Team:Edinburgh/Project/Results/Metal promoters Results
From 2013.igem.org
Hristianita (Talk | contribs) (Created page with "{{Team:Edinburgh/Menu}} {{Team:Edinburgh/Project_side_menu}} <div class='content'> </div> {{Team:Edinburgh/Footer}}") |
|||
| (44 intermediate revisions not shown) | |||
| Line 3: | Line 3: | ||
<div class='content'> | <div class='content'> | ||
| + | |||
| + | <h3> Fur Box Assembly </h3> | ||
| + | |||
| + | |||
| + | |||
| + | Using the protocol found in the [https://2013.igem.org/Team:Edinburgh/GenBrick/GenablerAppendix appendix], a 3-part construct was assembled with an acceptor vector pSB1C3 (BBa_K1122009), a Lac promoter and truncated gene called PLac_LacZ (BBa_J33207) optimised for GenBrick and the fluorescent protein GFP (BBa_K1122004). The assembly included a [https://2013.igem.org/Team:Edinburgh/Introduction/Metal_promoters fur box] in the linker between PLac_LacZ and GFP. More information on the structure of the assembly can be found in the [https://2013.igem.org/Team:Edinburgh/GenBrick/GenbrickAction GenBrick in action] section. When high intracellular iron concentrations are present, the [https://2013.igem.org/Team:Edinburgh/Project/Results/Metal_promoters_Results#Fur Fur transcription factor] binds to the fur box and represses transcription upstream. | ||
| + | |||
| + | |||
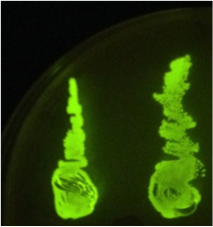
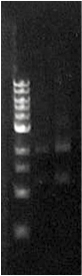
| + | When the assembly was finished, two transformations were made with competent ''E. coli'' JM109. These were then plated on IPTG + chloramphenicol plates and left overnight. In one plate, 4 out of 9 colonies were green fluorescent; the other five had re-ligated and were red fluorescent. Figure 1 shows this plate after two GFP+ colonies were taken and re-streaked; the red colonies exhibit RFP from the re-ligated vector. The other plate had a success rate of 8 out of 20 (the competent cells were concentrated x10 before plating). Figure 2 shows the assembly fluorescing under UV when no iron was present, which proves that GFP is successfully over-expressed. Figure 3 shows the restriction-digestion gel using PstI and XbaI. | ||
| + | |||
| + | |||
| + | |||
| + | [[File:APlateWithRFPandGFP.jpg|600px]] | ||
| + | |||
| + | ''' ''Figure 1.'' ''' This plate has chloramphenicol and IPTG. Originally it had 4 out of 9 colonies green fluorescent and the rest red fluorescent, however two green fluorescent colonies were taken and restreaked for the iron assay. | ||
| + | |||
| + | |||
| + | [https://2013.igem.org/Team:Edinburgh/Project/Results/Metal_promoters_Results#Top Back to top] | ||
| + | |||
| + | |||
| + | {|cellpadding="20" | ||
| + | |- | ||
| + | | | ||
| + | [[image:furbox1.png]] | ||
| + | | | ||
| + | [[image:furbox2.png]] | ||
| + | |||
| + | |- | ||
| + | | | ||
| + | ''' ''Figure 2.'' ''' Fur box Assembly. After transforming competent cells with the assembly and leaving overnight on an IPTG + chloramphenicol plate, 2 green fluorescent colonies were restreaked. A gel was performed to show the parts had correctly assembled. | ||
| + | | | ||
| + | ''' ''Figure 3.'' ''' Fur box digest. On the left is the ladder, the first column is the undigested assembly which shows only one band (the second lower band is an artifact due to leakage). The second column contains the digested assembly. The size of PLac_LacZ + linkers + GFP was correct. | ||
| + | |} | ||
| + | |||
| + | |||
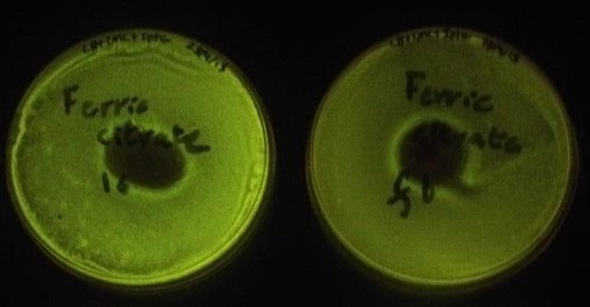
| + | Using the same GenBrick method, an assembly was made with the same parts but instead of the fur box linker, a new linker was designed ([https://2013.igem.org/Team:Edinburgh/GenBrick/Linker_Designer linker 6]). In effect, the new assembly was the same as the last one, except without a fur box and was used as a control for the iron assay. To test whether or not the fur box was affecting GFP production in the cell when iron concentration varied, a simple iron assay was devised with ferric citrate and ferrous sulfate. Four plates were lawn plated on IPTG and chloramphenicol plates; two plates were made with the fur box assembly (from the same original colony) and two plates were made with the linker 6 assembly (also from the same original colony). On a linker 6 and a fur box plate, 100 mg of ferric citrate was added in the middle of the plate (Figure 4). On the other plates, the same assay was made but with 100 mg of ferrous sulfate (Figure 5). | ||
| + | |||
| + | |||
| + | |||
| + | [[File:Ferric_Citrate.jpg]] | ||
| + | |||
| + | '' '''Figure 4.''' '' The plate on the left contained the assembly with linker 6 while the plate on the right contained the assembly with the fur box. The plate on the right was found to have a statistically significantly lower fluorescence than the plate on the left. The lower fluorescence can be explained by the fact that iron diffused out and created a gradient. Iron binds to the Fur protein which binds to the fur box and represses GFP production. | ||
| + | |||
| + | |||
| + | |||
| + | [[File:Ferrous_Sulphate.jpg]] | ||
| + | |||
| + | '' '''Figure 5.''' '' The plate on the left contained the assembly with linker 6 while the plate on the right contained the assembly with the fur box. Again, the plate on the right had a lower fluorescence than the plate on the left which confirms the results from Figure 4. | ||
| + | |||
| + | |||
| + | [https://2013.igem.org/Team:Edinburgh/Project/Results/Metal_promoters_Results#Top Back to top] | ||
| + | |||
| + | |||
| + | <div id="Fur"><h3> Fur transcription factor assay </h3></div> | ||
| + | |||
| + | |||
| + | To assay the [https://2013.igem.org/Team:Edinburgh/Introduction/Metal_promoters Fur gene] (BBa_K1122666) activity, PCR was first performed to amplify Fur genomic sequence using primers containing EarI restriction sites as well as EcoRI (GenBrick compatible primers). | ||
| + | |||
| + | The forward primer was: | ||
| + | |||
| + | C GAATTC CT TCTAG ATG GCC TCT TCT TCG AAC CGT ATT GAT CGT | ||
| + | |||
| + | The reverse complement primer was: | ||
| + | |||
| + | ATC CTGCAG CT AC TAG TAC TCT TCA GGC TTC AGT TTC TTT TCC | ||
| + | |||
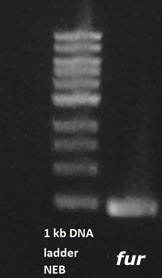
| + | An agarose gel was performed and showed that the Fur gene was properly amplified (figure 6). | ||
| + | |||
| + | |||
| + | |||
| + | [[File:AggregationResults2.png|center]] | ||
| + | |||
| + | '' '''Figure 6.''' '' Agarose gel of amplified Fur gene sequence. The Fur gene is normally 450 base pairs and the band shown is slightly lower than 500 base pairs. | ||
| + | |||
| + | |||
| + | |||
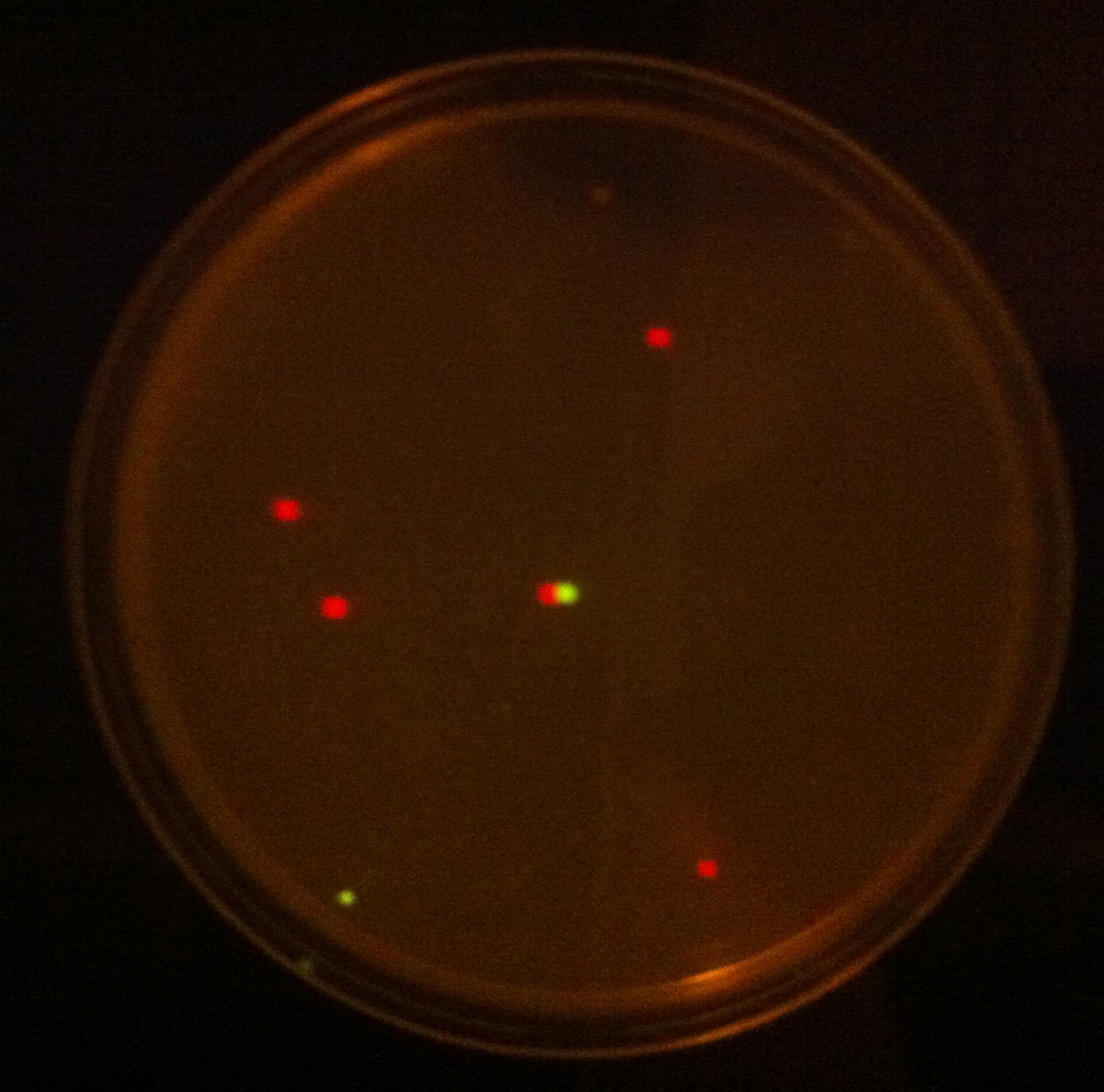
| + | Next, the Fur gene was restricted and ligated into pSB1C3 using XbaI and PstI. Once that was done, PLac_LacZ was placed upstream of the Fur gene to over-express it. It was assumed that over-expression of Fur could increase bacterial tolerance to increased iron concentration due to enhanced perception of environmental iron. This was analysed experimentally by culturing cells induced and un-induced to express Fur in LB medium supplemented with increasing concentrations of iron (Figure 7). | ||
| + | |||
| + | |||
| + | |||
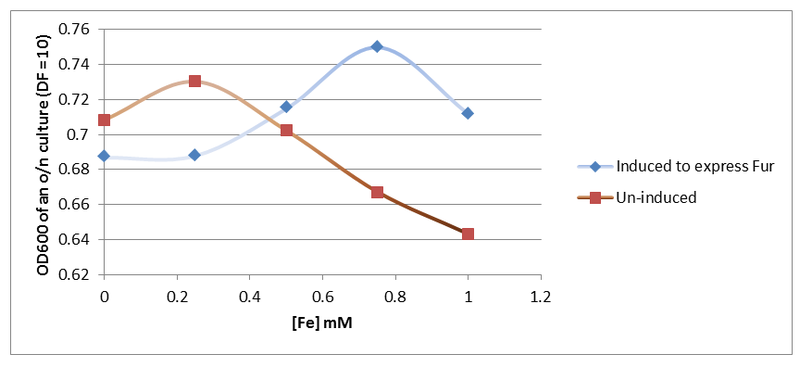
| + | [[File:800px-Fur_graph.png|600px]] | ||
| + | |||
| + | '' '''Figure 7.''' '' Results obtained (Figure 7) indicate that indeed the cells that contain more copies of Fur protein survive better in medium with increased iron levels. | ||
| + | |||
| + | |||
| + | |||
| + | [https://2013.igem.org/Team:Edinburgh/Project/Results/Metal_promoters_Results#Top Back to top] | ||
| + | |||
| + | |||
</div> | </div> | ||
{{Team:Edinburgh/Footer}} | {{Team:Edinburgh/Footer}} | ||
Latest revision as of 01:20, 5 October 2013
Fur Box Assembly
Using the protocol found in the appendix, a 3-part construct was assembled with an acceptor vector pSB1C3 (BBa_K1122009), a Lac promoter and truncated gene called PLac_LacZ (BBa_J33207) optimised for GenBrick and the fluorescent protein GFP (BBa_K1122004). The assembly included a fur box in the linker between PLac_LacZ and GFP. More information on the structure of the assembly can be found in the GenBrick in action section. When high intracellular iron concentrations are present, the Fur transcription factor binds to the fur box and represses transcription upstream.
When the assembly was finished, two transformations were made with competent E. coli JM109. These were then plated on IPTG + chloramphenicol plates and left overnight. In one plate, 4 out of 9 colonies were green fluorescent; the other five had re-ligated and were red fluorescent. Figure 1 shows this plate after two GFP+ colonies were taken and re-streaked; the red colonies exhibit RFP from the re-ligated vector. The other plate had a success rate of 8 out of 20 (the competent cells were concentrated x10 before plating). Figure 2 shows the assembly fluorescing under UV when no iron was present, which proves that GFP is successfully over-expressed. Figure 3 shows the restriction-digestion gel using PstI and XbaI.
Figure 1. This plate has chloramphenicol and IPTG. Originally it had 4 out of 9 colonies green fluorescent and the rest red fluorescent, however two green fluorescent colonies were taken and restreaked for the iron assay.
|
Figure 2. Fur box Assembly. After transforming competent cells with the assembly and leaving overnight on an IPTG + chloramphenicol plate, 2 green fluorescent colonies were restreaked. A gel was performed to show the parts had correctly assembled. |
Figure 3. Fur box digest. On the left is the ladder, the first column is the undigested assembly which shows only one band (the second lower band is an artifact due to leakage). The second column contains the digested assembly. The size of PLac_LacZ + linkers + GFP was correct. |
Using the same GenBrick method, an assembly was made with the same parts but instead of the fur box linker, a new linker was designed (linker 6). In effect, the new assembly was the same as the last one, except without a fur box and was used as a control for the iron assay. To test whether or not the fur box was affecting GFP production in the cell when iron concentration varied, a simple iron assay was devised with ferric citrate and ferrous sulfate. Four plates were lawn plated on IPTG and chloramphenicol plates; two plates were made with the fur box assembly (from the same original colony) and two plates were made with the linker 6 assembly (also from the same original colony). On a linker 6 and a fur box plate, 100 mg of ferric citrate was added in the middle of the plate (Figure 4). On the other plates, the same assay was made but with 100 mg of ferrous sulfate (Figure 5).
Figure 4. The plate on the left contained the assembly with linker 6 while the plate on the right contained the assembly with the fur box. The plate on the right was found to have a statistically significantly lower fluorescence than the plate on the left. The lower fluorescence can be explained by the fact that iron diffused out and created a gradient. Iron binds to the Fur protein which binds to the fur box and represses GFP production.
Figure 5. The plate on the left contained the assembly with linker 6 while the plate on the right contained the assembly with the fur box. Again, the plate on the right had a lower fluorescence than the plate on the left which confirms the results from Figure 4.
Fur transcription factor assay
To assay the Fur gene (BBa_K1122666) activity, PCR was first performed to amplify Fur genomic sequence using primers containing EarI restriction sites as well as EcoRI (GenBrick compatible primers).
The forward primer was:
C GAATTC CT TCTAG ATG GCC TCT TCT TCG AAC CGT ATT GAT CGT
The reverse complement primer was:
ATC CTGCAG CT AC TAG TAC TCT TCA GGC TTC AGT TTC TTT TCC
An agarose gel was performed and showed that the Fur gene was properly amplified (figure 6).
Figure 6. Agarose gel of amplified Fur gene sequence. The Fur gene is normally 450 base pairs and the band shown is slightly lower than 500 base pairs.
Next, the Fur gene was restricted and ligated into pSB1C3 using XbaI and PstI. Once that was done, PLac_LacZ was placed upstream of the Fur gene to over-express it. It was assumed that over-expression of Fur could increase bacterial tolerance to increased iron concentration due to enhanced perception of environmental iron. This was analysed experimentally by culturing cells induced and un-induced to express Fur in LB medium supplemented with increasing concentrations of iron (Figure 7).
Figure 7. Results obtained (Figure 7) indicate that indeed the cells that contain more copies of Fur protein survive better in medium with increased iron levels.

| 
| | | | 
|
| This iGEM team has been funded by the MSD Scottish Life Sciences Fund. The opinions expressed by this iGEM team are those of the team members and do not necessarily represent those of MSD | |||||
 "
"