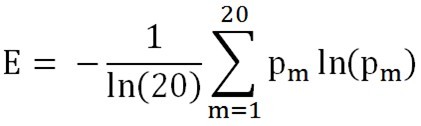Team:CAU China/Model2
From 2013.igem.org
(Difference between revisions)
| (3 intermediate revisions not shown) | |||
| Line 2: | Line 2: | ||
Content= | Content= | ||
__NOTOC__ | __NOTOC__ | ||
| - | + | <STRONG><p align=center><FONT style="FONT-SIZE:15pt;WIDTH: 100%;COLOR: #CD2626; LINE-HEIGHT: 120%; FONT-FAMILY:Arial">Model1</FONT></p></STRONG> | |
| + | ---- | ||
| - | |||
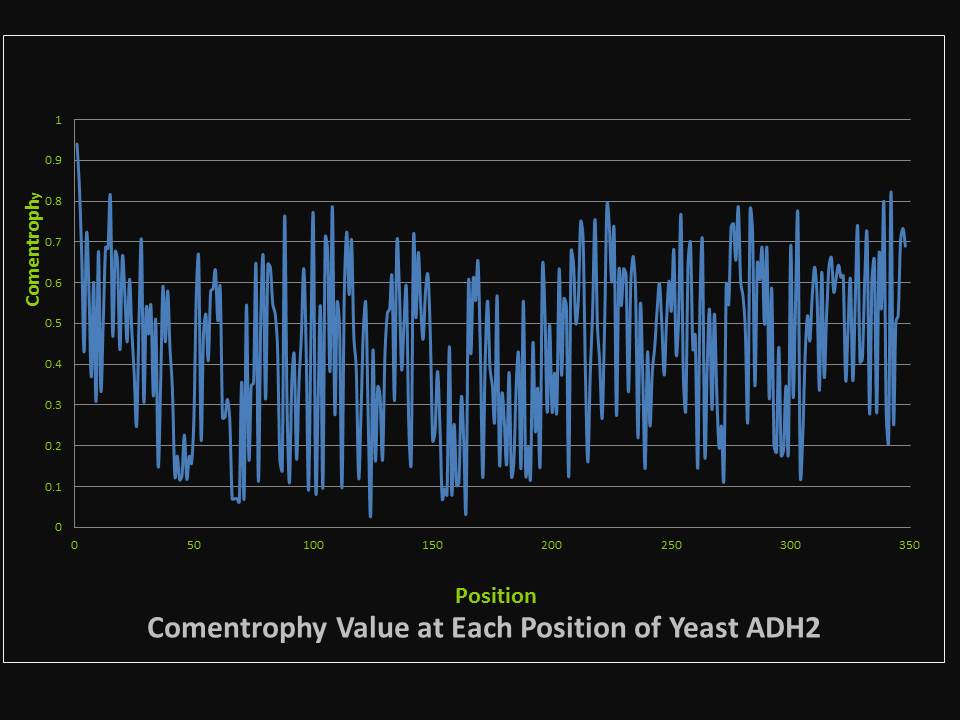
| + | We use comentrophy to evaluate the variation at each position of alcohol dehydrogenase, our engineering template. Comentrophy is a quantity with a value ranging from 0 to 1. The higher the value, the more variable the position is during the evolution. Here the figure describes the comentrophy value at each position of yeast alcohol dehydrogenase. | ||
| + | [[Image:CAU_project_md_2.png|center||500px]] | ||
| + | <p align=center>Where,pm is the possibility of the occurance of any amino acid at a specific position. | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | [[Image:CAU_project_md_1.png|center||500px]] | ||
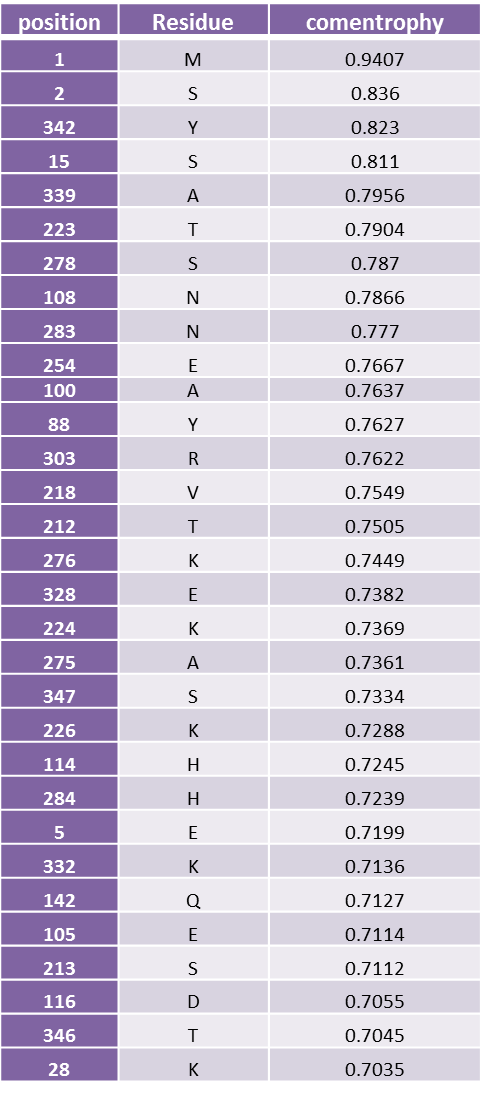
| + | Highly variable position of yeast alcohol dehydrogenase | ||
| + | [[Image:CAU_project_md_3.png|center||500px]] | ||
Latest revision as of 15:57, 27 September 2013

|
Model1
Where,pm is the possibility of the occurance of any amino acid at a specific position.
|
 "
"