Team:Imperial College/Modelling PLAdeg
From 2013.igem.org
| (17 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:Imperial_College/Templates:header}} | {{:Team:Imperial_College/Templates:header}} | ||
| - | <h1> | + | <h1>Polyhydroxybutyrate (P(3HB)) Synthesis Module </h1> |
| - | <h2> | + | <h2>Metabolic modelling</h2> |
| + | <h3>Introduction of the metabolic model</h3> | ||
<p align="justify"> | <p align="justify"> | ||
| - | The | + | The metabolic model predicts the interaction of our pathway with endogenous pathways. In synthetic biology, an ideal system is fully orthogonal. However, if the synthetic pathway uses some metabolites from the metabolism pathways, the metabolic fluxes (the rate of conversion between metabolites) will be affected. If the cell is overloaded by losing too much metabolites, it would either reject the synthetic pathway or burn out. As for the optimization of PHB production in our MAPLE system, metabolic analysis need to be carried out to estimate the effects on the cell metabolism from increases in the PHB production. </p> |
| - | < | + | <h3>Methods</h3> |
| - | <p>1. | + | <p align="justify">We altered the metabolic model by Angela Dixon in her paper “Predictive Mathematical Model for Polyhydroxybutyrate Synthesis in Escherichia coli”. [http://digitalcommons.usu.edu/engineering_datasets/1/] The metabolic model is built in Simbiology, which is an extension in MATLAB especially for biological system analysis. The model consists two metabolic pathways: Glycolysis and TCA cycle. The reactions in the metabolic pathways are defined by ordinary differential equations and kinetic parameters. The model also contains a synthetic pathway that produce P(3HB) from acetyl-coA. The pathway consists three enzymes which are PhaA, PhaB and PhaC. |
| - | <p> | + | The reasons we chose this model:</p> |
| - | <p> | + | <p>1. The model is based on ODEs instead of FBA (flux balance analysis) method, which is a common methods of metabolic analysis. However, FBA cannot determine the real time concentration of the metabolites whereas it can be done by models of ODEs.</p> |
| - | + | <p>2. Only single gene deletion or addition can be performed in FBA. More than that, the metabolic model here can perform modification on any single or multiple genes and reactions through Simbiology. </p> | |
| - | <p | + | <p>3. The FBA model is static, it cannot analyze the dynamic interactions between our synthetic pathways with the metabolic pathways. As both our P(3HB) synthesis model and the metabolic model are in the same platform. It was really easy to integrate our pathway into the metabolic model.</p> |
| - | < | + | <p><b>In our project, we have a different pathway:</b></p> |
| - | < | + | <p>1. We use 3HB as one of the input.</p> |
| - | + | <p>2. Existence of ato system in the E.coli MG1655 strain. </p> | |
| - | + | <p>3. We removed phaA in our case because we use 3HB as the major source to produce P(3HB) to avoid too much acetyl-coA be taken from the metabolic pathways.</p> | |
| - | + | <p align="justify">Therefore, we substitute the original pathway with our synthetic pathway. Moreover, we added our gene expression models for the enzymes into the metabolic model as well. In terms of the consistency of the model, we changed the units of our synthetic pathway into micro molar and seconds. </p> | |
| - | + | <p>Here is the combination of our model with the metabolic model</p> | |
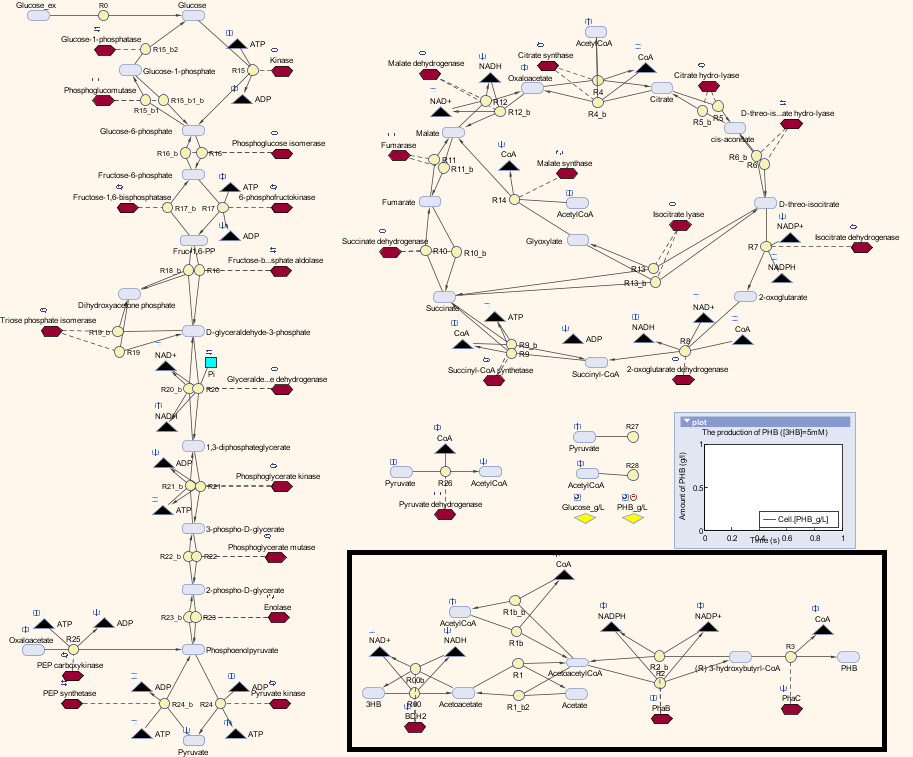
| - | < | + | [[File:MetaSyn.png|800px|centre]] |
| - | + | <p align="justify">We also carried out a parameter estimations of the initial concentrations of several substrates in our synthetic pathway. The reason is we have single-cell scaled magnitude for them originally (fM level), but there are much higher magnitudes in any of the species in the metabolic pathways (mM level). Therefore, we scaled up the pathway in order to maintain the consistency of the model. As for the metabolites which are involved in our pathway, we cloned the blocks of them from the metabolic pathway in Simbiology and deliberately put them into our pathway. That means our pathway dynamically interact with the metabolic pathways. </p> | |
| - | + | <p>Here is the table for the updated initial concentrations for the species in our synthetic pathway: | |
| - | + | All the kinetic data and initial concentrations of the metabolites in the metabolic pathways are referenced in the paper[http://digitalcommons.usu.edu/engineering_datasets/1/]: </p> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | <p> | + | |
| - | [[File: | + | |
| - | <p> | + | |
| - | < | + | |
| - | + | ||
<table border="2" bgcolor="#efefef"> | <table border="2" bgcolor="#efefef"> | ||
<tr> | <tr> | ||
| - | <th> | + | <th>Substrates</th> |
| - | <th> | + | <th>Concentration</th> |
| - | + | ||
<th>Units</th> | <th>Units</th> | ||
<th>Sources</th> | <th>Sources</th> | ||
| Line 40: | Line 33: | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <th> | + | <th>3HB</th> |
| - | + | ||
| - | <td> | + | <td>5000</td> |
| - | <td> | + | <td>uM</td> |
| - | <td> | + | <td>-</td> |
| - | <td> | + | <td>The initial input to the system</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <th> | + | <th>Acetoacetate</th> |
| - | <td> | + | |
| - | <td> | + | <td>1000</td> |
| - | <td> | + | <td>uM</td> |
| - | <td>[http:// | + | <td>-</td> |
| - | + | <td>The ato system in E.coli suggest a favourable conversion from acetoacetyl-coA to acetoacetate. Therefore, the concentration of acetoacetate is 10 times larger than acetoacetyl-coA[http://www.sciencedirect.com/science/article/pii/0003986177904969]</td> | |
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <th> | + | <th>Acetoacetyl-coA</th> |
| - | + | ||
| - | <td> | + | <td>100</td> |
| - | <td> | + | <td>uM</td> |
| - | <td>[http://www.ncbi.nlm.nih.gov/ | + | <td>[http://www.ncbi.nlm.nih.gov/pubmed/1103741]</td> |
| - | <td> | + | <td>The ato system in E.coli suggest a favourable conversion from acetoacetyl-coA to acetoacetate. [http://www.sciencedirect.com/science/article/pii/0003986177904969]</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <th> | + | <th>NADH</th> |
| - | + | ||
| - | <td> | + | <td>200</td> |
| - | <td> | + | <td>uM</td> |
| - | <td>[http:// | + | <td>[http://digitalcommons.usu.edu/engineering_datasets/1/]</td> |
| - | <td> | + | <td>-</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <th> | + | <th>NAD+</th> |
| - | <td> | + | |
| - | <td> | + | <td>1200</td> |
| - | <td> | + | <td>uM</td> |
| - | + | <td>[http://digitalcommons.usu.edu/engineering_datasets/1/]</td> | |
| - | <td> | + | <td>-</td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <th> | + | <th>Acetyl-coA</th> |
| - | + | ||
| - | <td> | + | <td>5000</td> |
| - | <td> | + | <td>uM</td> |
| - | <td> | + | <td>[http://digitalcommons.usu.edu/engineering_datasets/1/]</td> |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
<td>-</td> | <td>-</td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <th> | + | <th>Coenzyme A</th> |
| - | <td> | + | |
| + | <td>250</td> | ||
| + | <td>uM</td> | ||
| + | <td>[http://digitalcommons.usu.edu/engineering_datasets/1/]</td> | ||
<td>-</td> | <td>-</td> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <th> | + | <th>Acetate</th> |
| - | <td> | + | |
| - | <td> | + | <td>40000</td> |
| - | <td> | + | <td>uM</td> |
| - | <td> | + | <td>[http://www.ncbi.nlm.nih.gov/pubmed/1103741]</td> |
| - | + | <td>Assay data from the literature which the ato system efficiency was measured</td> | |
</tr> | </tr> | ||
| - | |||
<tr> | <tr> | ||
| - | <th> | + | <th>3HB-coA</th> |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | <td>0</td> | |
| - | + | <td>uM</td> | |
| - | + | ||
| - | <td> | + | |
| - | <td> | + | |
| - | + | ||
<td>-</td> | <td>-</td> | ||
| - | + | <td>One of the intermediate product of the pathway</td> | |
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <th> | + | <th>NADPH</th> |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | <td>200</td> | ||
| + | <td>uM</td> | ||
| + | <td>[http://digitalcommons.usu.edu/engineering_datasets/1/]</td> | ||
| + | <td>-</td> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <th> | + | <th>NADP+</th> |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | <td>1200</td> | ||
| + | <td>uM</td> | ||
| + | <td>[http://digitalcommons.usu.edu/engineering_datasets/1/]</td> | ||
| + | <td>-</td> | ||
</tr> | </tr> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| + | <tr> | ||
| + | <th>PHB</th> | ||
| + | |||
| + | <td>0</td> | ||
| + | <td>uM</td> | ||
| + | <td>-</td> | ||
| + | <td>The final product of the pathway</td> | ||
| + | </tr> | ||
| + | |||
| + | |||
| + | </table> | ||
| + | <h3>ATP analysis:</h3> | ||
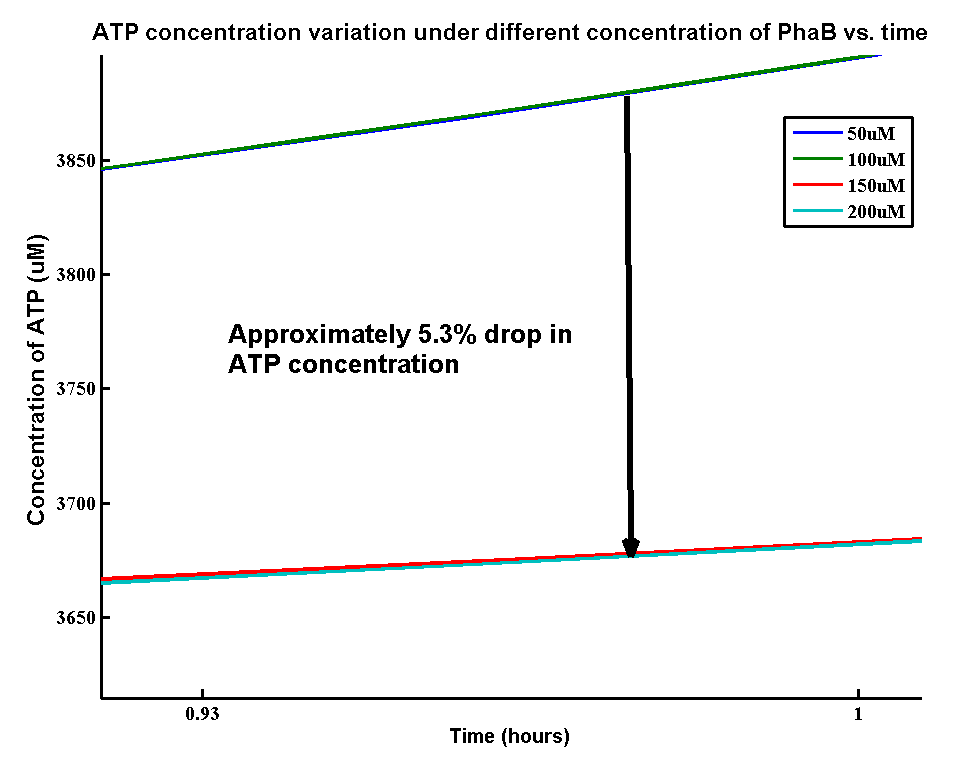
| + | <p>In our optimization of P(3HB) production module, we discovered that increasing in the PhaB expression can efficiently increase the yield of P(3HB). It’s important to see whether increase in the PhaB expression would dramatically change the fluxes in the metabolic pathways or not. As ATP is the essential energy carriers in the cell, we carried a parameter scan for the variation in ATP concentrations levels under different concentration of PhaB.</p> | ||
| + | [[File:ATPPHAB.png|600px|centre]] | ||
| + | <p>There is an approximately 5.3% drop in the concentration of ATP when increasing the phaB concentration from 50uM to 200uM. It suggests that increasing the efficiency in our pathway will reduce the efficiency of energy transports in the cell. However, it’s still unsure about the effect of 5.3% drop in ATP concentration on the cell growth.</p> | ||
| + | <h3>Large scale PHB production and implementation in MAPLE</h3> | ||
| + | <p>In our MAPLE system, the bioplastic Polyhydroxybutyrate P(3HB) is produced from both 3HB monomers and glucose. 3HB is the degradation product from the wasted P(3HB)plastic and glucose is the product from the degradation of mixed waste in Module 1. Therefore, the overall PHB production rates need to be defined by considering different situations:</p> | ||
| + | <p>1. In module 1, we have glucose as the only source to produce PHB.</p> | ||
| + | <p>2. In module 2, we have both glucose and 3HB as resources. </p> | ||
{{:Team:Imperial_College/Templates:footer}} | {{:Team:Imperial_College/Templates:footer}} | ||
Latest revision as of 13:41, 4 October 2013
Contents |
Polyhydroxybutyrate (P(3HB)) Synthesis Module
Metabolic modelling
Introduction of the metabolic model
The metabolic model predicts the interaction of our pathway with endogenous pathways. In synthetic biology, an ideal system is fully orthogonal. However, if the synthetic pathway uses some metabolites from the metabolism pathways, the metabolic fluxes (the rate of conversion between metabolites) will be affected. If the cell is overloaded by losing too much metabolites, it would either reject the synthetic pathway or burn out. As for the optimization of PHB production in our MAPLE system, metabolic analysis need to be carried out to estimate the effects on the cell metabolism from increases in the PHB production.
Methods
We altered the metabolic model by Angela Dixon in her paper “Predictive Mathematical Model for Polyhydroxybutyrate Synthesis in Escherichia coli”. [http://digitalcommons.usu.edu/engineering_datasets/1/] The metabolic model is built in Simbiology, which is an extension in MATLAB especially for biological system analysis. The model consists two metabolic pathways: Glycolysis and TCA cycle. The reactions in the metabolic pathways are defined by ordinary differential equations and kinetic parameters. The model also contains a synthetic pathway that produce P(3HB) from acetyl-coA. The pathway consists three enzymes which are PhaA, PhaB and PhaC. The reasons we chose this model:
1. The model is based on ODEs instead of FBA (flux balance analysis) method, which is a common methods of metabolic analysis. However, FBA cannot determine the real time concentration of the metabolites whereas it can be done by models of ODEs.
2. Only single gene deletion or addition can be performed in FBA. More than that, the metabolic model here can perform modification on any single or multiple genes and reactions through Simbiology.
3. The FBA model is static, it cannot analyze the dynamic interactions between our synthetic pathways with the metabolic pathways. As both our P(3HB) synthesis model and the metabolic model are in the same platform. It was really easy to integrate our pathway into the metabolic model.
In our project, we have a different pathway:
1. We use 3HB as one of the input.
2. Existence of ato system in the E.coli MG1655 strain.
3. We removed phaA in our case because we use 3HB as the major source to produce P(3HB) to avoid too much acetyl-coA be taken from the metabolic pathways.
Therefore, we substitute the original pathway with our synthetic pathway. Moreover, we added our gene expression models for the enzymes into the metabolic model as well. In terms of the consistency of the model, we changed the units of our synthetic pathway into micro molar and seconds.
Here is the combination of our model with the metabolic model
We also carried out a parameter estimations of the initial concentrations of several substrates in our synthetic pathway. The reason is we have single-cell scaled magnitude for them originally (fM level), but there are much higher magnitudes in any of the species in the metabolic pathways (mM level). Therefore, we scaled up the pathway in order to maintain the consistency of the model. As for the metabolites which are involved in our pathway, we cloned the blocks of them from the metabolic pathway in Simbiology and deliberately put them into our pathway. That means our pathway dynamically interact with the metabolic pathways.
Here is the table for the updated initial concentrations for the species in our synthetic pathway: All the kinetic data and initial concentrations of the metabolites in the metabolic pathways are referenced in the paper[http://digitalcommons.usu.edu/engineering_datasets/1/]:
| Substrates | Concentration | Units | Sources | Assumptions |
|---|---|---|---|---|
| 3HB | 5000 | uM | - | The initial input to the system |
| Acetoacetate | 1000 | uM | - | The ato system in E.coli suggest a favourable conversion from acetoacetyl-coA to acetoacetate. Therefore, the concentration of acetoacetate is 10 times larger than acetoacetyl-coA[http://www.sciencedirect.com/science/article/pii/0003986177904969] |
| Acetoacetyl-coA | 100 | uM | [http://www.ncbi.nlm.nih.gov/pubmed/1103741] | The ato system in E.coli suggest a favourable conversion from acetoacetyl-coA to acetoacetate. [http://www.sciencedirect.com/science/article/pii/0003986177904969] |
| NADH | 200 | uM | [http://digitalcommons.usu.edu/engineering_datasets/1/] | - |
| NAD+ | 1200 | uM | [http://digitalcommons.usu.edu/engineering_datasets/1/] | - |
| Acetyl-coA | 5000 | uM | [http://digitalcommons.usu.edu/engineering_datasets/1/] | - |
| Coenzyme A | 250 | uM | [http://digitalcommons.usu.edu/engineering_datasets/1/] | - |
| Acetate | 40000 | uM | [http://www.ncbi.nlm.nih.gov/pubmed/1103741] | Assay data from the literature which the ato system efficiency was measured |
| 3HB-coA | 0 | uM | - | One of the intermediate product of the pathway |
| NADPH | 200 | uM | [http://digitalcommons.usu.edu/engineering_datasets/1/] | - |
| NADP+ | 1200 | uM | [http://digitalcommons.usu.edu/engineering_datasets/1/] | - |
| PHB | 0 | uM | - | The final product of the pathway |
ATP analysis:
In our optimization of P(3HB) production module, we discovered that increasing in the PhaB expression can efficiently increase the yield of P(3HB). It’s important to see whether increase in the PhaB expression would dramatically change the fluxes in the metabolic pathways or not. As ATP is the essential energy carriers in the cell, we carried a parameter scan for the variation in ATP concentrations levels under different concentration of PhaB.
There is an approximately 5.3% drop in the concentration of ATP when increasing the phaB concentration from 50uM to 200uM. It suggests that increasing the efficiency in our pathway will reduce the efficiency of energy transports in the cell. However, it’s still unsure about the effect of 5.3% drop in ATP concentration on the cell growth.
Large scale PHB production and implementation in MAPLE
In our MAPLE system, the bioplastic Polyhydroxybutyrate P(3HB) is produced from both 3HB monomers and glucose. 3HB is the degradation product from the wasted P(3HB)plastic and glucose is the product from the degradation of mixed waste in Module 1. Therefore, the overall PHB production rates need to be defined by considering different situations:
1. In module 1, we have glucose as the only source to produce PHB.
2. In module 2, we have both glucose and 3HB as resources.
 "
"