Team:Goettingen/Project/OurProject
From 2013.igem.org
(→Diadenylate cyclase) |
|||
| (25 intermediate revisions not shown) | |||
| Line 7: | Line 7: | ||
</div><!--close header--> | </div><!--close header--> | ||
<br /> | <br /> | ||
| + | <img src="https://static.igem.org/mediawiki/2013/6/66/Goe-qNav.png" style="position: fixed;z-index: 199;bottom: 11%;right: 2.5%;_position: absolute;_top: expression(documentElement.scrollTop + documentElement.clientHeight-this.offsetHeight);cursor: pointer;opacity: 0.5;-moz-opacity: 0.5;-khtml-opacity: 0.5;-filter: alpha(opacity = 50 );" onclick="toggle(this.nextElementSibling || getnextElementSibling(this))" /> | ||
| + | <div style="position: fixed;z-index: 199;bottom: 19%;right: 2.5%;_position: absolute;_top: expression(documentElement.scrollTop + documentElement.clientHeight-this.offsetHeight);cursor: pointer;display: none;background: #fff;border: 1px solid #A3A3A3;padding: 20px; | ||
| + | border-radius: 5px;"> | ||
| + | <b>Project Overview:</b><br /> | ||
| + | <ul> | ||
| + | <li><a href="#Background">Background</a></li> | ||
| + | <ul> | ||
| + | <li><a href="#The_threat">The threat</a></li> | ||
| + | <li><a href="#c-di-AMP">c-di-AMP</a></li> | ||
| + | </ul> | ||
| + | <li><a href="#Our_Project">Our Project</a></li> | ||
| + | <ul> | ||
| + | <li><a href="#Reporter_systems">Reporter systems</a></li> | ||
| + | <li><a href="#Microarray">Microarray</a></li> | ||
| + | <li><a href="#Diadenylate_cyclase">Diadenylate cyclase</a></li> | ||
| + | </ul> | ||
| + | </ul> | ||
| + | </div> | ||
| - | |||
| - | |||
<div id="body"> | <div id="body"> | ||
<div class="f-box"> | <div class="f-box"> | ||
| Line 17: | Line 33: | ||
{{:Team:Goettingen/templates/navProject}} | {{:Team:Goettingen/templates/navProject}} | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
<html> | <html> | ||
</div><!--close nav--> | </div><!--close nav--> | ||
| Line 41: | Line 48: | ||
Further misuse will lead us back to medieval conditions not only for postoperative patients but even small infections will be able to cause severe problems and may cost many lives. Tragically, only four new classes of antibiotics made it to the market in the last 40 years as research in the field of antibiotics is getting less profitable due to high research and development costs and the poor chance of success to actually launch a new antibiotic (Cooper and Shlaes, 2011). | Further misuse will lead us back to medieval conditions not only for postoperative patients but even small infections will be able to cause severe problems and may cost many lives. Tragically, only four new classes of antibiotics made it to the market in the last 40 years as research in the field of antibiotics is getting less profitable due to high research and development costs and the poor chance of success to actually launch a new antibiotic (Cooper and Shlaes, 2011). | ||
| - | Therefore, we should better control the use of antibiotics in medicine as well as agriculture. Meanwhile, we need to develop new antibiotics, which can sufficiently eliminate the pathogenic bacteria without affecting beneficial and harmless bacteria such as the well-known gut bacterium Escherichia coli (Witte ''et al.'', 2008). | + | Therefore, we should better control the use of antibiotics in medicine as well as agriculture. Meanwhile, we need to develop new antibiotics, which can sufficiently eliminate the pathogenic bacteria without affecting beneficial and harmless bacteria such as the well-known gut bacterium ''Escherichia coli'' (Witte ''et al.'', 2008). |
==c-di-AMP== | ==c-di-AMP== | ||
| - | What a good antibiotic should target? The answer has been given by Dr. Sven Halbedel. From his view, a good antibacterial target should: | + | What a good antibiotic should target? The answer has been given by Dr. Sven Halbedel. From his point of view, a good antibacterial target should: |
<html><img src="https://static.igem.org/mediawiki/2013/c/c5/Goe-Numbers.png" width="200px" class="fl" style="margin-right:30px" /></html> | <html><img src="https://static.igem.org/mediawiki/2013/c/c5/Goe-Numbers.png" width="200px" class="fl" style="margin-right:30px" /></html> | ||
| - | be essential for growth/ virulence of pathogenic bacteria | + | ● be essential for growth/ virulence of pathogenic bacteria |
| - | + | ● have no homolog in humans | |
| - | be pleiotropic | + | ● be pleiotropic |
| - | + | ● have no suppressors | |
| - | be conserved among pathogenic bacteria | + | ● be conserved among pathogenic bacteria |
| - | Of all those possible targets for novel antibiotics, c-di-AMP is the "shining star". Though very recently discovered (in the year 2008), it has already caught a lot of attention. It is the only known essential signaling nucleotide. It exists and plays vital role in a wide range of Gram-positive bacteria like ''Bacillus, Listeria, Streptococcus and Staphylococcus''. Meanwhile its trace has never been found in Gram-negative bacteria nor human beings. With all these reasons, c-di-AMP and | + | Of all those possible targets for novel antibiotics, c-di-AMP is the "shining star". Though very recently discovered (in the year 2008), it has already caught a lot of attention. It is the only known essential signaling nucleotide. It exists and plays a vital role in a wide range of Gram-positive bacteria like ''Bacillus, Listeria, Streptococcus and Staphylococcus''. Meanwhile its trace has never been found in known beneficial Gram-negative bacteria nor human beings. With all these reasons, c-di-AMP and its regulatory compartments become the most heated targets of interest in the development of new antibacterial substances. |
| - | Therefore the iGEM Team Göttingen addresses the problem of multi-resistant bacteria by targeting c-di-AMP, the achilles heel of our enemy . | + | Therefore the iGEM Team Göttingen addresses the problem of multi-resistant bacteria by targeting c-di-AMP,the achilles heel of our enemy. |
| Line 77: | Line 84: | ||
<html> | <html> | ||
<img src="https://static.igem.org/mediawiki/2013/5/51/Goe-greenColi-labcoat.png" class="fr" width="170" /><p>Our project is aimed at finding a way to fight against multi-resistant bacteria by targeting c-di-AMP. We made three different approaches.</p> | <img src="https://static.igem.org/mediawiki/2013/5/51/Goe-greenColi-labcoat.png" class="fr" width="170" /><p>Our project is aimed at finding a way to fight against multi-resistant bacteria by targeting c-di-AMP. We made three different approaches.</p> | ||
| - | <p style="font-size:11pt;color:#7c7c7c"><strong>We built two reporter systems | + | <p style="font-size:11pt;color:#7c7c7c"><strong>We built two reporter systems which enable us to visualize the level of c-di-AMP (accomplished by <a href="https://2013.igem.org/Team:Goettingen/Team/Reporter" >Reporter Team</a>).</strong></p> |
| - | <p style="font-size:11pt;color:#7c7c7c"><strong>We also searched for the genes in <i>Bacillus | + | <p style="font-size:11pt;color:#7c7c7c"><strong>We also searched for the genes in <i>Bacillus subtilis</i>, whose expression level is affected by the level of c-di-AMP. We found <i>ydaO</i> and identified a Riboswitch upstream of its open reading frame, which responds to c-di-AMP. We used the <i>ydaO</i> Riboswitch directly in our second reporter system (accomplished by <a href="https://2013.igem.org/Team:Goettingen/Team/Array">Array Team</a>).</strong></p> |
| - | <p style="font-size:11pt;color:#7c7c7c"><strong>Last but not least, we | + | <p style="font-size:11pt;color:#7c7c7c"><strong>Last but not least, we characterized the diadenylate cyclase (DAC) from<i>Listeria monocytogenes</i>. We successfully expressed tagged truncated DAC (catalytic domain) in <i>E.coli</i> and purified it.We analyzed its enzymatic kinetics and crystallized it. In the end, we are able to determine the structure (accomplished by <a href="https://2013.igem.org/Team:Goettingen/Team/DAC">DAC Team</a>)</strong></p> |
<br /><br /> | <br /><br /> | ||
</html> | </html> | ||
| Line 87: | Line 94: | ||
==Reporter systems== | ==Reporter systems== | ||
<html> | <html> | ||
| - | <p>For the Reporter Team, our final goal is to build a screening system, which allows quick identification and characterization of substances which are able to disturb c-di-AMP homeostasis in pathogenic bacteria. We believe the accomplished screening system will be a great help for pharmaceutical industry worldwide in finding new and more effective antibiotics against Gram-positive pathogens, for which c-di-AMP homeostasis is crucial. To accomplish that goal, we first need a reporter system | + | <p>For the Reporter Team, our final goal is to build a screening system, which allows quick identification and characterization of substances which are able to disturb c-di-AMP homeostasis in pathogenic bacteria. We believe the accomplished screening system will be a great help for pharmaceutical industry worldwide in finding new and more effective antibiotics against Gram-positive pathogens, for which c-di-AMP homeostasis is crucial. To accomplish that goal, we first need a reporter system that can visualize different levels of c-di-AMP.<p> |
<img src="https://static.igem.org/mediawiki/2013/5/56/Goe-greenColi-reporter.png" class="fl" style="display:inline;width:130px" /> | <img src="https://static.igem.org/mediawiki/2013/5/56/Goe-greenColi-reporter.png" class="fl" style="display:inline;width:130px" /> | ||
| - | <p>We used a few existing | + | <p>We used a few existing BioBricks and also created new ones to build up our system. At first, we attempted to construct a reporter system which is controlled by DarR, a transcriptional inhibitor identified in <i>Mycobacterium smegmatis</i>. This reporter system consists of three parts, namely a constitutively active promoter, the operator sequence DarR binds and a reporter gene cassette. c-di-AMP is able to stimulate the binding of DarR and its operator, acting as a co-inhibitor. Therefore the level of c-di-AMP can be visualized by the fluorescence of GFP: the higher the c-di-AMP level is, the lower the GFP fluorescence becomes. The reporter system is transformed into E.coli, which produces no endogens c-di-AMP. Therefore, we are able to test the system by providing the <i>E.coli</i> with c-di-AMP.</p> |
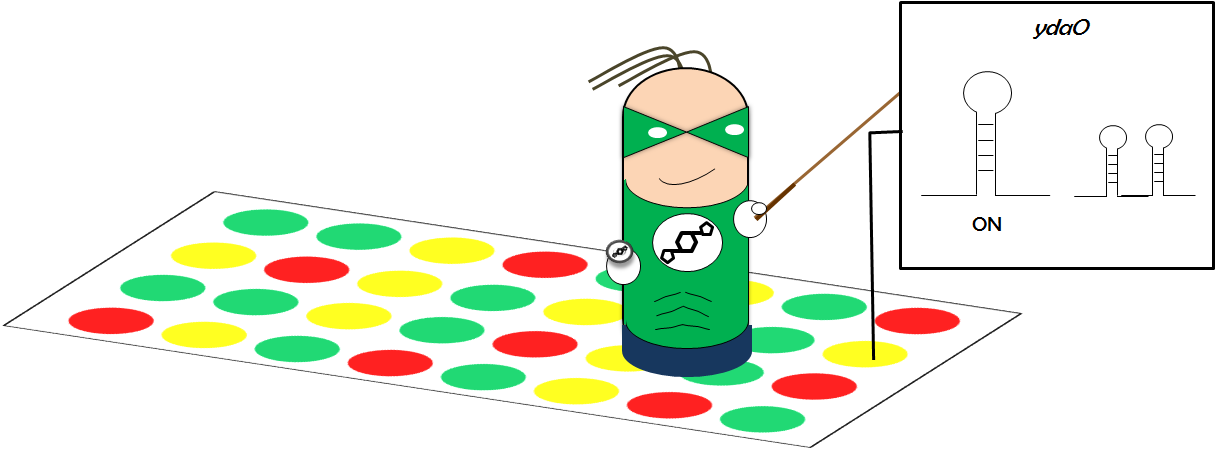
| - | <p>The second reporter system is based on the result of our Array Team. They found | + | <p>The second reporter system is based on the result of our Array Team. They found the gene <i>ydaO</i>, whose expression level is affected by the level of c-di-AMP. When the c-di-AMP level is low, the <i>ydaO</i> expression is up-regulated. We are able to identify a Riboswitch upstream of the <i>ydaO</i> open reading frame and used it in our second reporter system. The <i>ydaO</i> Riboswitch has two states: "ON" and "OFF". The switch between the two states depends on the presence of c-di-AMP: basically, when c-di-AMP is there, the Riboswitch is "OFF" and when there is no c-di-AMP, the Riboswitch is "ON". We cloned a reporter gene cassette CFP downstream of the native promoter + <i>ydaO</i> Riboswitch. This reporter system should act similarly to our first reporter system: when there is c-di-AMP, no signal is expected, but when there is no c-di-AMP, there will be a fluorescence signal.</p> |
| - | <p>We modified, improved and created several | + | <p>We modified, improved and created several BioBricks during the construct of our reporter system. To know more, please go to our <a href="/Team:Goettingen/Team/Reporter" >subteam page</a> and <a href="/Team:Goettingen/Parts">parts page</a>.</p> |
</html> | </html> | ||
| Line 98: | Line 105: | ||
==Microarray== | ==Microarray== | ||
| - | We | + | We focused mainly on the genes, whose expression level is regulated by c-di-AMP. Therefore, we compared the transcriptomic data of wild type with those of a hyperactive strain of Bacillus, which produces more c-di-AMP. |
By doing so, we are able to: first, find other compartments which also respond to c-di-AMP and therefore can be used to construct our reporter system, second we can shed light on the signaling web of c-di-AMP. | By doing so, we are able to: first, find other compartments which also respond to c-di-AMP and therefore can be used to construct our reporter system, second we can shed light on the signaling web of c-di-AMP. | ||
| - | + | In the collaboration with iGEM Team Groningen, we accomplished the microarray analysis, which has given us a first glimpse on all genes which could be regulated by c-di-AMP. Among those genes, ''ydaO'' caught most our attention. ''ydaO'' motif is reported to be a c-di-ATP-sensing Riboswitch in ''Bacillus substilis'' (unpublished data). We confirmed the array data with qRT-PCR. And the results consisted with array data. Therefore we believe we identified another genetic element that responds to c-di-AMP and we directly used it to build our second reporter system. | |
To know more about our work, please visit [[Team:Goettingen/Team/Array|subteam page]] and [[Team:Goettingen/Parts|parts page]]. | To know more about our work, please visit [[Team:Goettingen/Team/Array|subteam page]] and [[Team:Goettingen/Parts|parts page]]. | ||
| Line 110: | Line 117: | ||
==Diadenylate cyclase== | ==Diadenylate cyclase== | ||
<html> | <html> | ||
| - | <p>As the homeostasis of c-di-AMP is very important for <i>Listeria monocytogenes</i> (Witte <i>et al.</i>, 2013), a Gram-positive human pathogen, | + | <p>As the homeostasis of c-di-AMP is very important for <i>Listeria monocytogenes</i> (Witte <i>et al.</i>, 2013), a Gram-positive human pathogen, we are convinced that the DAC is a very promising target for the development of highly specific antibiotic substances which exclusively act on Gram-positive bacteria and are not harmful to Gram-negative ones, including the gut bacterium <i>Escherichia coli</i>. Therefore, we aimed to reveal the protein structure of a not yet characterized diadenylate cyclase domain. </p> |
<img src="https://static.igem.org/mediawiki/2013/6/6e/Goe-greenColi-crystal.png" class="fr" height="248" /> | <img src="https://static.igem.org/mediawiki/2013/6/6e/Goe-greenColi-crystal.png" class="fr" height="248" /> | ||
</html> | </html> | ||
| - | First we tried to express the complete CdaA protein of ''L. monocytogenes'' in E.coli. Unfortunately, we couldn't get any clone after several attempts. The full length protein might be toxic expressed in ''E.coli'', so we focused on a truncated version of the whole protein excluding the trans-membrane domains. The 173 amino acid protein fragment (100 – 273 of CdaA) harbors exclusively the catalytic domain of CdaA, also referred to as DacA. We attached an N-terminal ''Strep''-tag to the cyclase domain and expressed the protein driven by a T7-promoter. HPLC-MS/MS measurements determined the presence of c-di-AMP and, thus, the activity of the catalytic domain in also in the Gram-negative bacterium ''E.coli | + | First we tried to express the complete CdaA protein of ''L. monocytogenes'' in ''E.coli''. Unfortunately, we couldn't get any clone after several attempts. The full length protein might be toxic expressed in ''E.coli'', so we focused on a truncated version of the whole protein excluding the trans-membrane domains. The 173 amino acid protein fragment (100 – 273 of CdaA) harbors exclusively the catalytic domain of CdaA, also referred to as DacA. We attached an N-terminal ''Strep''-tag to the cyclase domain and expressed the protein driven by a T7-promoter. HPLC-MS/MS measurements determined the presence of c-di-AMP and, thus, the activity of the catalytic domain in also in the Gram-negative bacterium ''E.coli in vivo''. |
| - | Excluding the trans-membrane domains, the protein localizes to the cytoplasm and can be purified using streptavidin purification columns. After we purified the DAC protein, we wanted to | + | Excluding the trans-membrane domains, the protein localizes to the cytoplasm and can be purified using streptavidin purification columns. After we purified the DAC protein, we wanted to prove the functionality also ''in vitro''. The results showed that the purified catalytic domain of CdaA is still functional ''in vitro'' in proper conditions. Thus, the truncated diadenylate cyclase domain is qualified to construct a screening system with the ectopic production of c-di-AMP in ''E.coli'', as a further goal of our whole project. Also our experiments went on to our final step, crystallization and, finally, the determination of the 3D protein structure. |
| - | In collaboration with [[Team:Goettingen/NoteBook/Acknowlegement|Dr. Achim Dickmanns]], we successfully obtained protein crystals and with the help of [[Team:Goettingen/NoteBook/Acknowlegement|Dr. Piotr Neumann]], we managed to determine the 3D structure from | + | In collaboration with [[Team:Goettingen/NoteBook/Acknowlegement|Dr. Achim Dickmanns]], we successfully obtained protein crystals and with the help of [[Team:Goettingen/NoteBook/Acknowlegement|Dr. Piotr Neumann]], we managed to determine the 3D structure from the X-ray diffraction pattern. |
| - | We believe | + | We believe that the 3D structure of the diadenylate cyclase domain of ''Listeria'' can help pharmaceutical industry in developing novel antibiotics that interfere with DAC function. |
| - | In our progress we also created a new | + | In our progress we also created a new BioBrick, to know more, please go to our [[Team:Goettingen/Team/DAC|subteam page]] and [[Team:Goettingen/Parts|parts page]]. |
<html> | <html> | ||
Latest revision as of 13:51, 4 October 2013
 "
"





 ● be essential for growth/ virulence of pathogenic bacteria
● be essential for growth/ virulence of pathogenic bacteria