Team:Imperial College/Electron microscopy
From 2013.igem.org
Margarita K (Talk | contribs) |
Margarita K (Talk | contribs) |
||
| (25 intermediate revisions not shown) | |||
| Line 2: | Line 2: | ||
<h1>Scanning Electron Microscopy</h1> | <h1>Scanning Electron Microscopy</h1> | ||
| - | <p> A good way to observe the function of a degradation enzyme is to incubate it with the polymer and scan the surface after some time with an electron micrograph. This is a widely used method for studying plastic degradation and we are | + | <p> A good way to observe the function of a degradation enzyme is to incubate it with the polymer and scan the surface after some time with an electron micrograph. This is a widely used method for studying plastic degradation and we are applying it for characterising Proteinase K enzyme ([http://parts.igem.org/Part:BBa_K1149008 BBa_K1149008]) and recording how it "chews" on a PLA cup. </p> |
| - | <p> PLA can spontaneously hydrolyse in water. However, this only happens very slowly and at higher temperatures. | + | <p> PLA can spontaneously hydrolyse in water. However, this only happens very slowly and at higher temperatures. Below are SEM images of the process, taken from the literature (see references).</p> |
| - | + | ||
https://static.igem.org/mediawiki/parts/2/2d/PLA_hydrolisis_SEM.jpg | https://static.igem.org/mediawiki/parts/2/2d/PLA_hydrolisis_SEM.jpg | ||
| + | <br> SEM images of the surface of PLA incubated for 9 and 20 days in deionized water at 58 °C. | ||
| + | <p>We are expecting our enzymes to have at least as visible effect in 3 or less days.</p> | ||
<h5>method</h5> | <h5>method</h5> | ||
| - | We cut small pieces of the cup and washed them with ethanol and deionised water in order to remove any potential contamination. | + | We cut small pieces of the cup and washed them with ethanol and then with deionised water in order to remove any potential contamination. |
| + | <br> | ||
| + | |||
| + | https://static.igem.org/mediawiki/parts/7/7a/Pla_prapared_for_SEM.jpg | ||
| + | |||
| + | <p><b>Samples 1A, 2A and 3A </b> were incubated with 1mL proteinase K enzyme solution (purchased from Sigma Aldrich) for 1, 2 and 3 days. (0.667 mg enzyme in 1mL PK Buffer per sample) The negative control A is PLA piece incubated with PK Buffer at 37°C, shaking. </p> | ||
| + | <p><b>Samples 1B, 2B and 3B</b> were incubated with 1mL cell lysate-PK buffer solution (1:2) of E.coli MG1655 transformed with BBa_K1149008, for 1, 2 and 3 days. The negative control B was incubated with cell lysate solution (1:2) from MG1655 E.coli strain containing the empty vector. </p> | ||
| + | <p><b>Sample C0</b> was not treated with anything. </p> | ||
| + | (please see [https://2013.igem.org/Team:Imperial_College/Protocols protocols] for preparation of lysate and PK buffer) | ||
| + | |||
| + | All experiments were carried out at 37 degrees, shaking in a 1.5 mL tube. (summer in greece) We have used a scanning electron micrograph at the [http://www3.imperial.ac.uk/materials/facilities/em Harvey Flower Microstructural Characterisation Suite], Imperial College. | ||
| + | |||
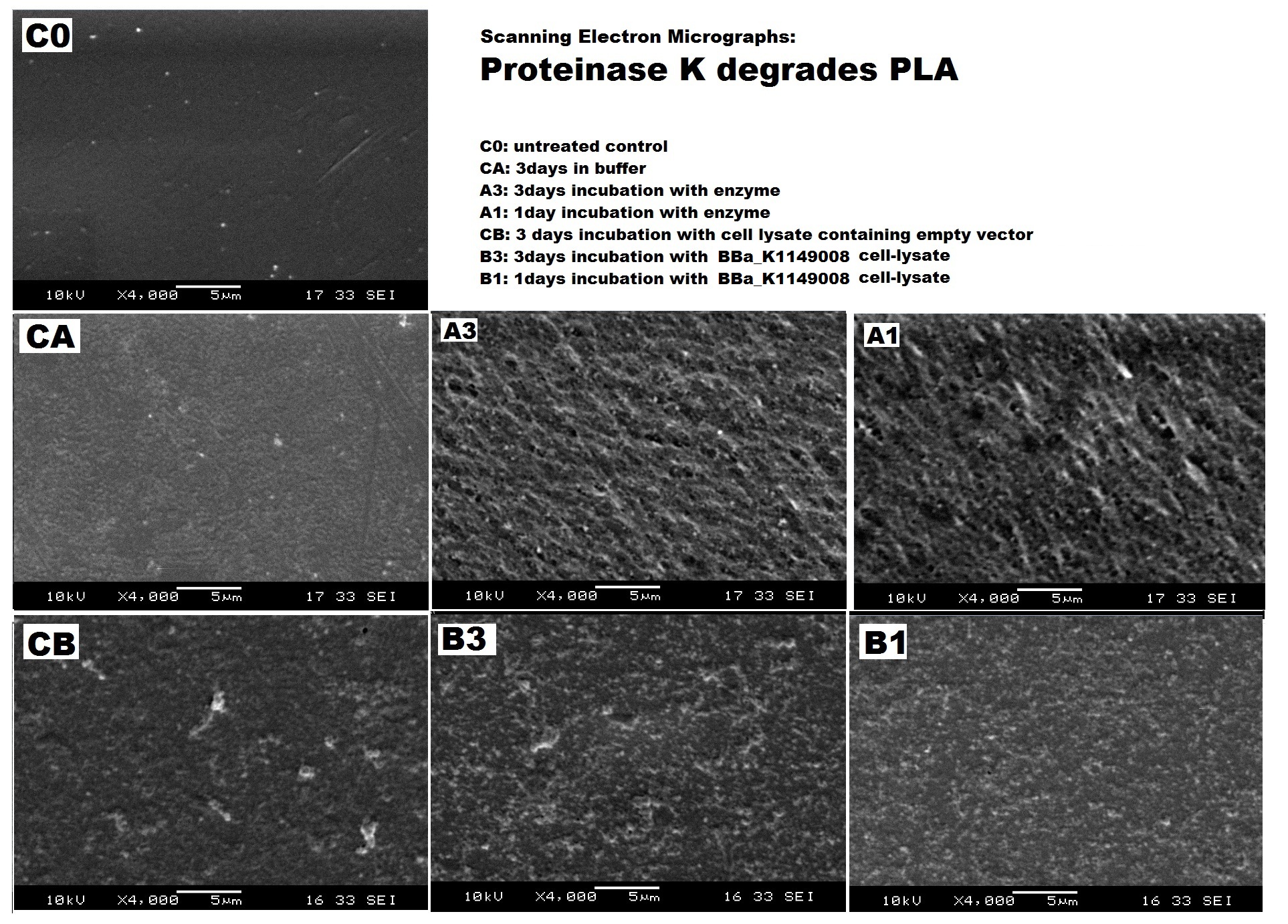
| + | <h5>results</h5> | ||
| + | |||
| + | [[File:PLA SEM all ICL.jpg|thumbnail|center|1000px|<b>Scanning Electron Micrographs show the increased pitting of PLA surface after enzyme and cell lysate treatment. (Images by Imperial College iGEM Team 3013)</b>]] | ||
| - | |||
| - | |||
| - | |||
<br> | <br> | ||
Latest revision as of 17:57, 4 October 2013
Contents |
Scanning Electron Microscopy
A good way to observe the function of a degradation enzyme is to incubate it with the polymer and scan the surface after some time with an electron micrograph. This is a widely used method for studying plastic degradation and we are applying it for characterising Proteinase K enzyme ([http://parts.igem.org/Part:BBa_K1149008 BBa_K1149008]) and recording how it "chews" on a PLA cup.
PLA can spontaneously hydrolyse in water. However, this only happens very slowly and at higher temperatures. Below are SEM images of the process, taken from the literature (see references).

SEM images of the surface of PLA incubated for 9 and 20 days in deionized water at 58 °C.
We are expecting our enzymes to have at least as visible effect in 3 or less days.
method
We cut small pieces of the cup and washed them with ethanol and then with deionised water in order to remove any potential contamination.

Samples 1A, 2A and 3A were incubated with 1mL proteinase K enzyme solution (purchased from Sigma Aldrich) for 1, 2 and 3 days. (0.667 mg enzyme in 1mL PK Buffer per sample) The negative control A is PLA piece incubated with PK Buffer at 37°C, shaking.
Samples 1B, 2B and 3B were incubated with 1mL cell lysate-PK buffer solution (1:2) of E.coli MG1655 transformed with BBa_K1149008, for 1, 2 and 3 days. The negative control B was incubated with cell lysate solution (1:2) from MG1655 E.coli strain containing the empty vector.
Sample C0 was not treated with anything.
(please see protocols for preparation of lysate and PK buffer)
All experiments were carried out at 37 degrees, shaking in a 1.5 mL tube. (summer in greece) We have used a scanning electron micrograph at the [http://www3.imperial.ac.uk/materials/facilities/em Harvey Flower Microstructural Characterisation Suite], Imperial College.
results
references
Effect of NR on the hydrolytic degradation of PLA, Huang et al., 2013
 "
"