Team:Imperial College/mainresults
From 2013.igem.org
| (44 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:Imperial_College/Templates:header}} | {{:Team:Imperial_College/Templates:header}} | ||
__NOTOC__ | __NOTOC__ | ||
| - | < | + | <h4>Main Results</h4> |
<html> | <html> | ||
| Line 8: | Line 8: | ||
<div id="TabbedPanels1" class="TabbedPanels"> | <div id="TabbedPanels1" class="TabbedPanels"> | ||
<ul class="TabbedPanelsTabGroup"> | <ul class="TabbedPanelsTabGroup"> | ||
| - | <li class="TabbedPanelsTab" tabindex="0">< | + | <li class="TabbedPanelsTab" tabindex="0"><h5>Resource-full Waste</h5></li> |
| - | <li class="TabbedPanelsTab" tabindex="0">< | + | <li class="TabbedPanelsTab" tabindex="0"><h5>Plastic Fantastic</h5></li> |
</ul> | </ul> | ||
| Line 17: | Line 17: | ||
<div id="CollapsiblePanelM11" class="CollapsiblePanel"> | <div id="CollapsiblePanelM11" class="CollapsiblePanel"> | ||
| - | <div class="CollapsiblePanelTab" tabindex="0"><h4> | + | <div class="CollapsiblePanelTab" tabindex="0"><h4>We made P3HB bioplastic </html><font size="1">▼</font size="1"><html></h4></div> |
<div class="CollapsiblePanelContent"> | <div class="CollapsiblePanelContent"> | ||
</html> | </html> | ||
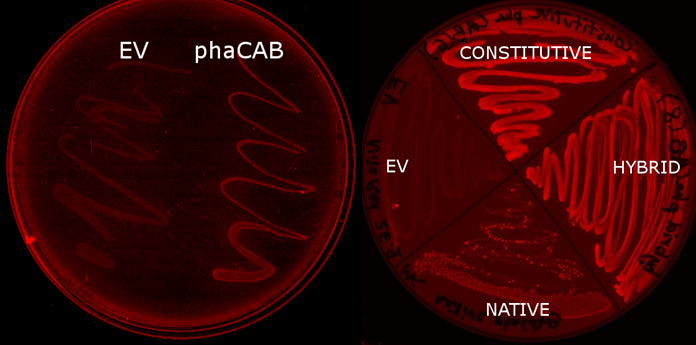
| - | <p>We | + | <p>We used <i>E. coli</i> that expresses PhaCAB enzymes to make P3HB bioplastic. P3HB produced in cells were stained with Nile Red for fluorescent microscope imaging, and strong fluorescence was detected at the area where PhaCAB-expressing cells were streaked.</p> |
| - | [[File: | + | {| class="wikitable" style="margin: 1em auto 1em auto;" |
| - | + | |[[File:Results-nile_red_plates.PNG|thumbnail|left|900px| <b> <i>E.coli</i> transformed with the phaCAB operon(native, hybrid and constitutive are different promoters expressing the operon) fluoresce when grown on LB-Agar plates with Nile Red stain. </b>The P3HB produced within them is stained. Those without the operon do not(EV- transformed with empty pSB1C3). When the promoter is changed from the native form to the hybrid or constitutive J23104 then the fluorescence is more intense, indicating increased P3HB production.]] | |
| + | |} | ||
| Line 30: | Line 31: | ||
</div> | </div> | ||
</div> | </div> | ||
| + | |||
<div id="CollapsiblePanelM12" class="CollapsiblePanel"> | <div id="CollapsiblePanelM12" class="CollapsiblePanel"> | ||
| - | <div class="CollapsiblePanelTab" tabindex="0"><h4> | + | <div class="CollapsiblePanelTab" tabindex="0"><h4>Our model predicts increased PhaB expression boosts P3HB production </html><font size="1">▼</font size="1"><html></h4></div> |
<div class="CollapsiblePanelContent"> | <div class="CollapsiblePanelContent"> | ||
</html> | </html> | ||
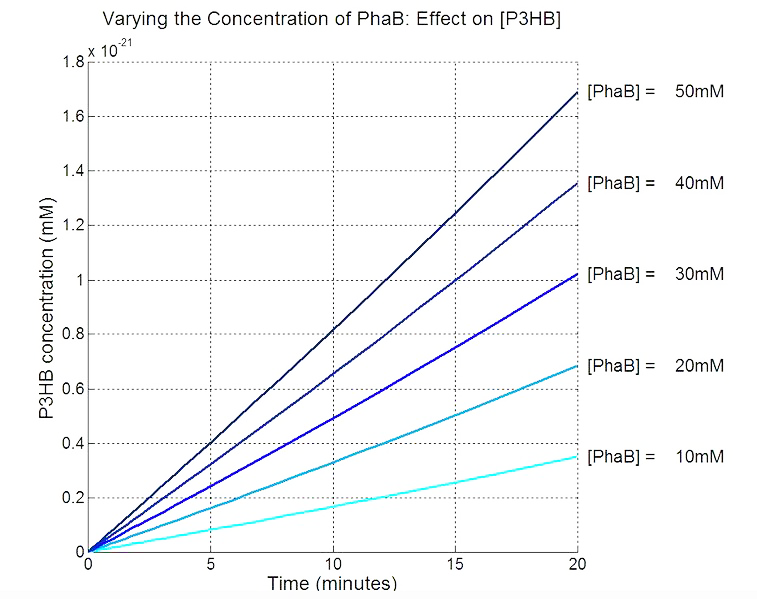
| - | <p align="justify> | + | <p align="justify">In order to realistically improve our experimental design, we produced P3HB synthesis model with considerations of central metabolic pathway within the cells. Our model predicts that during the synthesis of P3HB, the concentration of PhaB enzymes is an important rate limiting factor that can actually be regulated.</p> |
| - | + | ||
| + | [[File:Imperial College PhaB scan Single cellmM.JPG|thumbnail|center|700px|<b>PhaB concentration in the chassis is a rate limiting factor in P3HB synthesised.</b> The parameter values are from various sources, thus the simulation result should be interpreted as a qualitative perspective and the trend should be observed.]] | ||
| + | <p><h4>[https://2013.igem.org/Team:Imperial_College/mainresults Refresh] the page to see our video</h4></p> | ||
| + | <h5>Metabolic model preview</h5> <p>'''We simulated the interactions between metabolism pathways with our synthetic pathway'''</p> | ||
<html> | <html> | ||
| - | < | + | <iframe width="800" height="400" src="//www.youtube.com/embed/m2ZJUA-yw34" frameborder="0" allowfullscreen></iframe> |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | <h5>Preview to our GUI (Please refresh to see video):</h5> <p>'''We made a GUI so that the wet lab people could easily manipulate enzyme concentrations to see their effect on P(3HB) production.'''</p> | ||
| + | <iframe width="800" height="400" src="//www.youtube.com/embed/2K_lM6-U8fc" frameborder="0" allowfullscreen></iframe> | ||
</html> | </html> | ||
| - | |||
| - | |||
| - | |||
<html> | <html> | ||
</div> | </div> | ||
</div> | </div> | ||
| - | <div id=" | + | <div id="CollapsiblePanelM13" class="CollapsiblePanel"> |
| - | <div class="CollapsiblePanelTab" tabindex="0"><h4> | + | <div class="CollapsiblePanelTab" tabindex="0"><h4>We optimised P3HB bioplastic production </html><font size="1">▼</font size="1"><html></h4></div> |
<div class="CollapsiblePanelContent"> | <div class="CollapsiblePanelContent"> | ||
</html> | </html> | ||
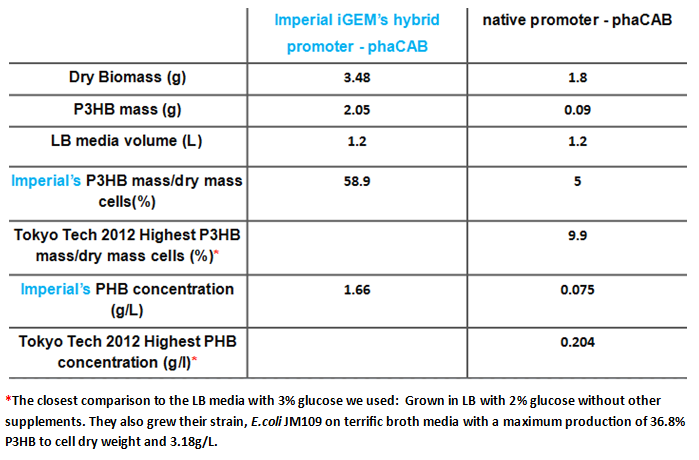
| - | <p align="justify">Since our model predicts that | + | <p align="justify">Since our model predicts that P3HB production can be significantly improved by expressing more P3HB polymerase PhaB, we designed a [http://parts.igem.org/Part:BBa_K1149051 hybrid promoter] which, consists of the J23104 constitutive promoter and the native promoter to optimise gene expression. Our results show that we have successfully produced <b>11-fold</b> more P3HB bioplastic compared with the native promoter. </p> |
{| class="wikitable" style="margin: 1em auto 1em auto;" | {| class="wikitable" style="margin: 1em auto 1em auto;" | ||
| - | |[[File: | + | |[[File:Moreplastic.JPG|thumbnail|left|700px|<b>Comparison of P3HB production </b>. <b> (left)</b> 1.5ml tube, natural phaCAB (BBa_K934001) <b>(right)</b> 5ml tube, phaCAB expressed from the hybrid promoter, (BBa_K1149051).]] |
|} | |} | ||
{| class="wikitable" style="margin: 1em auto 1em auto;" | {| class="wikitable" style="margin: 1em auto 1em auto;" | ||
| - | |[[File: | + | |[[File:PHB_production_table.PNG|thumbnail|left|900px| <b>A summary of the improved production of P3HB by our hybrid promoter-phaCAB construct(BBa_K1149051) over the native promoter-phaCAB.</b>]] |
|} | |} | ||
| + | |||
<html> | <html> | ||
| Line 76: | Line 75: | ||
</div> | </div> | ||
| - | <div id=" | + | <div id="CollapsiblePanelM14" class="CollapsiblePanel"> |
| - | <div class="CollapsiblePanelTab" tabindex="0"><h4> | + | <div class="CollapsiblePanelTab" tabindex="0"><h4>We made bioplastic from mixed waste </html><font size="1">▼</font size="1"><html></h4></div> |
<div class="CollapsiblePanelContent"> | <div class="CollapsiblePanelContent"> | ||
</html> | </html> | ||
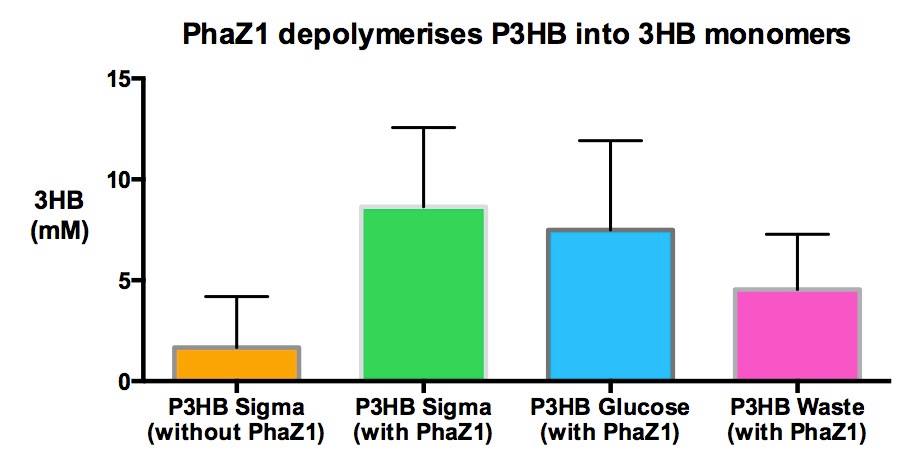
| - | <p align="justify">One of the objectives of [https://2013.igem.org/Team:Imperial_College/Waste_Degradation:_SRF Module 1] is to produce P3HB bioplastic from waste. | + | <p align="justify">One of the objectives of [https://2013.igem.org/Team:Imperial_College/Waste_Degradation:_SRF Module 1] is to produce P3HB bioplastic from waste. We compared P3HB degradation by measuring the concentration of 3HB monomers, using P3HB we bought from sigma, we produced from glucose, and we produced from the waste collected from Powerday. We found that there is no significant difference between P3HB purchased from Sigma and P3HB we made from waste (t-test p= 0.12). Thus <b>we conclude that we have made P3HB bioplastic from waste</b>.</p> |
| + | |||
{| class="wikitable" style="margin: 1em auto 1em auto;" | {| class="wikitable" style="margin: 1em auto 1em auto;" | ||
| - | |[[File: | + | |[[File:Bioplastic_from_waste.jpg|thumbnail|left|600px| <b>We have detected 3HB monomers for the chemical analysis of the produced bioplastic. </b> We synthesised P(3HB) using our improved Biobrick part (hybrid promoter phaCAB, BBa_K1149051). Our engineered bioplastic producing <i>E.coli</i> synthesised P(3HB) directly from waste. See data under [https://2013.igem.org/Team:Imperial_College/3HB_assay 3HB assay #2] Imperial iGEM data]] |
|} | |} | ||
| - | + | ||
| + | {| class="wikitable" style="margin: 1em auto 1em auto;" | ||
| + | |[[File:Imperial_P3HB_from_waste.jpg|thumbnail|left|600px|<b>Degradation of P3HB produced from various sources showing no significant difference between P3HB made from waste and P3HB made from other sources.</b>]] | ||
| + | |} | ||
| + | |||
| + | |||
| + | <html> | ||
| + | <iframe width="840" height="630" src="//www.youtube-nocookie.com/embed/4msf6EMNIrY?rel=0" frameborder="0" allowfullscreen></iframe> | ||
| + | </html> | ||
<html> | <html> | ||
| Line 98: | Line 106: | ||
| - | < | + | <h4 id="Plastic Fantastic"></h4> |
</div> | </div> | ||
| Line 104: | Line 112: | ||
<div id="CollapsiblePanelM21" class="CollapsiblePanel"> | <div id="CollapsiblePanelM21" class="CollapsiblePanel"> | ||
| - | <div class="CollapsiblePanelTab" tabindex="0"><h4> | + | <div class="CollapsiblePanelTab" tabindex="0"><h4>We degraded P3HB </html><font size="1">▼</font size="1"><html></h4></div> |
| - | <div class="CollapsiblePanelContent"> | + | <div class="CollapsiblePanelContent"> |
| + | |||
| + | </html> | ||
| + | |||
| + | <p align="justify">Our clearing zone assay indicates that P3HB depolymerase PhaZ1 started to fairly quickly degrade P3HB from the first day. After 3 days, there is evidently a clear zone around the well containing PhaZ1. <b>We are the first iGEM team to degrade bioplastics!</b></p> | ||
| + | |||
| + | [[File:Imperial_clearing_zone.jpg|thumbnail|centre|950px|<font size="2.7"><b>PhaZ1 showing P3HB degradation ability by generating a clear zone on a P3HB LB agar plate.</b> A. No difference between empty vector cell lysate and PhaZ1 cell lysate right after they were pipetted into the wells. B. PhaZ1 in the cell lysate started to clear P3HB around the well after 1 day. C. PhaZ1 created a clear zone around the well after 3 days, whereas it was still cloudy around the empty vector cell lysate well.]] | ||
| + | |||
| + | <html> | ||
</div> | </div> | ||
| Line 111: | Line 127: | ||
<div id="CollapsiblePanelM22" class="CollapsiblePanel"> | <div id="CollapsiblePanelM22" class="CollapsiblePanel"> | ||
| - | <div class="CollapsiblePanelTab" tabindex="0"><h4> | + | <div class="CollapsiblePanelTab" tabindex="0"><h4>We degraded P3HB we made from waste </html><font size="1">▼</font size="1"><html></h4></div> |
| - | <div class="CollapsiblePanelContent"> | + | <div class="CollapsiblePanelContent"> |
| - | </ | + | </html> |
| - | + | ||
| - | < | + | <p align="justify">Using [https://www.caymanchem.com/app/template/Product.vm/catalog/700190 3HB colourimetric assay kit], we have shown that we have degraded the P3HB made from waste into 3HB monomers <b>with our own enzyme</b>, phaZ1. In addition, there is no significant difference in 3HB concentration between different P3HB sources. This result proves that <b>we now have a closed loop for P3HB bioplastic recycling!</b></p> |
| - | + | ||
| - | + | ||
| - | < | + | {| class="wikitable" style="margin: 1em auto 1em auto;" |
| - | </ | + | |[[File:Imperial_P3HB_from_waste.jpg|thumbnail|left|600px|<b>Degradation of P3HB produced from various sources showing no significant difference between P3HB made from waste and P3HB made from other sources.</b>]] |
| + | |} | ||
| - | |||
| - | |||
| - | |||
| + | <html> | ||
</div> | </div> | ||
</div> | </div> | ||
| - | <div id=" | + | <div id="CollapsiblePanelM23" class="CollapsiblePanel"> |
| - | <div class="CollapsiblePanelTab" tabindex="0"><h4> | + | <div class="CollapsiblePanelTab" tabindex="0"><h4>3HB permease increases 3HB-uptake </html><font size="1">▼</font size="1"><html></h4></div> |
| - | <div class="CollapsiblePanelContent"> | + | <div class="CollapsiblePanelContent"> |
| + | </html> | ||
| + | In this module, we are looking at making P3HB bioplastic from 3HB monomers. It is essential for the cells to take up a relatively high concentration of 3HB. Here we show that having 3HB permease allow cells to grow better in M9 supplement media, where 3HB is the sole carbon source. | ||
| + | |||
| + | |||
| + | [[File:Imperial_permease.jpg|thumbnail|center|600px|x|<b> 3HB sole carbon source growth in M9S with 3HB permease.</b> MG1655 with 3HB permease can grow better in M9 supplement media using 3HB as the sole carbon source, compared to MG1655 empty vector. 3HB concentrations were tested at 4 levels - 1 mM, 10 mM, 50 mM and 100 mM. Growth inhibitory effect exhibited at 100 mM of 3HB. Data points show final time point after 6h growth for each concentration. Growth was at 37°C with shaking over 6h. Error bars are SEM, n=3. Figure made by Imperial College London 2013 iGEM.<br><br><b><span style="color:#FF8000"><font size="6">Performed after European Jamboree</font size="6"></span></b>]] | ||
| + | |||
| + | We also tested the concentration of 3HB in 3HB M9 supplement media to investigate if 3HB decreased significantly if cells express 3HB permease. Here we show that MG1655 with 3HB permease used significantly more 3HB in media than MG1655 empty vectors (p<0.01), given that the starting 3HB concentrations in the media are the same. | ||
| + | [[File:Imperial_permease2.jpg|thumbnail|center|600px|x|<b> 3HB permease enhances uptake of 3HB in media significantly.</b> 3HB permease enhances 3HB uptake because less 3HB was found in MG1655 with 3HB permease sample compared to MG1655 empty vector, given that the starting 3HB concentrations were the same. Data points show final time point after 6h growth at 37°C with shaking over 6h. Figure made by Imperial College London 2013 iGEM.<br><br><b><span style="color:#FF8000"><font size="6">Performed after European Jamboree</font size="6"></span></b>]] | ||
| + | <html> | ||
</div> | </div> | ||
</div> | </div> | ||
| Line 148: | Line 169: | ||
var CollapsiblePanelM11 = new Spry.Widget.CollapsiblePanel("CollapsiblePanelM11", {contentIsOpen:false}); | var CollapsiblePanelM11 = new Spry.Widget.CollapsiblePanel("CollapsiblePanelM11", {contentIsOpen:false}); | ||
var CollapsiblePanelM12 = new Spry.Widget.CollapsiblePanel("CollapsiblePanelM12", {contentIsOpen:false}); | var CollapsiblePanelM12 = new Spry.Widget.CollapsiblePanel("CollapsiblePanelM12", {contentIsOpen:false}); | ||
| - | var CollapsiblePanelM13 = new Spry.Widget.CollapsiblePanel("CollapsiblePanelM13", { | + | var CollapsiblePanelM13 = new Spry.Widget.CollapsiblePanel("CollapsiblePanelM13", {contentIsClose:false}); |
| - | var CollapsiblePanelM14 = new Spry.Widget.CollapsiblePanel("CollapsiblePanelM14", { | + | var CollapsiblePanelM14 = new Spry.Widget.CollapsiblePanel("CollapsiblePanelM14", {contentIsClose:false}); |
| - | + | ||
var CollapsiblePanelM21 = new Spry.Widget.CollapsiblePanel("CollapsiblePanelM21", {contentIsOpen:false}); | var CollapsiblePanelM21 = new Spry.Widget.CollapsiblePanel("CollapsiblePanelM21", {contentIsOpen:false}); | ||
| - | var CollapsiblePanelM22 = new Spry.Widget.CollapsiblePanel("CollapsiblePanelM22", { | + | var CollapsiblePanelM22 = new Spry.Widget.CollapsiblePanel("CollapsiblePanelM22", {contentIsClose:false}); |
| - | var CollapsiblePanelM23 = new Spry.Widget.CollapsiblePanel("CollapsiblePanelM23", { | + | var CollapsiblePanelM23 = new Spry.Widget.CollapsiblePanel("CollapsiblePanelM23", {contentIsClose:false}); |
| - | + | ||
| - | + | ||
</script> | </script> | ||
Latest revision as of 03:56, 29 October 2013
Main Results
Resource-full Waste
Plastic Fantastic
We made P3HB bioplastic ▼
We used E. coli that expresses PhaCAB enzymes to make P3HB bioplastic. P3HB produced in cells were stained with Nile Red for fluorescent microscope imaging, and strong fluorescence was detected at the area where PhaCAB-expressing cells were streaked.
Our model predicts increased PhaB expression boosts P3HB production ▼
In order to realistically improve our experimental design, we produced P3HB synthesis model with considerations of central metabolic pathway within the cells. Our model predicts that during the synthesis of P3HB, the concentration of PhaB enzymes is an important rate limiting factor that can actually be regulated.
Refresh the page to see our video
Metabolic model preview
We simulated the interactions between metabolism pathways with our synthetic pathway
Preview to our GUI (Please refresh to see video):
'''We made a GUI so that the wet lab people could easily manipulate enzyme concentrations to see their effect on P(3HB) production.'''
We optimised P3HB bioplastic production ▼
Since our model predicts that P3HB production can be significantly improved by expressing more P3HB polymerase PhaB, we designed a [http://parts.igem.org/Part:BBa_K1149051 hybrid promoter] which, consists of the J23104 constitutive promoter and the native promoter to optimise gene expression. Our results show that we have successfully produced 11-fold more P3HB bioplastic compared with the native promoter.
We made bioplastic from mixed waste ▼
One of the objectives of Module 1 is to produce P3HB bioplastic from waste. We compared P3HB degradation by measuring the concentration of 3HB monomers, using P3HB we bought from sigma, we produced from glucose, and we produced from the waste collected from Powerday. We found that there is no significant difference between P3HB purchased from Sigma and P3HB we made from waste (t-test p= 0.12). Thus we conclude that we have made P3HB bioplastic from waste.
 We have detected 3HB monomers for the chemical analysis of the produced bioplastic. We synthesised P(3HB) using our improved Biobrick part (hybrid promoter phaCAB, BBa_K1149051). Our engineered bioplastic producing E.coli synthesised P(3HB) directly from waste. See data under 3HB assay #2 Imperial iGEM data |
We degraded P3HB ▼
Our clearing zone assay indicates that P3HB depolymerase PhaZ1 started to fairly quickly degrade P3HB from the first day. After 3 days, there is evidently a clear zone around the well containing PhaZ1. We are the first iGEM team to degrade bioplastics!
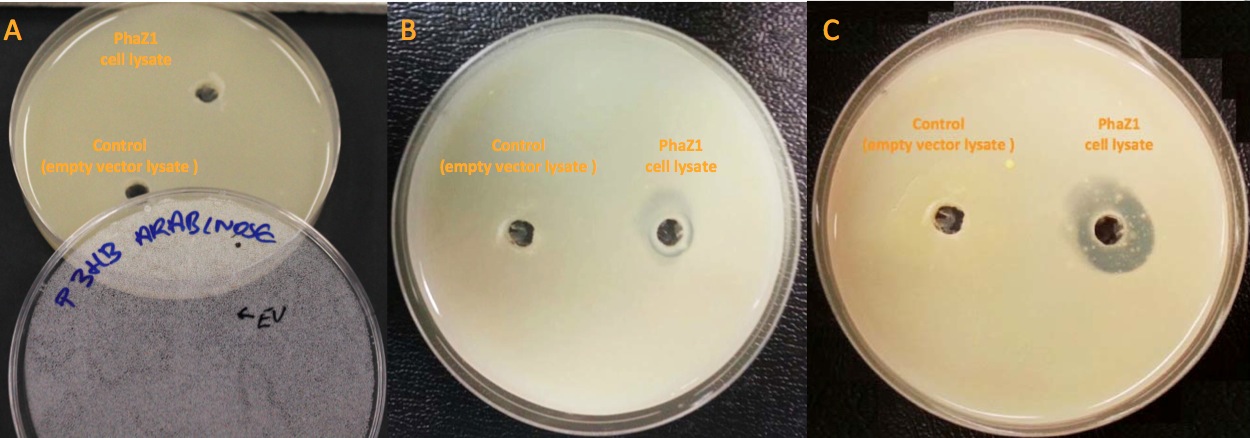
We degraded P3HB we made from waste ▼
Using 3HB colourimetric assay kit, we have shown that we have degraded the P3HB made from waste into 3HB monomers with our own enzyme, phaZ1. In addition, there is no significant difference in 3HB concentration between different P3HB sources. This result proves that we now have a closed loop for P3HB bioplastic recycling!
3HB permease increases 3HB-uptake ▼

Performed after European Jamboree
We also tested the concentration of 3HB in 3HB M9 supplement media to investigate if 3HB decreased significantly if cells express 3HB permease. Here we show that MG1655 with 3HB permease used significantly more 3HB in media than MG1655 empty vectors (p<0.01), given that the starting 3HB concentrations in the media are the same.
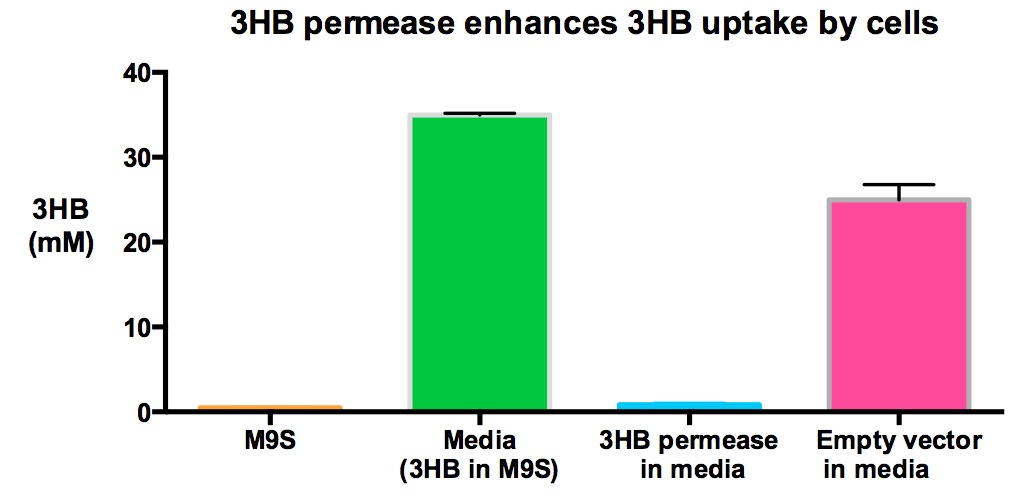
Performed after European Jamboree
 "
"