT, a plant enzyme, catalyzes the synthesis of iPRMP from DMAPP and AMP. According to the previous research, DMAPP and AMP are provided by the authentic metabolism in E. coli . It is also expected in the report that E. coli also has enzymes which catalyze the synthesis of iP or tZ (both sorts of cytokinin) from iPRMP.
4. Quantitative analysis for cytokinin by bioassay
Before constructing genetic parts for cytokinin synthesis, we learned methods for quantitative analysis for cytokinin through a bioassay of cucumber seed sprouts (Fletcher et al., 1971; Porra et al., 1989) and through using ultra-performance liquid chromatography (UPLC).
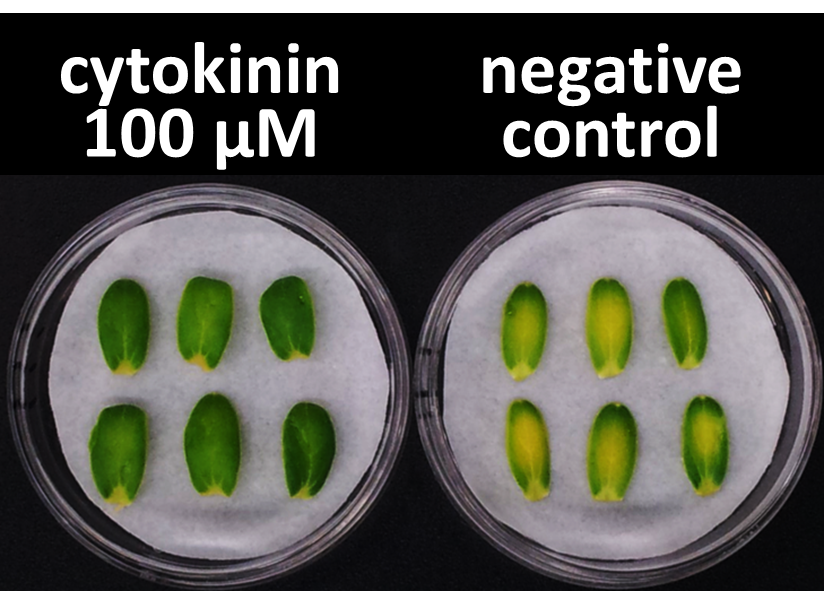
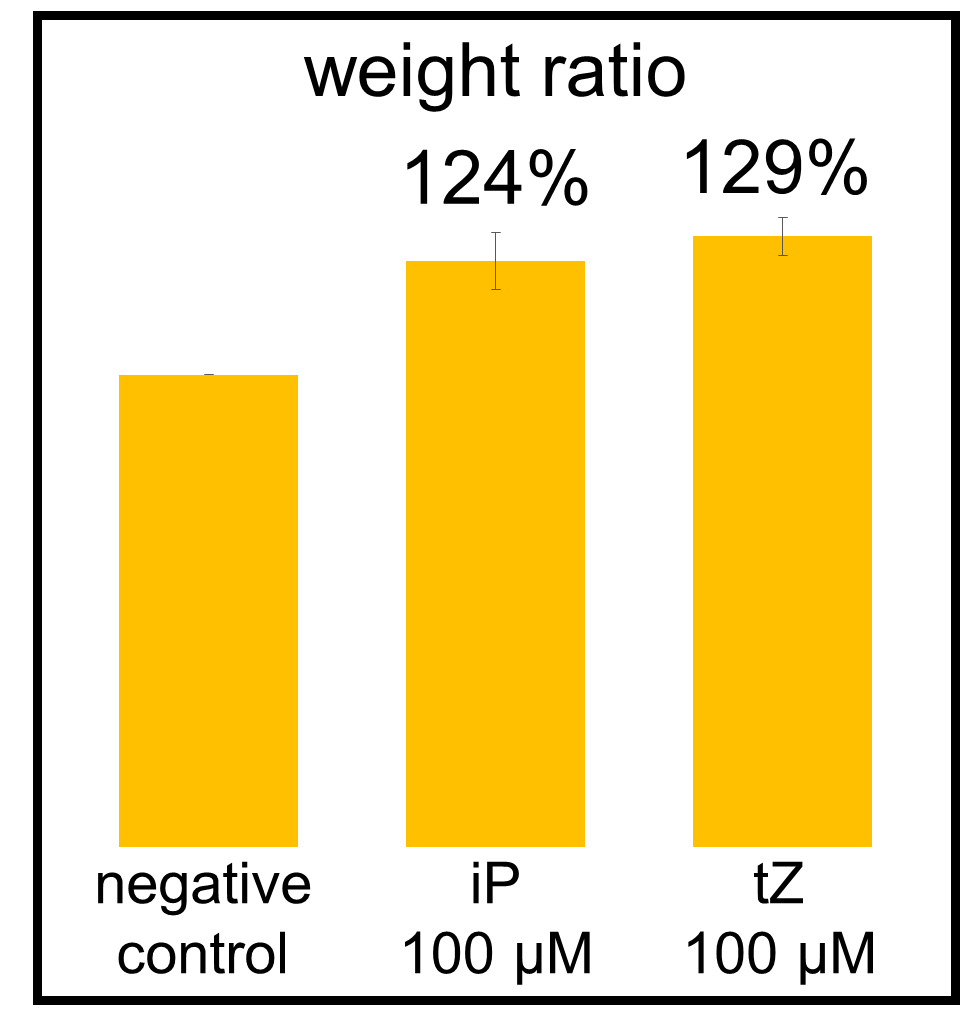
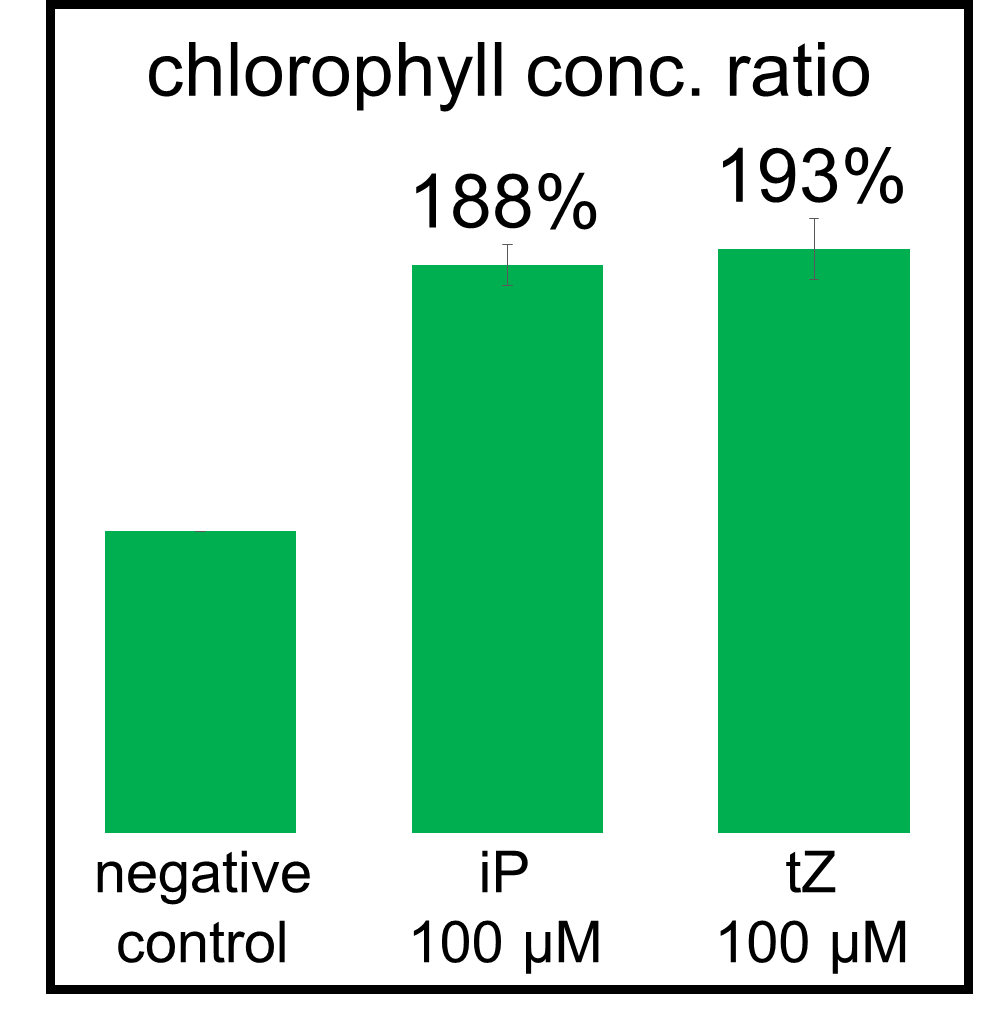
In our bioassay of cucumber seed sprouts, we learned how to detect cytokinin and found which concentration of cytokinin can act on plants. We planted the seeds and germinated for 5 days. Then, we cultivated the sprouts in standard cytokinin sample solutions. After 24 hours in the dark and 24 hours in the light, we measured the weight of the sprouts and also homogenized them to measure the concentration of chlorophyll (Fig. 2-3-12). Fig. 2-3-11 shows the result that the sprouts in the cytokinin solution are larger and greener than the negative controls, which are the sprouts in the solution without cytokinin. The quantitative results (Fig. 2-3-13 and Fig. 2-3-14) also show that cytokinin increases the weight of the sprouts and the concentration of chlorophyll (iP, tZ are both sorts of cytokinin). To know more about this assay, please see here.
iP : 6-(γ, γ-Dimethylallylamino) purine, tZ : trans-zeatin (both sorts of cytokinin)
The absorbances of the supernatant were read at 663.6 and 646.6 nm. Calculations of concentrations of the chlorophyll were carried out as described by Porra et al.,1989.
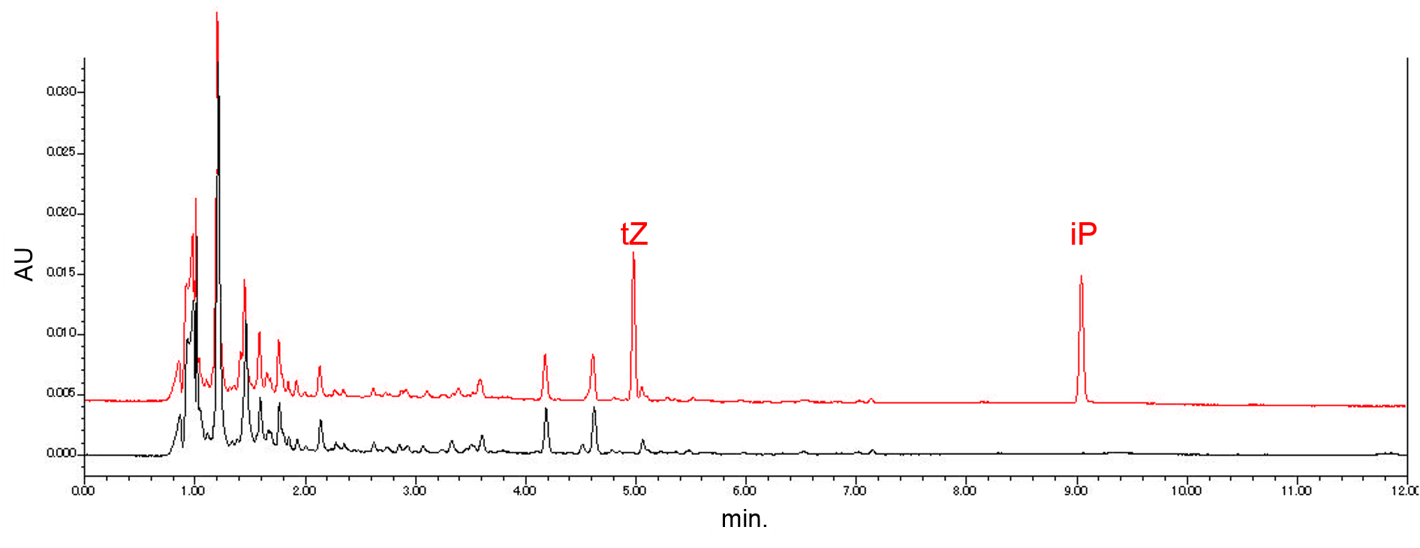
Through ultra-performance liquid chromatography (UPLC), we planned to confirm that our "Farmer E. coli" actually produces cytokinin. Before completing the system to produce cytokinin in E. coli, we determined the retention times of the authentic samples of iP and tZ by using UPLC. In addition, we confirmed that iP and tZ were able to be detected from the mixture of the E. coli culture medium and the cytokinin solution (Fig. 2-3-15). To see the detail method of this experiment, please click here.
5. Hybrid promoter for temporal pattern generation
In order to achieve the natural plants' temporal pattern for producing plant hormones in E. coli, we introduced an incoherent feed forward loop (Mangan et al., 2006) to our circuit, including a new hybrid promoter part ([http://parts.igem.org/Part:BBa_K1139150 BBa_K1139150]). Plants produce their hormones transiently rather than steadily (Takei et al., 2001). Moreover, continuous overexpression of hormones is harmful to plants (Thiman, 1937). Thus, we thought that it should be important to achieve this transient temporal pattern for producing plant hormones in E. coli, too. Our designed system with an incoherent feed forward loop is shown in Fig. 2-3-16. We newly developed the RM/lac hybrid promoter, which is activated by CI and repressed by LacI (Fig. 2-3-17). We planned to ligate a hormone synthase part downstream of this hybrid promoter. Our mathematical model (Fig. 2-3-18) shows the pulse wave which the temporal pattern for plant hormone production should achieve. While the RM/lac hybrid promoter activation by CI is a single-step reaction, the repression by LacI is a two-step reaction. Thus, the activation of the RM/lac hybrid promoter is faster than the repression. This time lag between the activation and the repression is important for generating a temporal pattern of plant hormone production (details about our mathematical model can be found here).
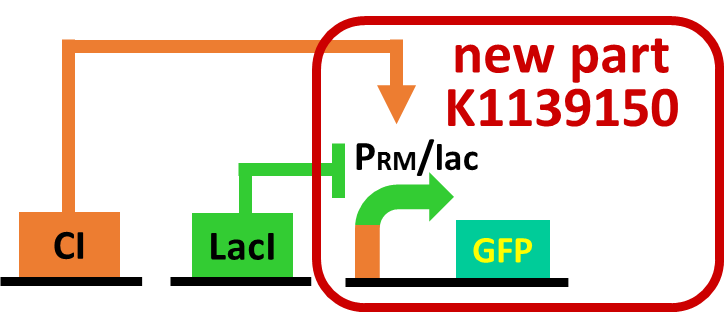
As a first step to achieve this system with an incoherent feed forward loop, we constructed a circuit shown in Fig. 2-3-19 to confirm that our new RM/lac hybrid promoter actually works. We set GFP as an output of the RM/lac hybrid promoter and introduced the circuit into E. coli . Through an induction assay (Fig. 2-3-20), we saw that our new RM/lac hybrid promoter was actually activated by CI and repressed by LacI. Details about this assay can be found here.
6. Applications
Our project can be applied to studying the plants' response to external plant hormones. By applying the incoherent feed forward loop to our circuit, we can achieve pulse-generation of the output (Basu et al., 2003). Moreover, the pulse wave can be customized with changing the parameters, for example, intensities of promoters or various interactions. Various artificial genetic circuits which can achieve the temporal pattern of the output, such as incoherent feed forward loop, have been reported (Elowitz et al., 2000; Fung et al., 2005). By combining plant hormone synthesis genes to these artificial genetic circuits, we will be able to make E. coli produce plant hormones in various temporal patterns. Analysis of the plants' response to plant hormones given in temporal change leads to elucidate the abilities of plants in signal transduction. Findings in plant science using this strategy will be important for contributing to new approaches in farming. We believe that such basic research can help to solve worldwide food shortages. We hope to meet public expectations by realizing agriculture aids by bacteria, though there may be some difficulties.
 "
"