Team:Edinburgh/GenBrick/GenablerAppendix
From 2013.igem.org
(Difference between revisions)
(→Genbrick Assembly) |
|||
| (One intermediate revision not shown) | |||
| Line 3: | Line 3: | ||
<div class='content'> | <div class='content'> | ||
| - | + | <h3>Genbrick Assembly</h3> | |
'''Design and Preparation - Linker and Segment | '''Design and Preparation - Linker and Segment | ||
| Line 47: | Line 47: | ||
;'''1. Incubation''' | ;'''1. Incubation''' | ||
| - | + | *Use 5 µL each E-P-H required for assembly | |
| - | + | *Add x µl 10X NEBuffer 4 to make 1X final concentration | |
| - | + | *Incubate 30-60 minutes at Room Temperature | |
| - | + | *3-part Example: | |
| - | + | :5 µL Acc_RFP E-P-H ([Acc_RFP -E]+[ Acc_RFP -P]+[GFP-H]) | |
| - | + | :5 µL PLac_LacZ E-P-H ([PLac_LacZ-E]+[PLac_LacZ-P]+[Acc_RFP-H) | |
| - | + | :5 µL GFP E-P-H ([GFP-E]+[GFP-P]+[PLac_LacZ-H]) | |
| - | + | :2 µl NEBuffer 4 | |
| - | + | :3 µL sterile water | |
| - | + | *If larger number of E-P-H in assembly, adjust 10X NEBuffer 4 and water accordingly | |
;'''2. Transformation''' | ;'''2. Transformation''' | ||
Latest revision as of 19:17, 4 October 2013
Genbrick Assembly
Design and Preparation - Linker and Segment
- 1. Overview
- Utilise the linker designer software from the EdiGEM 2013 wiki to design Eye and Hook linker oligo pairs
- Linker and Segment oligos can be custom-made as single-stranded, unphosphorylated DNA
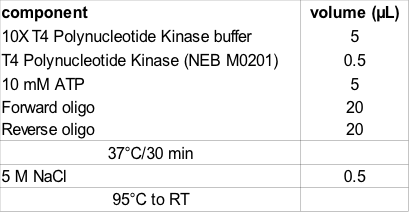
- Forward and reverse oligo pairs are mixed and phosphorylated prior to annealing
- 2. Preparation
- Re-suspend oligo in nuclease-free water to 100 µM (as per instructions)
- Phosphorylation reaction (not required if oligos with 5' phosphate are ordered)
- Annealing is achieved by addition of 5 µL 5 M NaCl (50 mM [final]) prior to heat denaturation at < 95°C and slow cooling to Room temperature.
Pre-assembly
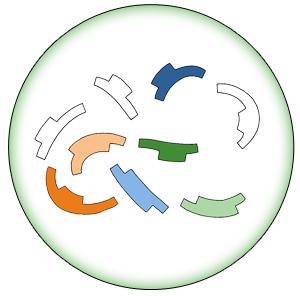
- 1. Eye-Part-Hook Preparation
- a) Using 3-part pathway as an example:
- Acceptor Vector cassette (+promoter); Acc_RFP
- LacZ truncated gene (+PLac);PLac_LacZ
- Green fluorescent protein; GFP
- b) Digestion-Ligation reaction of Eye-Part-Hook (E-P-H) (Overnight)
- Acc_RFP-E + Acc_RFP-P + GFP-H
- PLac_LacZ-E + PLac_LacZ-P + Acc_RFP-H
- GFP-E + GFP-P + PLac_LacZ-H
- b) c) Purification of Digestion-Ligation E-P-H product to remove non-ligated DNA/plasmid
- Run 50 µL E-P-H Digestion-Ligation reaction on 1% agarose gel
- Or: QIAquick PCR Purification kit can be used if Part plasmid does not carry resistance/marker
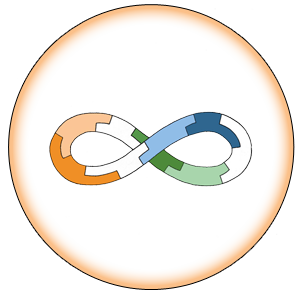
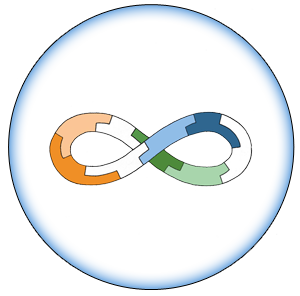
Assembly
- 1. Incubation
- Use 5 µL each E-P-H required for assembly
- Add x µl 10X NEBuffer 4 to make 1X final concentration
- Incubate 30-60 minutes at Room Temperature
- 3-part Example:
- 5 µL Acc_RFP E-P-H ([Acc_RFP -E]+[ Acc_RFP -P]+[GFP-H])
- 5 µL PLac_LacZ E-P-H ([PLac_LacZ-E]+[PLac_LacZ-P]+[Acc_RFP-H)
- 5 µL GFP E-P-H ([GFP-E]+[GFP-P]+[PLac_LacZ-H])
- 2 µl NEBuffer 4
- 3 µL sterile water
- If larger number of E-P-H in assembly, adjust 10X NEBuffer 4 and water accordingly
- 2. Transformation
- Use 10 µL assembly mix to transform 50 µL NEB 10-beta competent cells C3019H
- Culture above assembly example on LB agar plates with chloramphenicol and IPTG
- Incubate overnight at 37°C (further growth at RT if colonies require)

| 
| | | | 
|
| This iGEM team has been funded by the MSD Scottish Life Sciences Fund. The opinions expressed by this iGEM team are those of the team members and do not necessarily represent those of MSD | |||||
 "
"