Team:Dundee/Project/Notebook2
From 2013.igem.org
Kyleharrison (Talk | contribs) |
|||
| (11 intermediate revisions not shown) | |||
| Line 20: | Line 20: | ||
<div class="span6" style="text-align:justify"> | <div class="span6" style="text-align:justify"> | ||
<h2 style="margin-top:-10px;"> Week 6 </h2> | <h2 style="margin-top:-10px;"> Week 6 </h2> | ||
| - | <p> | + | <p> Since the cloning in to <em>B. subtilis</em> plasmid vector was proving fruitless, we decided to try a change of tack. In order to skip out the intermediate plasmid step we ordered primers to do a PCR which would add in restriction sites, allowing us to ligate our gene construct straight in to the <em>B. subtilis</em> plasmid vector pDR110. <br><br> |
| - | + | This week not much in the way of lab work was done as the team took a trip to London to meet up with other iGEM teams! It was brilliant to meet all of the other UK teams and discuss science together. We came back extremely motivated and full of new ideas for our human practices. | |
| - | + | ||
</p> | </p> | ||
<br> | <br> | ||
| Line 28: | Line 27: | ||
<div class="span6" style="margin-top:50px;"> | <div class="span6" style="margin-top:50px;"> | ||
| - | <img id="image-6" src="http:// | + | <img id="image-6" src="http://farm8.staticflickr.com/7408/9367934245_1b9a3fa747_o.jpg" width="600px" height="300px"> |
</div> | </div> | ||
</div> | </div> | ||
| Line 37: | Line 36: | ||
<div class="span6" style="text-align:justify"> | <div class="span6" style="text-align:justify"> | ||
<h2 style="margin-top:-10px;"> Week 7 </h2> | <h2 style="margin-top:-10px;"> Week 7 </h2> | ||
| - | <p> | + | <p> We did some more Western blots on our <i>tat</i> mutant (Δ<i>tat</i>) however the results are pretty rubbish, looks like we need a new sample!<br><br> |
| - | + | On the <em>Bacillus front</em>, we performed the PCR to add in the desired restriciton sites to the genetic construct and began attempting to clone it in to the <em>B. subtilis</em> vector.<br><br> | |
| - | + | This week the dry team also decided to split the work. Craig started setting up Matlab programmes for our stochastic modelling. Rachel and Nasir started getting to grips with NetLogo, modelling software, so we can visualise our bacterial mops and how the Sec and Tat pathways transport the mops. | |
</p> | </p> | ||
<br> | <br> | ||
| Line 45: | Line 44: | ||
<div class="span6" style="margin-top:50px;"> | <div class="span6" style="margin-top:50px;"> | ||
| - | <img id="image-6" src="http:// | + | <img id="image-6" src="http://farm6.staticflickr.com/5547/9552996651_6a20ba7dfa_o.png"> |
</div> | </div> | ||
</div> | </div> | ||
| Line 55: | Line 54: | ||
<div class="span6" style="text-align:justify"> | <div class="span6" style="text-align:justify"> | ||
<h2 style="margin-top:-10px;"> Week 8 </h2> | <h2 style="margin-top:-10px;"> Week 8 </h2> | ||
| - | <p> | + | <p> Blots of the <i>E. coli</i> mop this week suggest that PP1 is present in the Δ<i>tat</i> in the sphaeroplast and membrane fractions, but not in the cytoplasm or periplasm fractions, which is interesting. Wild type fractions are not blotting properly, may need to make fresh competent cells. <br><br> |
| - | + | The detector team also managed to clone PP1 gene downstream of <i>envZ</i> this week so we almost have a completed detector! Efforts to get the mop cloned into <i>B. subtilis</i> finally came to fruition. The construct was cloned in to the appropriate plasmid vector and transformed into <em>B. subtilis</em> cells. On with characterising it!<br><br> | |
| - | <br><br> | + | Craig finished our deterministic ODE model and we got some interesting information to help out the wet team as well as some nice graphs to show our data. The rest of the dry team were continuing as usual with Netlogo and the Moptopus.<br><br> |
| + | On the human practices side of things we met with our local Member of Scottish Parliament Joe Fitzpatrick, to discuss local and global policy regarding algal blooms, in addition to requesting that he contact the Scottish Environmental Protection Agency (SEPA) on our behalf. | ||
</p> | </p> | ||
<br> | <br> | ||
| Line 63: | Line 63: | ||
<div class="span6" style="margin-top:50px;"> | <div class="span6" style="margin-top:50px;"> | ||
| - | <img id="image-6" src="http:// | + | <img id="image-6" src="http://farm4.staticflickr.com/3776/9553290659_fd413a2d2d_o.jpg" width="600px"> |
</div> | </div> | ||
</div> | </div> | ||
| Line 73: | Line 73: | ||
<div class="span6" style="text-align:justify"> | <div class="span6" style="text-align:justify"> | ||
<h2 style="margin-top:-10px;"> Week 9 </h2> | <h2 style="margin-top:-10px;"> Week 9 </h2> | ||
| - | <p> | + | <p> Blots of the Δ<i>tat</i> strain expressing TorA-PP1 still have strong PP1 banding in the sphaeroplast and membrane fractions whereas the wild type expressing the same construct refuses to blot well.<br><br> |
| - | + | No PP1 antigen was detecting following Western blots of <em>B. subtilis</em> harbouring plasmid-encoded PrsA-PP1. Looks like a bad sample after Coomassie staining. | |
| - | + | ||
</p> | </p> | ||
<br> | <br> | ||
| Line 81: | Line 80: | ||
<div class="span6" style="margin-top:50px;"> | <div class="span6" style="margin-top:50px;"> | ||
| - | <img id="image-6" src="http:// | + | <img id="image-6" src="http://farm4.staticflickr.com/3679/9553643593_05c6ceea4a_o.jpg" width="600px"> |
</div> | </div> | ||
</div> | </div> | ||
| Line 90: | Line 89: | ||
<div class="span6" style="text-align:justify"> | <div class="span6" style="text-align:justify"> | ||
<h2 style="margin-top:-10px;"> Week 10 </h2> | <h2 style="margin-top:-10px;"> Week 10 </h2> | ||
| - | <p> | + | <p>We began testing ToxiMop this week! Although unfortunately one of our control experiments was found to actually contain PP1, as we found by Western blot. Never mind! The repeat experiment will actually have an empty plasmid vector next time! <br><br> |
| - | + | There is still no sign of PP1 production in <i>Bacillus</i> so we performed some PCRs to ensure that the insert DNA is definitely there. It bloody well isn’t, so we decided to focus our efforts elsewhere with the <i>E. coli</i> mop.<br><br> | |
| - | <br><br> | + | We successfuly cloned a reporter construct where the GFP gene was fused to the <i>ompC</i> promoter. This will serve as a reporter for the modified EnvZ detector. Hopefully we will have a working microcystin detector very soon - how exciting!<br><br> |
| + | NetLogo model for the Sec transporter was completed and tested online. To our luck, it worked! We will now begin our Tat pathway model. | ||
</p> | </p> | ||
<br> | <br> | ||
| Line 98: | Line 98: | ||
<div class="span6" style="margin-top:50px;"> | <div class="span6" style="margin-top:50px;"> | ||
| - | <img id="image-6" src="http:// | + | <img id="image-6" src="http://farm4.staticflickr.com/3675/9548806162_134fdca434_o.png"> |
</div> | </div> | ||
</div> | </div> | ||
Latest revision as of 22:08, 1 October 2013
Notebook
Week 6
Since the cloning in to B. subtilis plasmid vector was proving fruitless, we decided to try a change of tack. In order to skip out the intermediate plasmid step we ordered primers to do a PCR which would add in restriction sites, allowing us to ligate our gene construct straight in to the B. subtilis plasmid vector pDR110.
This week not much in the way of lab work was done as the team took a trip to London to meet up with other iGEM teams! It was brilliant to meet all of the other UK teams and discuss science together. We came back extremely motivated and full of new ideas for our human practices.

Week 7
We did some more Western blots on our tat mutant (Δtat) however the results are pretty rubbish, looks like we need a new sample!
On the Bacillus front, we performed the PCR to add in the desired restriciton sites to the genetic construct and began attempting to clone it in to the B. subtilis vector.
This week the dry team also decided to split the work. Craig started setting up Matlab programmes for our stochastic modelling. Rachel and Nasir started getting to grips with NetLogo, modelling software, so we can visualise our bacterial mops and how the Sec and Tat pathways transport the mops.

Week 8
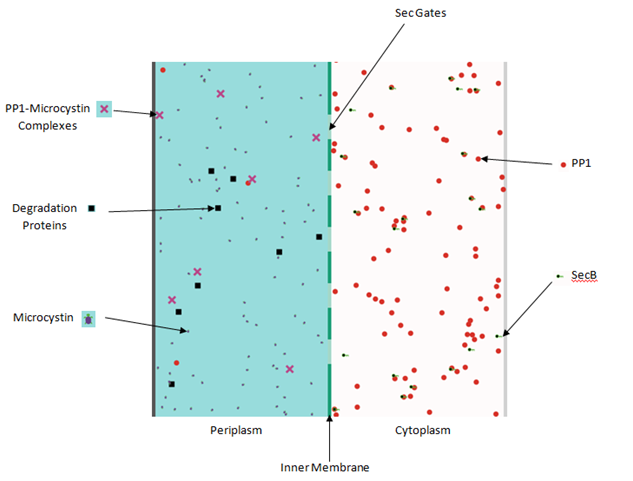
Blots of the E. coli mop this week suggest that PP1 is present in the Δtat in the sphaeroplast and membrane fractions, but not in the cytoplasm or periplasm fractions, which is interesting. Wild type fractions are not blotting properly, may need to make fresh competent cells.
The detector team also managed to clone PP1 gene downstream of envZ this week so we almost have a completed detector! Efforts to get the mop cloned into B. subtilis finally came to fruition. The construct was cloned in to the appropriate plasmid vector and transformed into B. subtilis cells. On with characterising it!
Craig finished our deterministic ODE model and we got some interesting information to help out the wet team as well as some nice graphs to show our data. The rest of the dry team were continuing as usual with Netlogo and the Moptopus.
On the human practices side of things we met with our local Member of Scottish Parliament Joe Fitzpatrick, to discuss local and global policy regarding algal blooms, in addition to requesting that he contact the Scottish Environmental Protection Agency (SEPA) on our behalf.

Week 9
Blots of the Δtat strain expressing TorA-PP1 still have strong PP1 banding in the sphaeroplast and membrane fractions whereas the wild type expressing the same construct refuses to blot well.
No PP1 antigen was detecting following Western blots of B. subtilis harbouring plasmid-encoded PrsA-PP1. Looks like a bad sample after Coomassie staining.

Week 10
We began testing ToxiMop this week! Although unfortunately one of our control experiments was found to actually contain PP1, as we found by Western blot. Never mind! The repeat experiment will actually have an empty plasmid vector next time!
There is still no sign of PP1 production in Bacillus so we performed some PCRs to ensure that the insert DNA is definitely there. It bloody well isn’t, so we decided to focus our efforts elsewhere with the E. coli mop.
We successfuly cloned a reporter construct where the GFP gene was fused to the ompC promoter. This will serve as a reporter for the modified EnvZ detector. Hopefully we will have a working microcystin detector very soon - how exciting!
NetLogo model for the Sec transporter was completed and tested online. To our luck, it worked! We will now begin our Tat pathway model.
 "
"
