Team:Imperial College/tour
From 2013.igem.org
Iain Bower (Talk | contribs) |
Iain Bower (Talk | contribs) |
||
| (61 intermediate revisions not shown) | |||
| Line 2: | Line 2: | ||
__NOTOC__ | __NOTOC__ | ||
| - | |||
| + | <h1>We have engineered bacteria to turn non-recyclable waste into bioplastic</h1> | ||
| + | [[File:Overviewdiagram.png|right|500px| System Overview]] | ||
<p> | <p> | ||
| - | We have taken an expensive byproduct of recycling facilities, a type of mixed waste and turned it into something quite amazing | + | We have taken an expensive byproduct of recycling facilities, a type of mixed waste and turned it into something quite amazing; a material that can be used for a diverse range of applications from making your lunchbox to a 3D printed tissue scaffold. This material is the bioplastic <b>poly-3-hydroxybutyric acid P(3HB)</b>. Our system is designed to maximise the recovery of resources from the waste and so we have also investigated how we can use the oil based plastics within it to produce the commodity chemical ethylene glycol. We are passionate about using synthetic biology to help us move towards a more sustainable economy. We have considered what happens to the material we produce after it is used. This led us to develop the first synthetic biology recycling system for P(3HB) and one of the first in the world. The other well known bioplastic, poly lactic acid (PLA) is also expanding in use and so we developed a system to turn it back into its constituent monomers so that it can also be resynthesised, chemically or in vivo.</p> |
| - | <p> Click [[Team:Imperial_College/Waste_Degradation:_SRF| | + | <p> Click [[Team:Imperial_College/Waste_Degradation:_SRF|here]] to find out how we made P3HB and broke down oil derived plastics and [[Team:Imperial_College/BioPlastic_Recycling:_PHB|here]] to see how we recycled P3HB and PLA.</p> |
| - | + | <p class="clear">See here how we convert trash to treasure, a longer more detailed version can be found [http://www.youtube.com/watch?v=1oXt_EfqmLU&feature=youtu.be here]</p> | |
| + | <html> | ||
| + | <iframe width="420" height="315" src="//www.youtube-nocookie.com/embed/4msf6EMNIrY?rel=0" frameborder="0" allowfullscreen></iframe> | ||
| + | </html> | ||
| + | <h2>Our System: Modular and Plastic Looping ''E. coli'' (M.A.P.L.E.)</h2> | ||
| + | <h3>Module 1: Resource-full Waste</h3> | ||
| + | [[File:Mod1diagram.png|800px|centre]] | ||
| - | < | + | <br><br> |
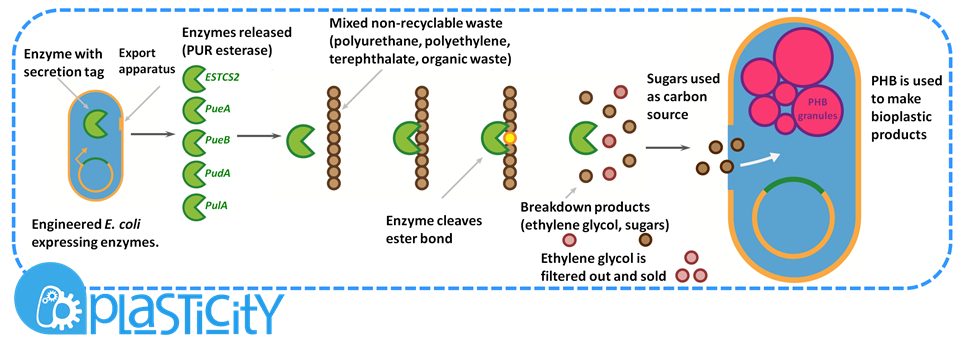
| + | <b>Non-recyclable waste is sourced from a recycling centre, placed in a bioreactor with our M.A.P.L.E system which breaks down the waste and synthesises the bioplastic P(3HB).</b> | ||
| - | |||
| - | [[File: | + | |
| - | <b> | + | [[File:Module_one_success.PNG|thumbnail|800px|center]] |
| + | |||
| + | <p><b>Click [[Team:Imperial_College/mainresults|here]] to see for yourself</b></p> | ||
| + | |||
<h3>Module 2: Plastic Fantastic</h3> | <h3>Module 2: Plastic Fantastic</h3> | ||
| - | [[File: | + | [[File:Mod2diagram.png|800px|centre]] |
<br><br><br><br> | <br><br><br><br> | ||
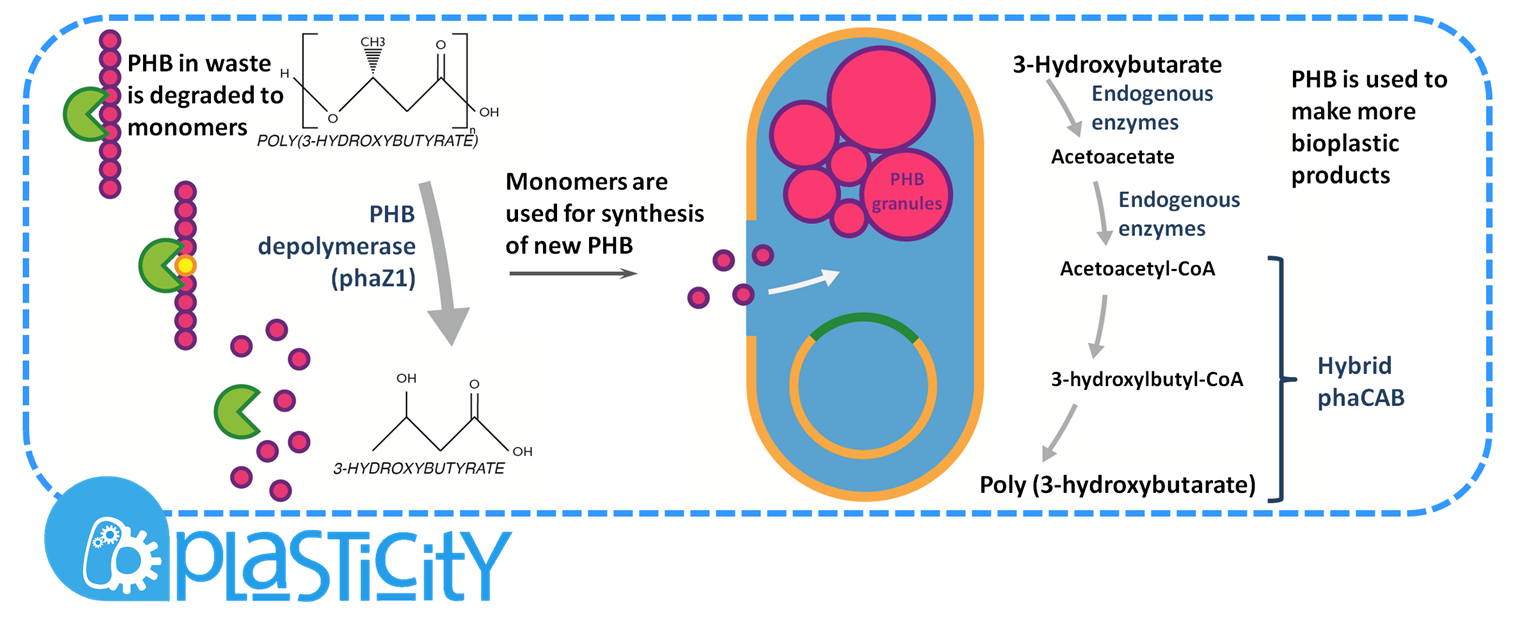
| - | <b>Plastic Fantastic is a complete P(3HB) bioplastic recycling platform, where P(3HB) | + | <b>Plastic Fantastic is a complete P(3HB) bioplastic recycling platform, where P(3HB) is degraded into monomeric form and then re-polymerised back into de novo P(3HB) for future applications.</b> |
| + | |||
| + | [[File:Module two success.PNG|thumbnail|800px|center]] | ||
| + | |||
| + | <p><b>Click [[Team:Imperial_College/mainresults|here]] to see for yourself</b></p> | ||
| + | |||
| + | |||
| + | <h1>Real world research guided the development of our project</h1> | ||
| + | [[File:POWERDAY VISIT.JPG|200px|right| Tour of POWERDAY]] [[File:Inside cityhall.JPG|200px|right| Meeting at London City Hall]] | ||
| + | We investigated how the system we have created would fit into the real world which is summarised [[Team:Imperial_College/Project_Evolution| here]]. We have identified a waste material which is an economic burden to those who produce it, this material is called Solid Recovered Fuel(SRF) but we will refer to it as mixed waste from now on. It is made up of plastics, wood and fibres. By using this waste we can not only make a [[Team:Imperial_College/Industrial_Implementation|profit]] but also offer a more environmentally sound option for dealing with the waste where the alternative is [[Team:Imperial_College/Background| polluting and resource inefficient]]. | ||
| + | |||
| + | The material we are producing has many great properties and is friendlier to the environment than petrochemical alternatives. It is biocompatible, industrially biodegradable and can be produced from sustainable resources. This material has a bright future, demonstrated by its many [[Team:Imperial_College/Applications| applications.]] | ||
| + | |||
| + | Trips to POWERDAY,(a materials recovery facility) and London City Hall highlighted the fact that recovering value from waste materials is one of the primary aims of waste management in London, the UK and Europe as a whole. Importantly it highlighted the need for social solutions combined with technical ones to reduce the waste we produce in the first place and then to maximise the recovery of as much of it as possible. It requires responsibility for our actions. Since we have created a method by which a new commodity can be produced, it would be wrong to not consider how it will be disposed of. This led to the creation of the [[Team:Imperial_College/BioPlastic_Recycling:_PHB|Plastic Fantastic]] recycling module. We encouraged people to reduce waste and promoted recycling on our university campus with [[Team:Imperial_College/Managing_Waste| posters]], highlighting 5 waste offenders. Through a strong [[Team:Imperial_College/Communication_work| social media campaign]], we provoked discussion by sharing interesting and sometimes shocking reports regarding the waste issue. | ||
| + | |||
| + | |||
| + | <br><br> | ||
| + | <h1>M.A.P.L.E. - 3D printing the future</h1> | ||
| + | |||
| + | [[File:Workbench1.jpeg|right|200px| Household M.A.P.L.E. appliance]] | ||
| + | <p>With our collaborators from the Royal College of Art, we have been considering how our system could be implemented in the future. We pictured how the technology could be used in different settings with various implications. Together we designed the futuristic company M.A.P.L.E. (Modular and Plastic Looping <i>E.coli</i>) with a range of appliances that transform domestic rubbish into new 3D printed bioplastic objects. In picturing this we were influenced by the 'DIY biology' culture, the 'maker' culture and the [[Team:Imperial_College/Applications| homebrew industry]]. We were intrigued by the prospect of coupling local bioplastic recycling with 3D printing and how people could start to see their waste as something valuable, something which could become beautiful and useful. You can find out more [[Team:Imperial_College/MAPLE|here]] about MAPLE's different incarnations.</p> | ||
| + | |||
| + | |||
| + | <h1>Communicating our Project</h1> | ||
| + | [[File:Z_The_Jack_Pine.jpg|200px|right| The Jack Pine]] [[File:Photo with adam.JPG|200px|right| Interview with Adam Rutherford for BBC Radio 4]] | ||
| + | <p>We have communicated our project at several events and across social media. As apart of our outreach, we have founded an art movement [https://2013.igem.org/Team:Imperial_College/Communication_work#Living_Art:_Bionouveau| BioNouveau]. Once we had produced a palette of different colours from various bacterial pigments, we set about recreating painting masterpieces using the humble media of E.coli on agar petri dishes. People loved these and it brought many visitors to our pages. They became so popular that people started putting in requests, with Calgary iGEM team giving us the inspiration for our bacterial 'The Jack Pine'. We were delighted to have are BioNouveau artwork featured in ATOMS Turkiye's SynArt show at the European Jamboree.</p> | ||
| + | |||
| + | <p>The Celebration of Science was a week's long spread of science events covering all sorts of topics. We presented our project as part of a series of lectures. Our talk was very well received as something of a contrast to the classical study of biology which had preceded us. The people we spoke to afterwards were particularly interested in the difficulties of engineering biology and also the capabilities of the technology.</p> | ||
| + | |||
| + | <p>We also featured on Dr. Adam Rutherford's Inside Science show on BBC Radio 4. We were delighted to have Adam visit us at Imperial College and it was great to talk to him about our project.</p> | ||
| + | |||
| + | |||
| + | |||
| + | <h1>Collaborations</h1> | ||
| + | |||
| + | <h2>Yale: PLA-degradation</h2> | ||
| + | <p align="justify">As soon as we found out [https://2013.igem.org/Team:Yale/Project_Collaboration Yale iGEM team] were synthesizing poly (lactic acid) (PLA) this year, we contacted them and sent them our PLA degradation parts to test on their product. We did this to further characterise the part and see if the Biobricks behave the same on both sides of the Atlantic! When Yale optimises PLA production, they will send some of their product to us to break down. Breaking down PLA is a second biological bioplastic recycling system we are working on.</p> | ||
| + | <h3>UCL:Beta-Amyloid Degradation</h3> | ||
| + | <p align="justify"> When we heard UCL's project was to degrade beta-amyloid plaques we were interested to see the results. Beta-amyloid is a component of amyloid plaques which are a pathological feature of Alzheimer's disease. It turns out that proteinase K does not only break down PLA, but also degrades beta-amyloid plaques. We built a degradation model to help the UCL team with their beta-amyloid degradation assays. The model, a MATLAB based extension called Simbiology, provides information about how efficient MMP-9 enzyme can degrade beta-amyloid plaques, which is informative for wetlab work.</p> | ||
| + | <h3>St. Paul's School iGEM team</h3> | ||
| + | [[File:Imperial_stpauls.jpg|thumbnail|left|350px|<b>From left to right: Sisi Fan (Imperial College), Iain Bower (Imperial College),Paul-Enguerrand Fady (St. Pauls), Margarita Kopniczky (Imperial College), Freddie Wilkinson (St. Pauls), and Guy Kirkpatrick (St. Pauls).</b>]] | ||
| + | <BR><BR><BR><BR> | ||
| + | <p align="justify">We met the team from St.Paul's School when we went to the Young Synthetic Biology event. We were impressed by their project of the previous year and got chatting to them. As school students they were keen to see some university labs and todiscuss their project further, so we invited them for a tour of the facilities at Imperial. We hope the tour was enjoyable and that the tips we gave to them come in useful this year.</p> | ||
| + | If you want to know more about our collaborations please see our [https://2013.igem.org/Team:Imperial_College/Collaboration collaboration page.] | ||
{{:Team:Imperial_College/Templates:footer}} | {{:Team:Imperial_College/Templates:footer}} | ||
Latest revision as of 01:58, 29 October 2013
We have engineered bacteria to turn non-recyclable waste into bioplastic
We have taken an expensive byproduct of recycling facilities, a type of mixed waste and turned it into something quite amazing; a material that can be used for a diverse range of applications from making your lunchbox to a 3D printed tissue scaffold. This material is the bioplastic poly-3-hydroxybutyric acid P(3HB). Our system is designed to maximise the recovery of resources from the waste and so we have also investigated how we can use the oil based plastics within it to produce the commodity chemical ethylene glycol. We are passionate about using synthetic biology to help us move towards a more sustainable economy. We have considered what happens to the material we produce after it is used. This led us to develop the first synthetic biology recycling system for P(3HB) and one of the first in the world. The other well known bioplastic, poly lactic acid (PLA) is also expanding in use and so we developed a system to turn it back into its constituent monomers so that it can also be resynthesised, chemically or in vivo.
Click here to find out how we made P3HB and broke down oil derived plastics and here to see how we recycled P3HB and PLA.
See here how we convert trash to treasure, a longer more detailed version can be found [http://www.youtube.com/watch?v=1oXt_EfqmLU&feature=youtu.be here]
Our System: Modular and Plastic Looping E. coli (M.A.P.L.E.)
Module 1: Resource-full Waste
Non-recyclable waste is sourced from a recycling centre, placed in a bioreactor with our M.A.P.L.E system which breaks down the waste and synthesises the bioplastic P(3HB).
Click here to see for yourself
Module 2: Plastic Fantastic
Plastic Fantastic is a complete P(3HB) bioplastic recycling platform, where P(3HB) is degraded into monomeric form and then re-polymerised back into de novo P(3HB) for future applications.
Click here to see for yourself
Real world research guided the development of our project
We investigated how the system we have created would fit into the real world which is summarised here. We have identified a waste material which is an economic burden to those who produce it, this material is called Solid Recovered Fuel(SRF) but we will refer to it as mixed waste from now on. It is made up of plastics, wood and fibres. By using this waste we can not only make a profit but also offer a more environmentally sound option for dealing with the waste where the alternative is polluting and resource inefficient.
The material we are producing has many great properties and is friendlier to the environment than petrochemical alternatives. It is biocompatible, industrially biodegradable and can be produced from sustainable resources. This material has a bright future, demonstrated by its many applications.
Trips to POWERDAY,(a materials recovery facility) and London City Hall highlighted the fact that recovering value from waste materials is one of the primary aims of waste management in London, the UK and Europe as a whole. Importantly it highlighted the need for social solutions combined with technical ones to reduce the waste we produce in the first place and then to maximise the recovery of as much of it as possible. It requires responsibility for our actions. Since we have created a method by which a new commodity can be produced, it would be wrong to not consider how it will be disposed of. This led to the creation of the Plastic Fantastic recycling module. We encouraged people to reduce waste and promoted recycling on our university campus with posters, highlighting 5 waste offenders. Through a strong social media campaign, we provoked discussion by sharing interesting and sometimes shocking reports regarding the waste issue.
M.A.P.L.E. - 3D printing the future
With our collaborators from the Royal College of Art, we have been considering how our system could be implemented in the future. We pictured how the technology could be used in different settings with various implications. Together we designed the futuristic company M.A.P.L.E. (Modular and Plastic Looping E.coli) with a range of appliances that transform domestic rubbish into new 3D printed bioplastic objects. In picturing this we were influenced by the 'DIY biology' culture, the 'maker' culture and the homebrew industry. We were intrigued by the prospect of coupling local bioplastic recycling with 3D printing and how people could start to see their waste as something valuable, something which could become beautiful and useful. You can find out more here about MAPLE's different incarnations.
Communicating our Project
We have communicated our project at several events and across social media. As apart of our outreach, we have founded an art movement BioNouveau. Once we had produced a palette of different colours from various bacterial pigments, we set about recreating painting masterpieces using the humble media of E.coli on agar petri dishes. People loved these and it brought many visitors to our pages. They became so popular that people started putting in requests, with Calgary iGEM team giving us the inspiration for our bacterial 'The Jack Pine'. We were delighted to have are BioNouveau artwork featured in ATOMS Turkiye's SynArt show at the European Jamboree.
The Celebration of Science was a week's long spread of science events covering all sorts of topics. We presented our project as part of a series of lectures. Our talk was very well received as something of a contrast to the classical study of biology which had preceded us. The people we spoke to afterwards were particularly interested in the difficulties of engineering biology and also the capabilities of the technology.
We also featured on Dr. Adam Rutherford's Inside Science show on BBC Radio 4. We were delighted to have Adam visit us at Imperial College and it was great to talk to him about our project.
Collaborations
Yale: PLA-degradation
As soon as we found out Yale iGEM team were synthesizing poly (lactic acid) (PLA) this year, we contacted them and sent them our PLA degradation parts to test on their product. We did this to further characterise the part and see if the Biobricks behave the same on both sides of the Atlantic! When Yale optimises PLA production, they will send some of their product to us to break down. Breaking down PLA is a second biological bioplastic recycling system we are working on.
UCL:Beta-Amyloid Degradation
When we heard UCL's project was to degrade beta-amyloid plaques we were interested to see the results. Beta-amyloid is a component of amyloid plaques which are a pathological feature of Alzheimer's disease. It turns out that proteinase K does not only break down PLA, but also degrades beta-amyloid plaques. We built a degradation model to help the UCL team with their beta-amyloid degradation assays. The model, a MATLAB based extension called Simbiology, provides information about how efficient MMP-9 enzyme can degrade beta-amyloid plaques, which is informative for wetlab work.
St. Paul's School iGEM team
We met the team from St.Paul's School when we went to the Young Synthetic Biology event. We were impressed by their project of the previous year and got chatting to them. As school students they were keen to see some university labs and todiscuss their project further, so we invited them for a tour of the facilities at Imperial. We hope the tour was enjoyable and that the tips we gave to them come in useful this year.
If you want to know more about our collaborations please see our collaboration page.
 "
"