Team:Newcastle/Notebook/calendar
From 2013.igem.org
| (175 intermediate revisions not shown) | |||
| Line 14: | Line 14: | ||
====Training==== | ====Training==== | ||
| - | + | The teams split into small groups to discuss individual themes. They did some research into multiple aspects of the project including: wiki layout, t-shirt design and possible team logos. We also looked at past iGEM team wikis for inspirational ideas. | |
| - | [[File:P1050457.JPG|frameless| | + | [[File:P1050457.JPG|frameless|300px]],[[File:P1050459.JPG|frameless|300px]] |
| - | + | A talk on Computational Modelling and BioBrick concepts was given. This talk led the team to start building the biophysical model of the lipid membrane of ''Bacillus subtilis'' and modelling of lipid synthetic pathway. Later in the day, the team met to discuss initial aims and progress of our work so far. | |
| - | [[File:P1050460.JPG|frameless| | + | [[File:P1050460.JPG|frameless|300px]] |
====Wiki==== | ====Wiki==== | ||
| Line 40: | Line 40: | ||
We started off working on a Modelling Exercise to identify components and species of the subtilin biosynthesis pathway. We also sketched down some potential logo ideas and created a shortlist of favorites. | We started off working on a Modelling Exercise to identify components and species of the subtilin biosynthesis pathway. We also sketched down some potential logo ideas and created a shortlist of favorites. | ||
| - | [[File:P1050461.JPG|frameless| | + | [[File:P1050461.JPG|frameless|300px]] |
| - | We | + | We went on to learn about the differences and similarities between stochastic and deterministic modeling. We also used rule based modeling such as RuleBender and BioNetGen. Using the Michaelis-Menten kinetics model as an example, we alter the parameters and rates of reaction to observe the effects. |
| - | + | ||
| - | + | ||
====Wiki==== | ====Wiki==== | ||
| Line 62: | Line 60: | ||
==== Modelling Training==== | ==== Modelling Training==== | ||
| - | Today we continued learning about stochastic and deterministic modelling and looked at converting the rate parameters in RuleBender. We | + | Today we continued learning about stochastic and deterministic modelling and looked at converting the rate parameters in RuleBender. We made the logo more detailed and narrowed it down to two ideas. |
| - | [[File:Logo2.jpg|frameless| | + | [[File:Logo2.jpg|frameless|300px]][[File:P1050467.JPG|frameless|300px]] |
====GUS cloning strategy==== | ====GUS cloning strategy==== | ||
| Line 83: | Line 81: | ||
====Basic Lab Skills==== | ====Basic Lab Skills==== | ||
| - | + | Before we started work in the lab, we were briefed on various lab health and safety policies to ensure we maintained a safe working environment. We were then given a tour around the lab to inform us of the layout including waste disposal procedures, location of chemicals and use of equipment. | |
| - | We | + | We were then taught how to calibrate pipettes and learned how to operate them. We are also shown how to inoculate bacteria to a liquid culture and how to prepare bacteria streak plates. |
| - | [[File:P1050469.JPG|frameless| | + | [[File:P1050469.JPG|frameless|260px]] [[File:P1050472.JPG|frameless|260px]] [[File:P1050475.JPG|frameless|260px]] |
| + | |||
| + | ====Transformation of GFP and RFP bricks into E. coli ''Competent Cells''==== | ||
| + | |||
| + | '''Aim:''' | ||
| + | |||
| + | The aim of this experiment is to transform GFP and RFP BioBricks into Escherichia coli. | ||
| + | |||
| + | '''Methods:''' | ||
| + | |||
| + | Please refer to the [https://2013.igem.org/Team:Newcastle/Notebook/protocols#Escherichia_coli_Competent_Cell_Preparation ''Escherichia coli'' competent cells protocol] and the [https://2013.igem.org/Team:Newcastle/Notebook/protocols#Escherichia_coli_Transformation ''E.coli'' Transformation Protocol ]. | ||
| + | |||
| + | We then plated those cells onto LB with 5ug/ml chloramphenicol plates. | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | Please see <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#14/6/13" target="_blank">June 14th</a> </html> for results | ||
| - | |||
</div> | </div> | ||
| Line 97: | Line 111: | ||
==== Basic Lab Skills 2 ==== | ==== Basic Lab Skills 2 ==== | ||
<div> | <div> | ||
| - | * | + | *Transformation of GFP and RFP bricks into E. coli ''Competent Cells'' Results |
*Wiki | *Wiki | ||
</div> | </div> | ||
==== Details ==== | ==== Details ==== | ||
<div> | <div> | ||
| - | ==== | + | ====Transformation of GFP and RFP bricks into E. coli ''Competent Cells'' Results==== |
| - | + | '''Aim:''' | |
| + | |||
| + | Please see <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#14/6/13" target="_blank">June 14th</a> </html> for aim. | ||
| + | |||
| + | '''Methods:''' | ||
| + | |||
| + | Please see <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#14/6/13" target="_blank">June 14th</a> </html> for methods. | ||
| + | |||
| + | '''Results:''' | ||
| - | + | We checked the plates to see if anything grew. The RFP plates showed a lot of colonies, while the GFP plates did not work as expected. The next thing we did was to view the plates under the fluorescent inverted microscope. In the afternoon,we had an introductory lecture on basic molecular biology and biobrick design. | |
| + | [[File:P1050483.JPG|frameless|260px]] [[File:P1050490.JPG|frameless|260px]] [[File:P1050488.JPG|frameless|260px]] | ||
====Wiki==== | ====Wiki==== | ||
| Line 130: | Line 155: | ||
<div> | <div> | ||
*Basic Lab Skills | *Basic Lab Skills | ||
| + | *Day Overview | ||
*BioGame | *BioGame | ||
*HBsu BioBrick clonning strategy | *HBsu BioBrick clonning strategy | ||
| Line 137: | Line 163: | ||
====Basic Lab Skills==== | ====Basic Lab Skills==== | ||
| - | + | '''Aim:''' | |
| - | + | To familiarize with miniprep, nanodrop, restriction digest and gel electrophoresis. | |
| + | '''Methods:''' | ||
| + | |||
| + | We transfer colonies of bacteria into LB and extracted the plasmids via the [https://2013.igem.org/Team:Newcastle/Notebook/protocols#QIAprep_Spin_Miniprep_Kit miniprep protocol], and checked the concentration of plasmids through the use of [https://2013.igem.org/Team:Newcastle/Notebook/protocols#Nanodrop_quantification_of_DNA_extracts nanodrop spectrophotometry]. We then prepared a 1% Agarose gel and performed a single digest with XbaI and PstI and a double digest with both restriction enzymes. We loaded our samples to the gel with the DNA ladder and control (Figure 1). | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | [[File:P1050504.JPG|frameless|300px]] | ||
| + | |||
| + | Figure 1 | ||
| + | |||
| + | ====Day Overview==== | ||
| + | The group started thinking about their cloning strategies for each theme of the project. We also discussed ideas for human practices and came up with the idea of making an educational game based on synthetic biology. At the end of the day we had a group meeting to discuss our progress and our ideas for the modelling workshop for the UCL meet up. | ||
====BioGame==== | ====BioGame==== | ||
| - | Work began on the [[Team:Newcastle/Outreach/BioGame|BioGame]]. Largely contained to simply choosing the best method of implementation for the game. | + | Work began on the [[Team:Newcastle/Outreach/BioGame|BioGame]]. Largely contained to simply choosing the best method of implementation for the game. The following options were considered: |
*Android SDK (with no game engine/plug ins) | *Android SDK (with no game engine/plug ins) | ||
*LibGDX | *LibGDX | ||
| Line 149: | Line 187: | ||
Ultimately the choice was made to use LibGDX due to the documentation available on it, the ease at which games can be developed and the consistent structure it provides with standard gaming development. After this decision was made, a series of tutorials were found and followed to provide a base of knowledge going into developing the application itself. | Ultimately the choice was made to use LibGDX due to the documentation available on it, the ease at which games can be developed and the consistent structure it provides with standard gaming development. After this decision was made, a series of tutorials were found and followed to provide a base of knowledge going into developing the application itself. | ||
| - | ====HBsu BioBrick Cloning Strategy==== | + | ====HBsu- (x)fp BioBrick Cloning Strategy==== |
The team has been looking in literature for a protein that binds non-specifically to DNA. This will aid the visualization of Genome Shuffling among L-forms by enabling the team to see sections of DNA that have come from two different Bacteria. | The team has been looking in literature for a protein that binds non-specifically to DNA. This will aid the visualization of Genome Shuffling among L-forms by enabling the team to see sections of DNA that have come from two different Bacteria. | ||
The decision was made to use the HBsu protein which is encoded by the ''hbs'' gene in ''B.subtilis'' because it is well characterised and should remain associated with the DNA for a long period of time. | The decision was made to use the HBsu protein which is encoded by the ''hbs'' gene in ''B.subtilis'' because it is well characterised and should remain associated with the DNA for a long period of time. | ||
| - | The HBsu will be conjugated with a | + | The HBsu will be conjugated with a florescent protein such as GFP and RFP. |
</div> | </div> | ||
| Line 167: | Line 205: | ||
We came up with a script, story board and a list of materials needed for the introductory video on our wiki. We had fun learning to crochet in order to create characters for the video. Furthermore, we updated the team profile page on the wiki by adding individual pictures of all team members. | We came up with a script, story board and a list of materials needed for the introductory video on our wiki. We had fun learning to crochet in order to create characters for the video. Furthermore, we updated the team profile page on the wiki by adding individual pictures of all team members. | ||
| - | [[File:P1050589.JPG|frameless| | + | [[File:P1050589.JPG|frameless|300px]] |
| - | + | ||
====HBsu BioBrick Cloning Strategy==== | ====HBsu BioBrick Cloning Strategy==== | ||
| Line 185: | Line 222: | ||
==== Details ==== | ==== Details ==== | ||
<div> | <div> | ||
| - | |||
| - | |||
| - | |||
====Gus Reporter BioBrick==== | ====Gus Reporter BioBrick==== | ||
| Line 193: | Line 227: | ||
====BioGame==== | ====BioGame==== | ||
| - | Today the game project was set up, created using the LibGDX-setup GUI in order to make projects for Android, Desktop and HTML | + | Today the game project was set up, created using the LibGDX-setup GUI in order to make projects for Android, Desktop and HTML deployment, all utilising LibGDX's built in features to run off a single "core" project. This was a big deciding factor in the choice to go with LIBGdx for developing the game as it allows us to quickly deploy across multiple platforms should we wish to do so. |
With the project set up, we have started to create assets such as textures and fonts to be loaded in and used within the game. | With the project set up, we have started to create assets such as textures and fonts to be loaded in and used within the game. | ||
| - | |||
====HBsu BioBrick Cloning Strategy==== | ====HBsu BioBrick Cloning Strategy==== | ||
| - | + | We plan to clone it into [http://parts.igem.org/wiki/index.php/Part:pSB1C3 pSB1C3] then PCR the insert before integrating it into the amyE integration plasmid (pSB<sub>Bs</sub>1C). We discovered that the Kanamycin resistance is an ''E.coli'' version and wouldn't work in the ''B. subtilis''. Different antibiotic markers that would work in ''B.subtilis'' are trying to be found. | |
</div> | </div> | ||
| Line 214: | Line 247: | ||
We continued work on the cloning strategies as well as starting modelling the gus reporter system using BioNetGen. We produced a preliminary model of the gus reporter system. However, this model needs to be modified by adding parameters. We have also looked through some papers to find parameters required for the model. | We continued work on the cloning strategies as well as starting modelling the gus reporter system using BioNetGen. We produced a preliminary model of the gus reporter system. However, this model needs to be modified by adding parameters. We have also looked through some papers to find parameters required for the model. | ||
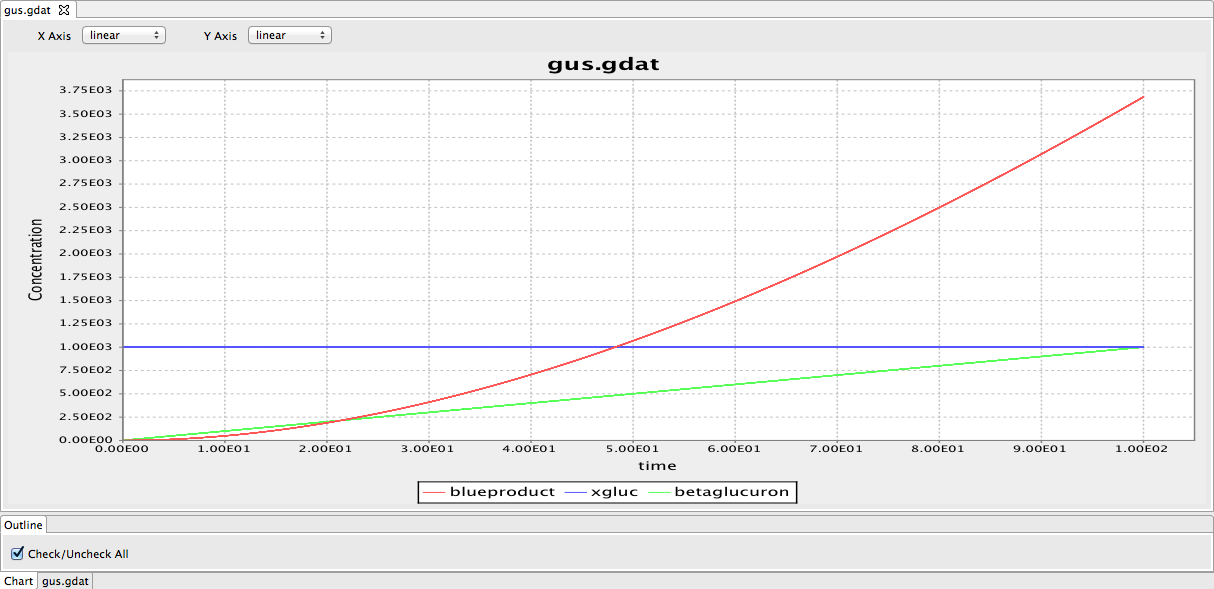
| - | [[File:Gus model.png|frameless| | + | [[File:Gus model.png|frameless|600px]] |
| + | |||
| + | Figure 1: The framework of the GUS model. The red line indicates the blue product in the GUS reporter system. The blue line represents substrate 5-bromo-4-chloro-3indolyl glucuronide (X-Gluc). The green line represents the beta B-glucuronidase. | ||
====BioGame==== | ====BioGame==== | ||
| Line 220: | Line 255: | ||
====HBsu BioBrick Cloning Strategy==== | ====HBsu BioBrick Cloning Strategy==== | ||
| - | Designed Primers to be used in the Golden Braid assembly, and checked the melting temperature and self-annealing to make sure they work perfectly. Uses RFP (BBa_E1010) and GFP (BBa_E0040) to conjugate it to the HBsu protein in order to observe the recombination. | + | Designed Primers to be used in the Golden Braid assembly, and checked the melting temperature and self-annealing to make sure they work perfectly. Uses RFP (BBa_E1010) and GFP (BBa_E0040) to conjugate it to the HBsu protein in order to observe the recombination. |
| - | + | ||
</div> | </div> | ||
| Line 233: | Line 267: | ||
==== Details ==== | ==== Details ==== | ||
<div> | <div> | ||
| - | ====GUS | + | ====GUS Reporter Gene Cloning Strategy==== |
| - | + | We further developed our cloning strategies and decided on the basic design of our construct. We made a preliminary design of the construct using Gene Designer to give a visual representation of how our construct should look like. From this image (Figure 1), we could see the positioning of all the parts, which will be more convenient for us when we want to devise a method to produce this. | |
| - | + | We also improved the logo so it's ready for T-shirt printing. (picture from Alina) | |
| - | + | ||
| - | We also improved the logo so it's ready for T-shirt printing. | + | |
====HBsu BioBrick Cloning Strategy==== | ====HBsu BioBrick Cloning Strategy==== | ||
| - | A discussion with | + | A discussion with our supervisor revealed that the assembly system might be very time consuming. So it was decided that DNA 2.0 would be used to synthesis the HBsu BioBrick. |
</div> | </div> | ||
| - | |||
== 6 == | == 6 == | ||
| Line 257: | Line 288: | ||
<div> | <div> | ||
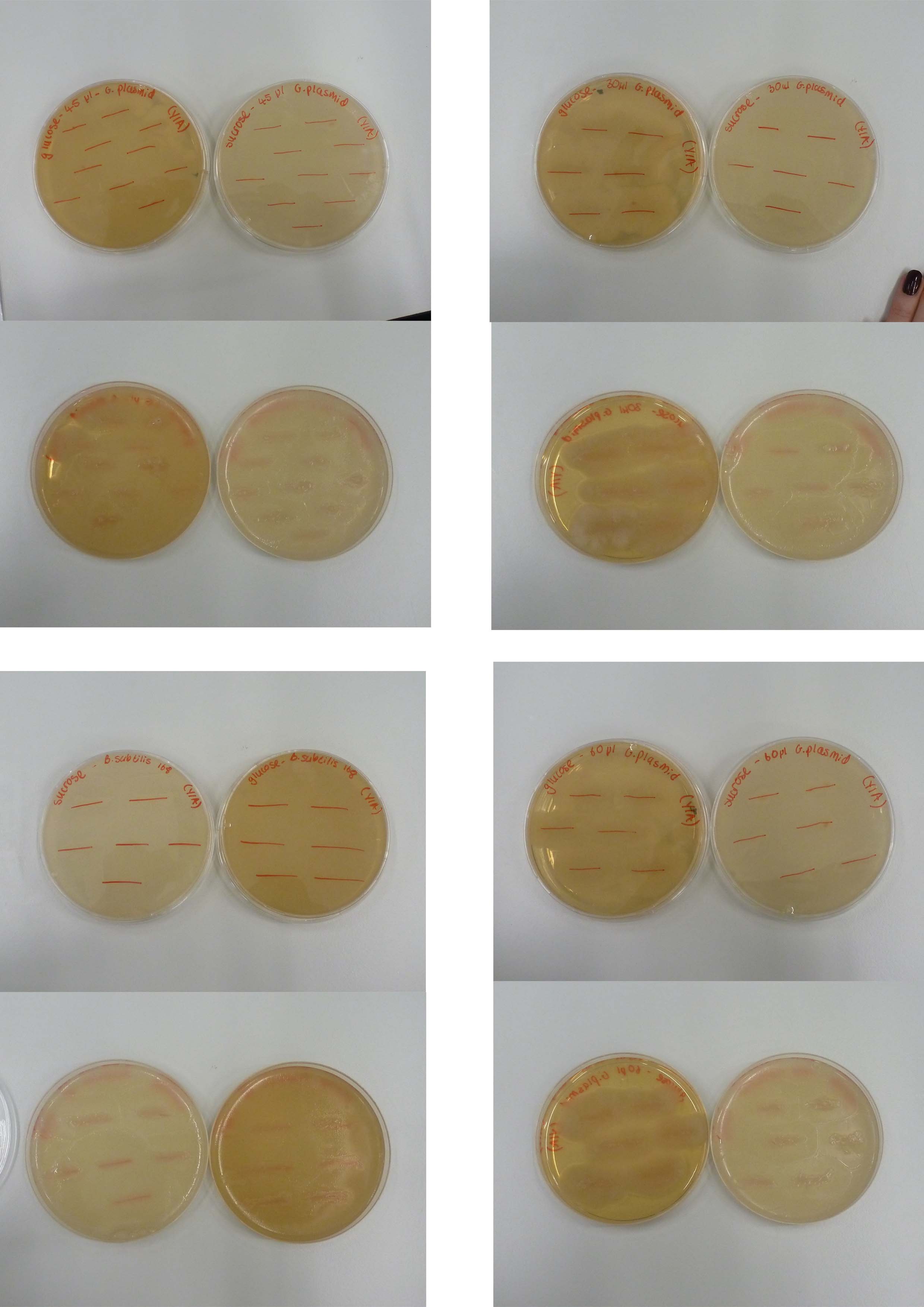
====Groningen plasmid Mini-prep==== | ====Groningen plasmid Mini-prep==== | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| + | '''Aim:''' | ||
| - | + | To isolate the groningen plasmid. | |
| - | + | ||
| + | '''Methods:''' | ||
| + | |||
| + | We performed a [https://2013.igem.org/Team:Newcastle/Notebook/protocols#QIAprep_Spin_Miniprep_Kit mini prep] of the Groningen 2012 integration plasmid DNA grown in ''E.coli'' overnight at 37 ̊C. The purity was tested using [https://2013.igem.org/Team:Newcastle/Notebook/protocols#Nanodrop_quantification_of_DNA_extracts NanoDrop]. | ||
| + | |||
| + | We confirmed the insert size by first cut with fast digest XbaI and BamHI and then running the product on a gel to confirm the presence of the plasmid in the purified pDNA. | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | [[File:Groningen plasmid uncut,xba1,bamh1,both.jpg |frameless|500px]] | ||
| + | |||
| + | Figure 1: Gel electrophoresis of Groningen plasmid. | ||
| + | |||
| + | Lane 1: DNA Ladder | ||
| + | |||
| + | Lane 2: Uncut plasmid | ||
| + | |||
| + | Lane 3: XbaI | ||
| + | |||
| + | Lane 4: BamHI | ||
| + | |||
| + | Lane 5: XbaI and BamHI | ||
| + | |||
| + | '''Discussion:''' | ||
| + | |||
| + | The gel shows that in lane 5 where the plasmid is cut with both XbaI and BamHI, two fragments are present. This is what we expected to have happened as the plasmid is cut at two places, isolating an insert of interest. The smaller fragment is the insert (RFP reporter gene) while the bigger fragment is the plasmid backbone. | ||
| + | |||
| + | ====BioGame==== | ||
| + | Further work on the game architecture has been done today. The main BioBrick class been created to provide common methods that all BioBricks will use. A very basic BioBrick has been created, and can be added upon touch to the Bacteria. This is simply to demonstrate what will happen when a BioBrick is added and the possibilities therein. | ||
====HBsu BioBrick Cloning Strategy==== | ====HBsu BioBrick Cloning Strategy==== | ||
| - | Due to the high cost of sequencing, the integration method of the HBsu BioBrick is now to clone the gene construct into the pMutin4 plasmid and use a single cross-over via the amyE gene to integrate it into the bacteria genome. Also the | + | Due to the high cost of sequencing, the integration method of the HBsu BioBrick is now to clone the gene construct into the pMutin4 plasmid and use a single cross-over via the amyE gene to integrate it into the bacteria genome. Also the antibiotic marker and promoter is no longer being used because after SCO the whole plasmid will be integrated and as such will contain the promoter Pspac and Kanamycin cassette. |
</div> | </div> | ||
| - | |||
== 6 == | == 6 == | ||
| Line 285: | Line 338: | ||
<div> | <div> | ||
====Preparation for UCL==== | ====Preparation for UCL==== | ||
| - | We started preparing for our project presentation | + | We started preparing for our project presentation for UCL, and have started to prepare a presentation for our workshop about BioNetGen modelling. |
====''B.subtilis'' Transformation==== | ====''B.subtilis'' Transformation==== | ||
| - | A single colony of ''B. subtilis'' strain 168 was picked and transferred into [https://2013.igem.org/Team:Newcastle/Notebook/protocols#Bacillus_subtilis_168_Competent_cell_Prep_and_Transformation MM media] and left to grow at | + | '''Aim:''' |
| + | |||
| + | To transform the bacteria with Groningen plasmid. | ||
| + | |||
| + | '''Methods:''' | ||
| + | |||
| + | A single colony of ''B.subtilis'' strain 168 was picked and transferred into [https://2013.igem.org/Team:Newcastle/Notebook/protocols#Bacillus_subtilis_168_Competent_cell_Prep_and_Transformation MM media] and left to grow at 37 ̊C overnight. We continued the protocol on | ||
| + | <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#26/6/13" target="_blank">June 26th</a> </html>. | ||
====Microfluidics Introduction==== | ====Microfluidics Introduction==== | ||
| - | Our iGEM team attended a talk about microfluidics given by Sunny Park. Microfluidics is vital to some of our sub-projects, therefore we looked into mechanism of operation | + | Our iGEM team attended a talk about microfluidics given by Sunny Park. Microfluidics is vital to some of our sub-projects ([https://2013.igem.org/Team:Newcastle/Project/shuffling_endosymbiosis Genome Shuffling] and [https://2013.igem.org/Team:Newcastle/Project/shape_shifting Shape Shifting]), therefore we looked into the mechanism of operation, possible uses and creation of design for master moulds on the silicon wafer. |
====HBsu BioBrick Cloning Strategy==== | ====HBsu BioBrick Cloning Strategy==== | ||
| - | + | The HBsu-GFP and HBsu-RFP BioBricks were sent off for synthesis. We then created an overnight liquid culture of LR2 rod ''B.subtilis cells'' in [https://2013.igem.org/Team:Newcastle/Notebook/protocols#L-form_Media_Components NB/MSM] media, ready to be used to make L-form plates to obtain microscopic pictures. | |
</div> | </div> | ||
| Line 302: | Line 363: | ||
<div> | <div> | ||
*Preparation for UCL | *Preparation for UCL | ||
| - | *Transformation | + | *''B.subtilis'' Transformation |
</div> | </div> | ||
==== Details ==== | ==== Details ==== | ||
<div> | <div> | ||
====Preparation for UCL==== | ====Preparation for UCL==== | ||
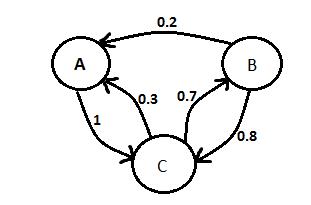
| - | The team continued work on our modelling presentation for the UCL meetup. We added information about ordinary differential equation vs stochastic modelling and about Markov chain. | + | The team continued work on our modelling presentation for the UCL meetup. We added information about ordinary differential equation vs stochastic modelling and about Markov chain (Figure 1). |
| - | [[File:Geoff_working.jpg |frameless| | + | [[File:Geoff_working.jpg |frameless|200px]] |
| - | [[File:Markov.png |frameless| | + | [[File:Markov.png |frameless|500px]] |
| - | + | Figure 1: Markov Chain | |
| - | + | ||
| - | + | ||
| - | + | ====''B.subtilis'' Transformation==== | |
| + | '''Aim:''' | ||
| + | |||
| + | To transform the bacteria with Groningen plasmid. | ||
| + | |||
| + | '''Methods:''' | ||
| + | |||
| + | Please refer to <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#25/6/13" target="_blank">June 25th</a> </html> for the first part of the protocol. | ||
| + | |||
| + | We transferred 1ml of the prepared ''B.subtilis'' str. 168 into fresh MM media. We then tranformed the bacteria with the Groningen plasmid following the [https://2013.igem.org/Team:Newcastle/Notebook/protocols#Bacillus_subtilis_168_Competent_cell_Prep_and_Transformation ''B.subtilis'' str. 168 transformation protocol]. We also made some frozen competent ''B.subtilis'' str. 168 in glycerol. | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | Please refer to <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#27/6/13" target="_blank">June 27th</a> </html>. | ||
| + | |||
| + | |||
| + | </div> | ||
== 6 == | == 6 == | ||
| Line 332: | Line 409: | ||
We began brainstorming what we should cover in our [https://2013.igem.org/Team:Newcastle/Outreach/Workshop modelling workshop] presentation for the [https://2013.igem.org/Team:Newcastle/Outreach/uk_meet YSB 1.0]. We decided to first describe biological modelling and talk about stochastic vs differential models. Before introducing BioNetGen,talking about it's benefits and covering the basics of this language. | We began brainstorming what we should cover in our [https://2013.igem.org/Team:Newcastle/Outreach/Workshop modelling workshop] presentation for the [https://2013.igem.org/Team:Newcastle/Outreach/uk_meet YSB 1.0]. We decided to first describe biological modelling and talk about stochastic vs differential models. Before introducing BioNetGen,talking about it's benefits and covering the basics of this language. | ||
| - | ====Transformation | + | ====''B.subtilis'' Transformation==== |
| - | + | ||
| - | + | '''Aim and Methods:''' | |
| + | |||
| + | For Aim and Protocol, please refer to <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#25/6/13" target="_blank">June 25th</a> </html> and <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#26/6/13" target="_blank">June 26th</a> </html>. | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | No growth has been observed on the chloramphenicol plates apart from a single colony which might be just background growth. | ||
</div> | </div> | ||
| Line 349: | Line 434: | ||
====Groningen plasmid characterisation==== | ====Groningen plasmid characterisation==== | ||
| - | |||
| - | |||
| + | '''Aim:''' | ||
| + | |||
| + | To characterise the [http://parts.igem.org/wiki/index.php?title=Part:BBa_K818000 Groningen 2012 integration plasmid]. | ||
| + | |||
| + | '''Methods:''' | ||
| + | |||
| + | The transformation was done using the [https://2013.igem.org/Team:Newcastle/Notebook/protocols#Bacillus_subtilis_168_Competent_cell_Prep_and_Transformation protocol] for competent ''Bacillus subtilis''. | ||
| + | |||
| + | Positive control: pGFPrrnB plasmid | ||
| + | |||
| + | Negative control: water | ||
| + | |||
| + | We then plated the transformants into LB + 5ug/ml Chloramphenicol plates. | ||
| + | |||
| + | '''Results and Discussion''' | ||
| + | |||
| + | Please refer <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#1/7/13" target="_blank">July 1st</a> </html>. | ||
| + | |||
| + | </div> | ||
== 7 == | == 7 == | ||
| Line 358: | Line 461: | ||
<div> | <div> | ||
*Grow ''B.subtilis 168'' for transformation | *Grow ''B.subtilis 168'' for transformation | ||
| - | * | + | *Sucrose test |
</div> | </div> | ||
| Line 365: | Line 468: | ||
<div> | <div> | ||
| - | ==== | + | ==== Results from Transformation ==== |
| - | + | '''Aim and Methods:''' | |
| - | ==== | + | Please refer <html> |
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#28/6/13" target="_blank">June 28th</a> </html>. | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | We checked the transformation plates from the <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#28/6/13" target="_blank">June 28th</a> </html> and found colonies have grown. We decided to conduct the sucrose test to check whether the cell contained actual Groningen plasmid. (see below) | ||
| + | |||
| + | ==== Sucrose test ==== | ||
| + | |||
| + | '''Aim:''' | ||
| + | |||
| + | To check whether the colonies on the Cm plate with ''B.subtilis 168'' containing Groningen 2012 integration plasmid actually contained transformant cells rather than a random mutants/contaminant. | ||
| + | |||
| + | '''Methods:''' | ||
| + | |||
| + | Transfer the culture of into LB + 1% sucrose media and leaving it to grow over night. Protocol to be continued on <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#2/7/13" target="_blank">July 2nd</a> </html>. | ||
| + | |||
| + | ''' Results and Discussion:''' | ||
| + | |||
| + | Please refer <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#2/7/13" target="_blank">July 2nd</a> </html>. | ||
| + | |||
| + | ==== Transform ''B.subtilis 168'' with Groningen 2012 plasmid and RFP==== | ||
| + | |||
| + | '''Aim:''' | ||
| + | |||
| + | In case if the transformation of Groningen plasmid into B. subtilis did not work, we decided to inoculated 5 ml of freshly prepared MM media with ''B.subtilis'' str.168 colonies to grow overnight in preparation to repeat the protocol. | ||
| + | |||
| + | '''Methods''' | ||
| + | |||
| + | A single colony of B.subtilis strain 168 was picked and transferred into MM media and left to grow at 37 ̊C overnight. We continued the protocol on <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#2/7/13" target="_blank">July 2nd</a> </html> . | ||
| + | |||
| + | ''' Results and Discussion:''' | ||
| + | |||
| + | Please refer <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#3/7/13" target="_blank">July 3rd</a> </html>. | ||
| - | |||
</div> | </div> | ||
| Line 387: | Line 527: | ||
====Sucrose Test==== | ====Sucrose Test==== | ||
| - | + | '''Aim:''' | |
| - | The results were | + | |
| + | To check whether the colonies on the Cm plate with B.subtilis 168 containing Groningen 2012 integration plasmid actually contained transformant cells rather than a random mutants/contaminant. | ||
| + | |||
| + | '''Methods:''' | ||
| + | |||
| + | Following from the protocol indicated on <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#1/7/13" target="_blank">July 1st</a> </html>, we checked the pH of the solution of cells from the Cm plate with Groningen 2012 integration plasmid incubated in sucrose rich media overnight to check for presence of acidic metabolites of sucrose and hence the activity of AmyE. | ||
| + | |||
| + | '''Results''' | ||
| + | |||
| + | The results were as hoped, as the pH of both the samples we prepared was the same as the pH of the +ve control (''Bacillus 168'' cells):4.7, 4.8, 5.04. | ||
====Transformation==== | ====Transformation==== | ||
| - | We | + | '''Aim:''' |
| + | |||
| + | To transform B. subtilis with Groningen 2012 integration plasmid. | ||
| + | |||
| + | '''Methods:''' | ||
| + | |||
| + | We transformed ''B.subtilis 168'' cells with Groningen 2012 integration plasmid and with pGMP (+ve control) and no plasmid at all (-ve control). Plated the colonies out according to the protocol and left to grow over night. | ||
| + | |||
| + | '''Results and Discussion:''' | ||
| + | |||
| + | Please see <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#3/7/13" target="_blank">July 3rd</a> </html> for results and discussion. | ||
| + | |||
</div> | </div> | ||
| Line 405: | Line 567: | ||
<div> | <div> | ||
| - | ====Transformation Results==== | + | ====Groningen Plasmid Transformation Results==== |
| - | + | ||
| + | '''Aim and Methods:''' | ||
| + | |||
| + | Please refer to <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#2/7/13" target="_blank">July 2nd</a> </html> | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | No colonies were found on the chloramphenicol plates ''B.subtilis 168'' with Groningen 2012 plasmid, however there were a few colonies on the positive control, ''B.subtilis 168'' with pGMP (Figure 1) integration plasmid plates and no colonies on ''B.subtilis 168'' LB + chloramphenicol plates. | ||
| + | |||
| + | [[File:P1050607.JPG|frameless|500px]] | ||
| + | |||
| + | Figure 1: ''B. subtilis 168'' + pGMP on LB + Cm plate. Although a few colonies are found, it is not confirmed whether the pGMP insert is present. | ||
====Amylase Test==== | ====Amylase Test==== | ||
| - | + | '''Aim:''' | |
| + | |||
| + | To confirm that the pGMP plasmid was integrated in all of the colonies. | ||
| + | |||
| + | '''Methods:''' | ||
| + | |||
| + | Transfer a few colonies onto a freshly made LB + Starch agar plates. Check the amylase function using the iodine test for starch on <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#4/7/13" target="_blank">July 4th</a> </html>. | ||
</div> | </div> | ||
| + | |||
| + | '''Results and Discussion''' | ||
| + | |||
| + | Please refer to <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#4/7/13" target="_blank">July 4th</a> </html> | ||
== 7 == | == 7 == | ||
| Line 425: | Line 611: | ||
We made a few changes to our modelling presentation and practised our delivery in preparation for the presentation to members of the school of bioinformatics. | We made a few changes to our modelling presentation and practised our delivery in preparation for the presentation to members of the school of bioinformatics. | ||
| - | ==== | + | ====Amylase Test==== |
| - | The starch plates | + | |
| + | '''Aim and Methods:''' | ||
| + | |||
| + | Please refer to <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#3/7/13" target="_blank">July 3rd</a> </html> | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | The starch plates from yesterday were checked and it was found that they have overgrown. | ||
| + | |||
| + | [[File:P1050610.JPG|frameless|500px]] | ||
| + | |||
| + | Figure 1: Overgrown starch plates. | ||
| + | |||
| + | '''Discussion:''' | ||
| + | |||
| + | The experiment will be repeated. | ||
| + | |||
| + | ====Repeating Amylase Test==== | ||
| + | |||
| + | '''Aim:''' | ||
| + | |||
| + | Please refer to <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#3/7/13" target="_blank">July 3rd</a> </html> | ||
| + | |||
| + | '''Methods:''' | ||
| + | |||
| + | Starch agar plates were made including the antibiotic chloramphenicol (Cm) in the media. After these plates were made, colonies were transferred directly onto the plates and were left overnight. | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | Please refer to <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#5/7/13" target="_blank">July 5th</a> </html> | ||
| - | |||
</div> | </div> | ||
| Line 443: | Line 660: | ||
We gave a workshop practice presentation to members of the school of bioinformatics. It was decided that we shouldn't talk about the subtilin two-components system model, because it might be too complex for our workshop. | We gave a workshop practice presentation to members of the school of bioinformatics. It was decided that we shouldn't talk about the subtilin two-components system model, because it might be too complex for our workshop. | ||
It was suggested that our workshop needed to be a little less like a presentation, with more interaction for the participants. | It was suggested that our workshop needed to be a little less like a presentation, with more interaction for the participants. | ||
| + | |||
====Starch test==== | ====Starch test==== | ||
| - | |||
| - | [[File: | + | ''' Aim and Methods:''' |
| + | |||
| + | Please refer to <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#3/7/13" target="_blank">July 3rd</a> </html> | ||
| + | |||
| + | '''Results:''' | ||
| + | The starch plates were checked and a lawn of bacteria was found. The starch test was done anyway using vaporised iodine. The test showed a positive result for pGFP as it turned black (Figure 1: left plate) showing no amylase was present and there for it had transformed. The B.subtilis (positive control) did not go black because the starch was consumed by the bacteria as expected (Figure 1: white plate). Also E.coli was left with the Groningen plasmid in 10ml of LB over the weekend for purification. This was done because stocks of the Groningen plasmid were running low. | ||
| + | |||
| + | [[File:Barecillus IodineTest.jpg|400px]] | ||
| + | |||
| + | Figure 1: Results from Iodine Test. Black plate contains pGFP, white plate contains B. subtilis and crystalized plate is just iodine crystals | ||
| + | |||
| + | '''Discussion''' | ||
| + | |||
| + | From the results, we can confirm that pGFP rrnB has an unfunctional amyE and hence it is unable to digest the starch, resulting in a black plate. The B. subtilis plate is white because there is a functional amyE and therefore the starch has been digested. From this experiment, we can tell that pGFP rrnB has been integrated into the correct region while there is no insert in that region in the B.subtilis plate. | ||
| + | |||
</div> | </div> | ||
| Line 460: | Line 692: | ||
====SGM tutorial==== | ====SGM tutorial==== | ||
| - | We | + | We added a modelling section to the SGM tutorial for potential funding. Deciding what information to include was a challenge due to the time constraint. We considered some of the exercises in the workshop for the tutorial. |
| - | |||
====UCL Presentation Practice==== | ====UCL Presentation Practice==== | ||
| - | We | + | We practiced the UCL presentation to everyone on the team before everyone on the team gave feedback and constructive criticism which was recorded and addressed. |
| + | |||
| + | [[File:UCLpresentation.jpeg |frameless|400px]] | ||
==== Workshop presentation==== | ==== Workshop presentation==== | ||
| - | The slides for the workshop were edited | + | The slides for the workshop were edited according to criticism from the feedback session on the 5th. We also began producing a handout with exercises to complete during the workshop. |
| - | We also began producing a handout with exercises to complete during the workshop. | + | |
| - | |||
</div> | </div> | ||
| Line 478: | Line 709: | ||
<div> | <div> | ||
*Modelling Workshop | *Modelling Workshop | ||
| - | * | + | *Groningen plasmid Mini-prep |
| + | *''B.subtilis'' Transformation | ||
</div> | </div> | ||
==== Details ==== | ==== Details ==== | ||
| Line 484: | Line 716: | ||
====Modelling Workshop Feedback Form==== | ====Modelling Workshop Feedback Form==== | ||
We produced a feedback form for attendees of our Workshop. This will be useful to access our workshops success and to improve its content for any future occasions we deliver a modelling workshop. | We produced a feedback form for attendees of our Workshop. This will be useful to access our workshops success and to improve its content for any future occasions we deliver a modelling workshop. | ||
| - | ==== | + | ====Groningen plasmid Mini-prep==== |
| - | + | ||
| - | + | ||
| - | + | ||
| + | '''Aim and Methods:''' | ||
| + | |||
| + | Please refer to <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#24/6/13" target="_blank">June 24th</a> </html> | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | The Groningen plasmid was purified using plasmid [https://2013.igem.org/Team:Newcastle/Notebook/protocols#QIAprep_Spin_Miniprep_Kit Miniprep] of previously prepared ''E.coli''. The purity was checked using the [https://2013.igem.org/Team:Newcastle/Notebook/protocols#Nanodrop_quantification_of_DNA_extracts NanoDrop] and a result of 183.7 was obtained meaning the DNA was usable in subsequent protocols. | ||
| + | |||
| + | ====''B.subtilis'' Transformation==== | ||
| + | '''Aim:''' | ||
| + | |||
| + | To transform the bacteria with Groningen plasmid. | ||
| + | |||
| + | '''Methods:''' | ||
| + | |||
| + | A single colony of ''B.subtilis'' strain 168 was picked and transferred into [https://2013.igem.org/Team:Newcastle/Notebook/protocols#Bacillus_subtilis_168_Competent_cell_Prep_and_Transformation MM media] and left to grow at 37 ̊C overnight. We continued the protocol on | ||
| + | <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#10/7/13" target="_blank">July 10th</a> </html>. | ||
| + | |||
| + | |||
| + | </div> | ||
==7== | ==7== | ||
| Line 500: | Line 751: | ||
<div> | <div> | ||
====''B.subtilis'' transformation==== | ====''B.subtilis'' transformation==== | ||
| + | |||
| + | '''Aim:''' | ||
| + | |||
| + | To transform the bacteria with Groningen plasmid. | ||
| + | |||
| + | '''Methods:''' | ||
| + | |||
| + | Please refer to <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#9/7/13" target="_blank">July 9th</a> </html> for the first part of the protocol. | ||
| + | |||
The ''B.subtilis'' transformation was carried out again to see if it would successfully integrate the Groningen plasmid when a lower concentration of Cm antibiotic is used in the plates. The plates used include: | The ''B.subtilis'' transformation was carried out again to see if it would successfully integrate the Groningen plasmid when a lower concentration of Cm antibiotic is used in the plates. The plates used include: | ||
*''B.subtilis'' transformed with 10µl Groningen plasmid on 5µg/ml Cm, 12.5µg/ml Cm, Amp and LB | *''B.subtilis'' transformed with 10µl Groningen plasmid on 5µg/ml Cm, 12.5µg/ml Cm, Amp and LB | ||
| Line 505: | Line 766: | ||
*''B.subtilis'' transformed with 10µl pGFP (positive control) on 5µg/ml Cm, 12.5µg/ml Cm | *''B.subtilis'' transformed with 10µl pGFP (positive control) on 5µg/ml Cm, 12.5µg/ml Cm | ||
*''B.subtilis'' with water (negative control) on 5µg/ml Cm, 12.5µg/ml Cm and Amp | *''B.subtilis'' with water (negative control) on 5µg/ml Cm, 12.5µg/ml Cm and Amp | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | Please refer to <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#11/7/13" target="_blank">July 11th</a> </html>. | ||
====Receiving ''B.subtilis'' L-Form==== | ====Receiving ''B.subtilis'' L-Form==== | ||
| Line 516: | Line 782: | ||
==== Titles ==== | ==== Titles ==== | ||
<div> | <div> | ||
| - | * | + | *''B.subtilis'' transformation |
| + | *Sucrose test | ||
*Chromosomal Extraction | *Chromosomal Extraction | ||
</div> | </div> | ||
==== Details ==== | ==== Details ==== | ||
<div> | <div> | ||
| - | ==== | + | ====''B.subtilis'' transformation==== |
| - | + | ||
| - | + | '''Aim and Methods:''' | |
| + | |||
| + | Please refer to <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#9/7/13" target="_blank">July 9th</a> </html> and <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#10/7/13" target="_blank">July 10th</a> </html>. | ||
| + | |||
| + | '''Results:''' | ||
| + | The following are pictures of the plates: | ||
| + | |||
| + | [[File:P1050613.JPG |frameless|500px]] | ||
| + | |||
| + | Figure 1: ''B.subtilis'' transformed with 10µl Groningen plasmid on 5µg/ml Cm, 12.5µg/ml Cm, Amp and LB | ||
| + | |||
| + | [[File:P1050615.JPG |frameless|500px]] | ||
| + | |||
| + | Figure 2: ''B.subtilis'' transformed with 10µl pGFP (positive control) on 5µg/ml Cm, 12.5µg/ml Cm | ||
| + | |||
| + | ====Sucrose test==== | ||
| + | |||
| + | '''Aim:''' | ||
| + | |||
| + | To check whether the colonies on the Cm plate with ''B.subtilis 168'' containing Groningen 2012 integration plasmid actually contained transformant cells rather than a random mutants/contaminant. | ||
| + | |||
| + | '''Methods:''' | ||
| + | |||
| + | Both sucrose and glucose agar was made, left to cool, plated and left to set. Then the culture was transfered into LB + 1% sucrose media and leaving it to grow over night. | ||
| + | |||
| + | ''' Results and Discussion:''' | ||
| + | |||
| + | Please refer <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#16/7/13" target="_blank">July 16th</a> </html>. | ||
| + | |||
====Extracting Chromosome from the ''B. subtilis'' received==== | ====Extracting Chromosome from the ''B. subtilis'' received==== | ||
| Line 540: | Line 838: | ||
==== YSB 1.0 Conference ==== | ==== YSB 1.0 Conference ==== | ||
| - | [[File:Newcastle2013ysbgroup.JPG | | + | [[File:Newcastle2013ysbgroup.JPG |500px| link=https://2013.igem.org/Team:Newcastle/Outreach/uk_meet|alt=YSB meetup]] |
| - | The team travelled down to London for the Young Synthetic Biologists conference with the other UK iGEM teams. On arrival at the conference, the team put up their poster for display and proceeded to listen to other teams giving presentations about their projects. After lunch Yana, Geoff, Justas and James gave a modelling workshop on [https://2013.igem.org/Team:Newcastle/Outreach/Workshop Rule Based Modelling with BioNetGen]. During this time other team members attended various workshops about BioArt and public engagement. Discussions were ongoing throughout the day regarding teams posters and our own project. | + | The team travelled down to London for the Young Synthetic Biologists (YSB1.0) conference with the other UK iGEM teams. On arrival at the conference, the team put up their poster for display and proceeded to listen to other teams giving presentations about their projects. After lunch Yana, Geoff, Justas and James gave a modelling workshop on [https://2013.igem.org/Team:Newcastle/Outreach/Workshop Rule Based Modelling with BioNetGen]. During this time other team members attended various workshops about BioArt and public engagement. Discussions were ongoing throughout the day regarding teams posters and our own project. |
More details can be found on the [https://2013.igem.org/Team:Newcastle/Outreach/uk_meet YSB1.0 page]. | More details can be found on the [https://2013.igem.org/Team:Newcastle/Outreach/uk_meet YSB1.0 page]. | ||
| Line 558: | Line 856: | ||
==== YSB 1.0 Conference ==== | ==== YSB 1.0 Conference ==== | ||
<div style="text-align:center;"> | <div style="text-align:center;"> | ||
| - | [[File:BareCillus_UCL_Presentation_1.jpg | left | | + | [[File:BareCillus_UCL_Presentation_1.jpg | left | 400px]] |
| - | [[File:BareCillus_UCL_Presentation_2.jpg | left | | + | [[File:BareCillus_UCL_Presentation_2.jpg | left | 400px]] |
</div> | </div> | ||
<div class="clear"></div> | <div class="clear"></div> | ||
| - | On arrival at the second day of the conference the team had the opportunity to view the posters and projects of other teams. A 10 minute [https://2013.igem.org/Team:Newcastle/Outreach/uk_meet presentation] about our project was also given, followed by 5 minutes of questions. The presentation was very well received, resulting in many interesting questions and generating a lot of interest | + | On arrival at the second day of the conference the team had the opportunity to view the posters and projects of other teams. A 10 minute [https://2013.igem.org/Team:Newcastle/Outreach/uk_meet presentation] about our project was also given, followed by 5 minutes of questions. The presentation was very well received, resulting in many interesting questions and generating a lot of interest about the project. The entire conference split into groups after the presentations to discuss topics including modelling, wiki design, parts characterisation, human practices and SynBioSoc. |
More details can be found on the [https://2013.igem.org/Team:Newcastle/Outreach/uk_meet YSB1.0 page]. | More details can be found on the [https://2013.igem.org/Team:Newcastle/Outreach/uk_meet YSB1.0 page]. | ||
| Line 578: | Line 876: | ||
====Switch Brick Sequence Analysis==== | ====Switch Brick Sequence Analysis==== | ||
| - | We looked through the chromosomal sequence of the L-form ''B.subtilis'' we acquired from Dr. Ling Juan Wu, and | + | We looked through the chromosomal sequence of the L-form ''B.subtilis'' we acquired from Dr. Ling Juan Wu, and located the region (ranging from the pbpb up to the murE gene) and sequence of the Switch Brick. Inside the strain that we received, the Pxyl and the Chloramphenicol resistance gene were already present in the region so it saved us a bit of work as we didn't have to clone it in. However, the sequence contains multiple illegal restriction sites for the BioBrick (EcoRI and PstI). To fix this issue, we decided to make a to synonymous change (no AA change) to the sequence where the restriction sites are located. The Switch Brick sequence is then sent off to DNA 2.0 for synthesis. Whilst waiting for the synthesised BioBrick to arrive, we will be using the current Switch BioBrick which contain EcoRI and PstI for our experiments. |
====Primer design for switch BioBrick==== | ====Primer design for switch BioBrick==== | ||
| Line 586: | Line 884: | ||
The primers were as follows: | The primers were as follows: | ||
| - | BBa_K1185000_1_Forward Primer | + | BBa_K1185000_1_Forward Primer (20bp + XbaI tag + 'AAA') |
| - | + | 5’ AAATTCTAGAGCAAAAGAAAGCAAAAGAGGA 3’ | |
| + | |||
| + | BBa_K1185000_1_Reverse Primer (19bp + SpeI tag + 'AAA') | ||
| + | |||
| + | 5’ AAATACTAGTATTGTCCCTGTTATCCCAAT 3’ | ||
</div> | </div> | ||
| Line 597: | Line 899: | ||
<div> | <div> | ||
*Making Plant Agar Plates | *Making Plant Agar Plates | ||
| - | *Sucrose test | + | *Sucrose test |
</div> | </div> | ||
==== Details ==== | ==== Details ==== | ||
| Line 603: | Line 905: | ||
====Making Agar Plates for our Plants==== | ====Making Agar Plates for our Plants==== | ||
| - | + | '''Aim:''' | |
| - | After the solutions | + | |
| + | Make some M&S Agar Plates for growing plants. | ||
| + | |||
| + | '''Methods:''' | ||
| + | |||
| + | Some Murashige and Skoog Medium (M&S Medium) was made. This medium was ordered in powder form and water had to be added to it to be made into a solution. The solutions were then autoclaved to be used to make the agar for growing plants. | ||
| + | |||
| + | After the solutions have been autoclaved, we made some agar with the M&S Medium. We weighed out 3.0g of agar no.2 into 2x500mL bottles. 250mL of M&S Medium was then added and mixed. The agar was then autoclaved and then cooled at 50 ̊C. When it is ready to be poured, we poured the agar onto 25 plates and left it to set. | ||
[[File:BareCillus MandS.jpg | 300px]] | [[File:BareCillus MandS.jpg | 300px]] | ||
| - | ====Sucrose test | + | |
| - | + | ====Sucrose test==== | |
| + | |||
| + | '''Aim and Methods:''' | ||
| + | |||
| + | Please refer to <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#11/7/13" target="_blank">July 11th</a> </html>. | ||
| + | |||
| + | '''Results and Discussion:''' | ||
| + | |||
| + | Inconclusive results were found as the agar was cloudy and individual colonies were not clear. For this reason a sucrose test was set up to see if the cells containing the Groningen plasmid from the plates had integrated the plasmid, consequently interrupting the sacA gene and so failing to metabolise sucrose and showing as more basic. For a positive control we used ''B.subtilis'' and sucrose (this was expected to be acidic) and our negative control was LB and sucrose (this was expected to be more basic as no metabolism). These cultures were left overnight. To see results please refer to <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#17/7/13" target="_blank">July 17th</a> </html>. | ||
| + | |||
</div> | </div> | ||
| Line 618: | Line 938: | ||
<div> | <div> | ||
*Sucrose test | *Sucrose test | ||
| - | * | + | *Transforming ''B.subtilis 168'' |
*Planting Seeds | *Planting Seeds | ||
*Shape-shifting modelling | *Shape-shifting modelling | ||
| Line 625: | Line 945: | ||
<div> | <div> | ||
====Sucrose test==== | ====Sucrose test==== | ||
| - | The sucrose test was carried out on the | + | |
| + | '''Aim and Method: | ||
| + | Please refer to <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#11/7/13" target="_blank">July 11th</a> </html>. | ||
| + | |||
| + | '''Results and Discussion:''' | ||
| + | The sucrose test was carried out on the cultures using a PH meter. The results observed were as follows: | ||
*LB + sucrose + ''B.subtilis'' = 5.15 | *LB + sucrose + ''B.subtilis'' = 5.15 | ||
| Line 634: | Line 960: | ||
These results show that cells with the Groningen plasmid had functional sucrase showing the plasmid has not been integrated so colonies must have been contaminants. | These results show that cells with the Groningen plasmid had functional sucrase showing the plasmid has not been integrated so colonies must have been contaminants. | ||
| - | |||
| - | |||
| - | ==== Planting Seeds ==== | + | ====Transforming ''B.subtilis 168''==== |
| + | |||
| + | '''Aim:''' | ||
| + | |||
| + | To transform B. subtilis with Groningen 2012 integration plasmid. | ||
| + | |||
| + | '''Methods:''' | ||
| + | |||
| + | Preparation for ''B.subtilis'' transformation was carried out; 5 ml of freshly prepared MM media was inoculated with ''B.subtilis'' 168 colonies to grow overnight. Also E.coli with Groningen plasmid was put in LB media and left to grow overnight. Protocol continued on <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#18/7/13" target="_blank">July 18th</a> </html>. | ||
| + | |||
| + | '''Results and Discussion:''' | ||
| + | |||
| + | Please see <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#19/7/13" target="_blank">July 19th</a> </html> for results and discussion. | ||
| + | |||
| + | |||
| + | ==== Planting Chinese Cabbage Seeds ==== | ||
| + | |||
| + | '''Aim:''' | ||
| + | |||
| + | Plant some Chinese Cabbage seeds to prepare for washing with L-forms. | ||
| + | |||
| + | '''Methods:''' | ||
| + | |||
| + | We washed and planted cabbage and french dwarf bean seeds in preparation for the introduction of L-forms into seedlings following the [https://2013.igem.org/Team:Newcastle/Notebook/protocols#Production_of_L-form_containing_Plants Production of L-form containing Plants protocol] up to and including step four. The seeds were planted in Murishage and Skoog containing petri dishes and placed in a dark cupboard to germinate. | ||
| - | + | The washing of plant seeds with L-forms will be done on <html> | |
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#18/7/13" target="_blank">July 18th</a> </html>. | ||
[[File:Bare_Cillus_seedz.jpg | 400px]] | [[File:Bare_Cillus_seedz.jpg | 400px]] | ||
| Line 667: | Line 1,017: | ||
==== Innoculating Plants with L-forms ==== | ==== Innoculating Plants with L-forms ==== | ||
| - | + | '''Aim and Methods:''' | |
| + | |||
| + | The seeds that were planted on <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#17/7/13" target="_blank">July 17th</a> </html> were checked and it was observed that the cabbage seeds had begun to germinate. This meant that they were ready to absorb L-forms. We soaked 5 cabbage seeds for 3 hours in 5ml of L-form solution and another 5 seeds in 5%(w/v) mannitol solution to act as a control. The seeds were then washed with distilled water (to lyse extracellular L-forms) and replated on Murishage and Skoog agar plates. This was done following the [https://2013.igem.org/Team:Newcastle/Notebook/protocols#Production_of_L-form_containing_Plants Production of L-form containing Plants protocol] from stage five onwards. | ||
[[File:BareCillusL-forms n seeds.jpg|200px]] | [[File:BareCillusL-forms n seeds.jpg|200px]] | ||
====Groningen Plasmid Mini-prep==== | ====Groningen Plasmid Mini-prep==== | ||
| - | We performed a mini prep of the Groningen 2012 integration plasmid DNA grown in E.coli overnight at 37 | + | |
| + | '''Aim:''' | ||
| + | |||
| + | To isolate the groningen plasmid. | ||
| + | |||
| + | '''Methods:''' | ||
| + | |||
| + | We performed a [https://2013.igem.org/Team:Newcastle/Notebook/protocols#QIAprep_Spin_Miniprep_Kit mini prep] of the Groningen 2012 integration plasmid DNA grown in ''E.coli'' overnight at 37 ̊C. The purity was tested using [https://2013.igem.org/Team:Newcastle/Notebook/protocols#Nanodrop_quantification_of_DNA_extracts NanoDrop]. We confirmed the insert size by first cut with fast digest XbaI and BamHI and then running the product on a gel to confirm the presence of the plasmid in the purified pDNA. | ||
| + | |||
| + | '''Results''' | ||
| + | |||
| + | This was done in 8 micro centrifuge tubes. The purity of each was tested using a NanoDrop.The average purity of the 8 tubes was 324ng/µl. | ||
====''B.subtilis'' Transformation==== | ====''B.subtilis'' Transformation==== | ||
| - | The B.subtilis transformation was carried out again to see if it would successfully integrate the Groningen plasmid when a higher concentration of the plasmid was used. The plates used include: | + | |
| + | '''Aim:''' | ||
| + | |||
| + | To transform B. subtilis with Groningen 2012 integration plasmid. | ||
| + | |||
| + | '''Methods:''' | ||
| + | |||
| + | Please refer to <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#17/7/13" target="_blank">July 17th</a> </html> for the first part of the protocol. The B.subtilis transformation was carried out again to see if it would successfully integrate the Groningen plasmid when a higher concentration of the plasmid was used. The plates used include: | ||
*pGFP (positive control)on 5µg/ml Cm and 12.5µg/ml Cm | *pGFP (positive control)on 5µg/ml Cm and 12.5µg/ml Cm | ||
| Line 686: | Line 1,058: | ||
*Water (negative control) on 5µg/ml Cm and 12.5µg/ml Cm | *Water (negative control) on 5µg/ml Cm and 12.5µg/ml Cm | ||
| + | |||
| + | '''Results and Discussion:''' | ||
| + | |||
| + | Please see <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#19/7/13" target="_blank">July 19th</a> </html> for results and discussion. | ||
| + | |||
| + | |||
</div> | </div> | ||
| Line 692: | Line 1,071: | ||
==== Titles ==== | ==== Titles ==== | ||
<div> | <div> | ||
| - | * | + | *''B.subtilis'' Transformation |
*Modelling | *Modelling | ||
</div> | </div> | ||
==== Details ==== | ==== Details ==== | ||
<div> | <div> | ||
| - | ====Transformation | + | ====''B.subtilis'' Transformation==== |
| + | |||
| + | '''Aim and Methods:''' | ||
| + | Please refer to <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#17/7/13" target="_blank">July 17th</a> </html>. | ||
| + | |||
| + | '''Results and Discussion:''' | ||
The results of the last transformation of ''B.subtilis'' with Groningen 2012 integration plasmid look promising. Everything has worked as expected. The insignificant difference in the number of colonies grown on plate with cells inoculated with different amount of plasmid indicates that the transformation occurred as efficiently as it could at around 9ug of DNA per reaction tube. The reason why nothing grew on the plates with 12.5ug/ml of chloramphenicol may be the inefficiency of the promoter used. The colony counts are recorded in a table below. | The results of the last transformation of ''B.subtilis'' with Groningen 2012 integration plasmid look promising. Everything has worked as expected. The insignificant difference in the number of colonies grown on plate with cells inoculated with different amount of plasmid indicates that the transformation occurred as efficiently as it could at around 9ug of DNA per reaction tube. The reason why nothing grew on the plates with 12.5ug/ml of chloramphenicol may be the inefficiency of the promoter used. The colony counts are recorded in a table below. | ||
| Line 755: | Line 1,140: | ||
====First trial on creating ''B.subtilis'' L-form==== | ====First trial on creating ''B.subtilis'' L-form==== | ||
| - | + | ||
| + | '''Aim:''' | ||
| + | |||
| + | To create L-forms. | ||
| + | |||
| + | '''Methods:''' | ||
| + | |||
| + | We used the plate of B.subtilis rod cells (containg the switch BioBrick) that [https://2013.igem.org/Team:Newcastle/Team Dr. Ling Juan Wu] provided us on the 15th July. This was carried out using both; [https://2013.igem.org/Team:Newcastle/Notebook/protocols#Creating_L-forms Direct Method] and [https://2013.igem.org/Team:Newcastle/Notebook/protocols#Protoplasting_to_generate_L-form Protoplasting to generate L-form]. | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | Both protocols were successful in creating L-forms. | ||
[[File:IMG-20130727-WA0000.jpg|200px]] | [[File:IMG-20130727-WA0000.jpg|200px]] | ||
| Line 786: | Line 1,182: | ||
[[File:L-form (growth curve).jpg|200px]] | [[File:L-form (growth curve).jpg|200px]] | ||
| + | |||
====Sucrose and Glucose test==== | ====Sucrose and Glucose test==== | ||
| - | The agar was melted on | + | '''Aim:''' |
| + | |||
| + | To check whether the colonies on the Cm plate with B.subtilis 168 containing Groningen 2012 integration plasmid actually contained transformed cells rather than a random mutants/contaminant. | ||
| + | |||
| + | '''Methods:''' | ||
| + | |||
| + | The agar was melted on heat plates for half an hour, left to cool, plated and left to set in the fume cupboard. Then the colonies from the transformation plates were plated out onto the agar. Both ''B.subtilis'' with the Groningen plasmid at concentrations of 30, 45 and 60 µl and ''B.subtilis'' alone (negative control) were put onto both sucrose and glucose plates. | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | Please refer to <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#23/7/13" target="_blank">July 23th</a> </html>. | ||
| + | |||
====Improving the poster design==== | ====Improving the poster design==== | ||
| Line 806: | Line 1,215: | ||
<div> | <div> | ||
====Sucrose and Glucose plates==== | ====Sucrose and Glucose plates==== | ||
| + | |||
| + | '''Aim and Methods:''' | ||
| + | |||
| + | Please refer to <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#23/7/13" target="_blank">July 23th</a> </html>. | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
Both the Sucrose and Glucose plates overgrew which means that the Groningen plasmid is not integrated. our next step is to consider whether to sequence the plasmid using the BioBrick primers. | Both the Sucrose and Glucose plates overgrew which means that the Groningen plasmid is not integrated. our next step is to consider whether to sequence the plasmid using the BioBrick primers. | ||
| - | [[File:24th.jpg| | + | [[File:24th.jpg|500px]] |
| + | |||
====Poster Design==== | ====Poster Design==== | ||
Today, the layout was developed further and shown to the group. The next steps would be to introduce the text, logos and illustrations. | Today, the layout was developed further and shown to the group. The next steps would be to introduce the text, logos and illustrations. | ||
| - | [[File:New poster layout.jpg| | + | [[File:New poster layout.jpg|500px]] |
====BioGame==== | ====BioGame==== | ||
A loading bar has now been added for the BioGame, providing the user graphical representation of how long they will be waiting for the game to load. This then opens into the main game screen where your Bacterium can be seen. The next stage is to add the User Interface allowing the user to "take care" of their Bacterium and install BioBricks. | A loading bar has now been added for the BioGame, providing the user graphical representation of how long they will be waiting for the game to load. This then opens into the main game screen where your Bacterium can be seen. The next stage is to add the User Interface allowing the user to "take care" of their Bacterium and install BioBricks. | ||
| Line 828: | Line 1,246: | ||
====PCR Switch BioBrick==== | ====PCR Switch BioBrick==== | ||
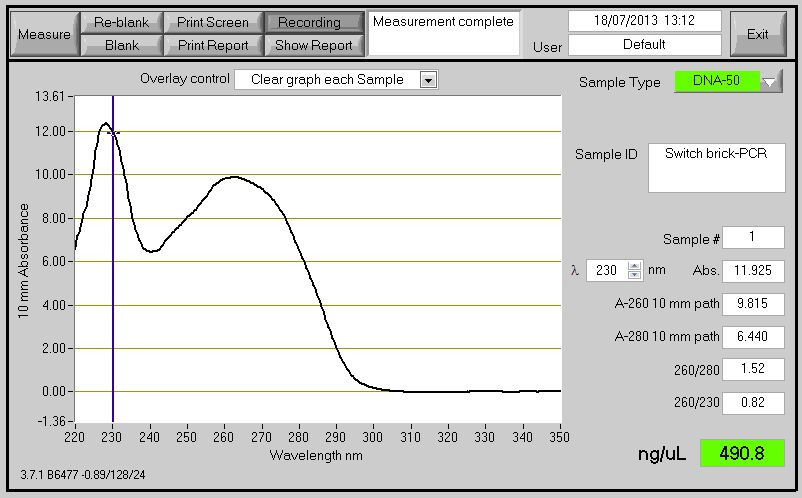
| - | The Primers ordered to clone the Switch BioBrick arrived. Using the chromosome extracted from the B. subtilis which contain the Switch BioBrick we perform PCR to amplify the BioBrick. The spectophotometer reading for the PCR product showed good results, however when running it on agarose gel to check for the size it didn't show (expected product around 2,666bp). | + | The Primers ordered to clone the Switch BioBrick arrived. Using the chromosome extracted from the ''B. subtilis'' which contain the Switch BioBrick we perform PCR to amplify the BioBrick. The spectophotometer reading for the PCR product showed good results (Figure 1) , however when running it on agarose gel to check for the size it didn't show (expected product around 2,666bp)(Figure 2). |
| - | [[File:Swirtch_Brick.jpg|frameless| | + | [[File:Swirtch_Brick.jpg|frameless|500px]] |
| + | |||
| + | Figure 1: DNA concentration reading by conducting Nanodrop spectrophotometry | ||
| + | |||
| + | [[File:IspA_current_version_all_can%27t_see_2.jpg|frameless||500px]] | ||
| + | |||
| + | Figure 2:Gel to check the size of the BioBrick (To label) | ||
====Planting Seeds==== | ====Planting Seeds==== | ||
| - | We washed and planted chinese cabbage seeds in preparation for the introduction of L-forms into seedlings following the [https://2013.igem.org/Team:Newcastle/Notebook/protocols#Production_of_L-form_containing_Plants Production of L-form containing Plants protocol] up to and including step four. The seeds were planted in Murishage and Skoog containing petri dishes and placed in a dark | + | '''Aim:''' |
| + | |||
| + | To plant seeds in order to wash them with L-forms and visualized by confocal microscopy. | ||
| + | |||
| + | '''Methods:''' | ||
| + | |||
| + | We washed and planted chinese cabbage seeds in preparation for the introduction of L-forms into seedlings following the [https://2013.igem.org/Team:Newcastle/Notebook/protocols#Production_of_L-form_containing_Plants Production of L-form containing Plants protocol] up to and including step four. The seeds were planted in Murishage and Skoog containing petri dishes and placed in a dark cupboard to germinate. | ||
| + | |||
| + | '''Results:''' | ||
| + | Due to contamination did not get results. Please refer to <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#26/7/13" target="_blank">July 26th</a> </html> for more detail | ||
| + | |||
</div> | </div> | ||
| Line 851: | Line 1,286: | ||
====PCR Switch Brick==== | ====PCR Switch Brick==== | ||
| - | |||
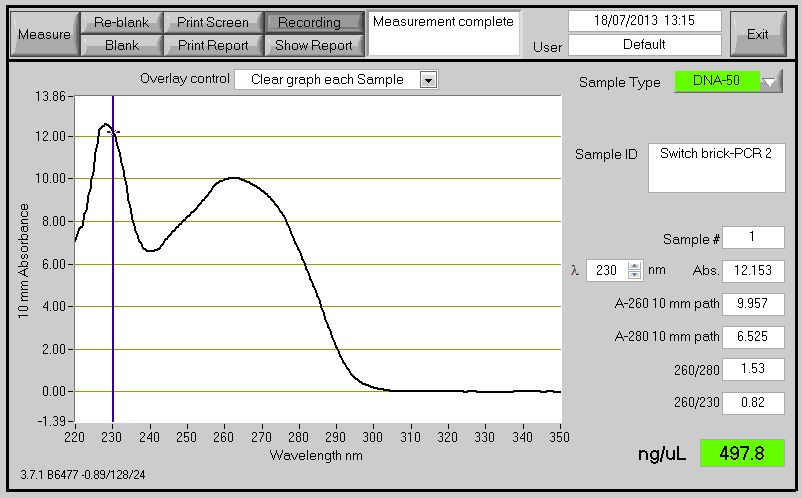
| - | [[File:Switch_brick-PCR_2.jpg| | + | '''Aim:'' |
| + | |||
| + | To confirm the size of the switch brick. | ||
| + | |||
| + | '''Methods:''' | ||
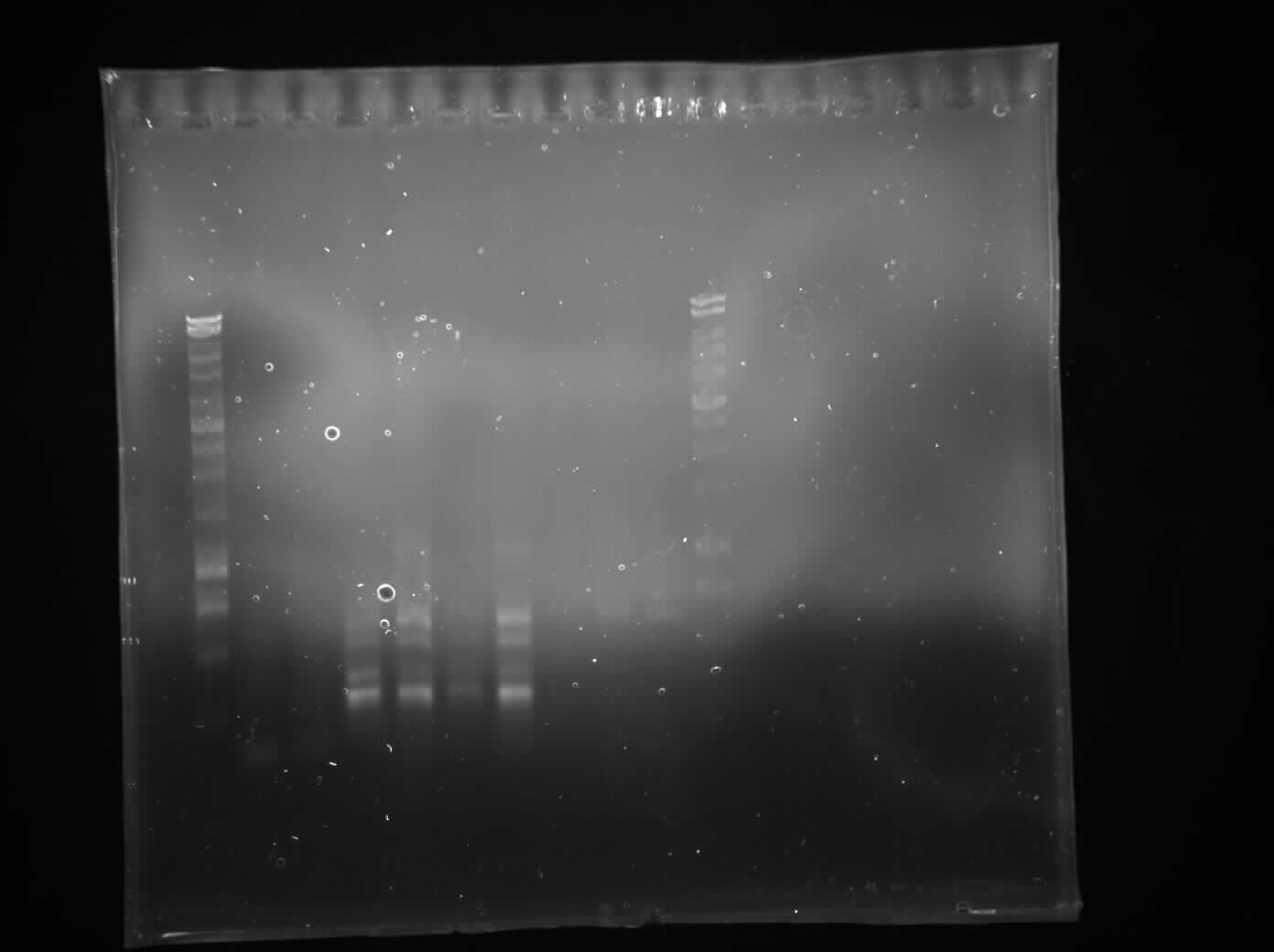
| + | |||
| + | We performed another round of PCR to amplify the Switch BioBrick from the extracted chromosome. The spec reading show good results (Figure 1), and we run an agarose gel to confirm its present. From the gel we can see a ban around 2,000-3,000bp region (Figure 2), and our BioBrick is around 2,600bp this conclude that we have successfully amplified our BioBrick. | ||
| + | |||
| + | [[File:Switch_brick-PCR_2.jpg|500px]] | ||
| + | |||
| + | Figure 1: Switch brick spectrophotometer reading with Nanodrop | ||
| + | |||
| + | [[File:IspA_its_there.jpg|500px]] | ||
| + | |||
| + | Figure 2: Gel electrophoresis to confirm size of switch brick size(To label) | ||
====Travel Arrangements==== | ====Travel Arrangements==== | ||
Research was done into the most convenient way to travel to the regional jamboree in Lyon in October. A spread sheet was made of the different travel options so a balance could be found between cost and time efficiency. This included timings, prices and duration of trains, flights and buses. | Research was done into the most convenient way to travel to the regional jamboree in Lyon in October. A spread sheet was made of the different travel options so a balance could be found between cost and time efficiency. This included timings, prices and duration of trains, flights and buses. | ||
| - | ==== | + | ====Contamination by ''Staphylococcus aureus''==== |
We checked our stocks L-form culture under the inverted microscopes and found out that all of the culture have been contaminated by ''S. aureus''. | We checked our stocks L-form culture under the inverted microscopes and found out that all of the culture have been contaminated by ''S. aureus''. | ||
====Treating Plants with L-forms==== | ====Treating Plants with L-forms==== | ||
| - | The above mentioned ''S. aureus'' contamination means we do not have any L-forms to wash our seedlings in and so | + | The above mentioned ''S. aureus'' contamination means we do not have any L-forms to wash our seedlings in and so cannot continue with the [https://2013.igem.org/Team:Newcastle/Notebook/protocols#Production_of_L-form_containing_Plants Production of L-form containing Plants protocol]. We will have to grow new seedlings once we have some L-forms free from contamination. |
</div> | </div> | ||
| Line 870: | Line 1,318: | ||
<div> | <div> | ||
*Creating L-form | *Creating L-form | ||
| + | *Transformation of ''B. subtilis'' with the switch biobrick | ||
*MM media innoculation | *MM media innoculation | ||
*Cell wall synthesis modelling | *Cell wall synthesis modelling | ||
| Line 876: | Line 1,325: | ||
<div> | <div> | ||
====Creating L-form culture==== | ====Creating L-form culture==== | ||
| - | |||
| - | ==== | + | '''Aim:''' |
| - | + | ||
| + | To make an L-form culture. | ||
| + | |||
| + | '''Methods:''' | ||
| + | |||
| + | Using the [https://2013.igem.org/Team:Newcastle/Notebook/protocols#Protoplasting_to_generate_L-form Protoplasting protocol], we converted both of the L-form strains we possess (GFP and RFP marker) into L-form. We left them to grow for 2 days in 30 ̊C incubator. | ||
| + | |||
| + | ====Transformation of ''B. subtilis'' with the switch biobrick==== | ||
| + | |||
| + | '''Aim:''' | ||
| + | |||
| + | To transform ''B.subtilis'' str.168 with the switch brick. | ||
| + | |||
| + | '''Methods:''' | ||
| + | |||
| + | Inoculate 10ml of MM medium with ''B.subtilis'' str.168 and left to grow overnight in 37 ̊C for preparation of Transformation. | ||
| + | |||
| + | The protocol to be continued on <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#30/7/13" target="_blank">July 30th</a> </html> | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#31/7/13" target="_blank">July 31st</a> </html> | ||
====Cell wall synthesis modelling==== | ====Cell wall synthesis modelling==== | ||
| Line 891: | Line 1,362: | ||
==== Titles ==== | ==== Titles ==== | ||
<div> | <div> | ||
| - | |||
*Cell Shape Model | *Cell Shape Model | ||
| + | *Transformation of ''B. subtilis'' with the switch biobrick | ||
</div> | </div> | ||
==== Details ==== | ==== Details ==== | ||
| Line 899: | Line 1,370: | ||
Substantial progress was made towards the understanding of fundamental approaches to mathematical modelling. Dr. David Swailes from the Department of Mechanical Engineering has kindly agreed to help us with our task. It was thought sensible to divide the project into three parts. First model the growth of the vesicle which is not limited by anything, then model the process of growth in a confined environment from the time just after the vesicle grows to touch the sides of the chamber, followed by a model of the transition between the two phases. Tasks for Wednesday: research more on the physiology of the vesicle growth. | Substantial progress was made towards the understanding of fundamental approaches to mathematical modelling. Dr. David Swailes from the Department of Mechanical Engineering has kindly agreed to help us with our task. It was thought sensible to divide the project into three parts. First model the growth of the vesicle which is not limited by anything, then model the process of growth in a confined environment from the time just after the vesicle grows to touch the sides of the chamber, followed by a model of the transition between the two phases. Tasks for Wednesday: research more on the physiology of the vesicle growth. | ||
| - | ====''B. subtilis'' | + | ====Transformation of ''B. subtilis'' with the switch biobrick==== |
| - | Using the overnight ''B.subtilis'' strain.168 grown in MM media we carried out the [https://2013.igem.org/Team:Newcastle/Notebook/protocols#Bacillus_subtilis_168_Competent_cell_Prep_and_Transformation transformation protocol] and transformed them with our naked PCR switch brick product and pGFPrrnb (as a control). We then plated the ''B.subtilis'' transformed with pGFPrrnb onto LB agar + Chloramphenicol (5ug/ml) plates, and plated out the B.subtilis with the switch brick onto LB agar + Chloramphenicol (5ug/ml) + Xyl 0.8% plates. | + | |
| + | '''Aim:''' | ||
| + | |||
| + | To transform ''B.subtilis'' str.168 with the switch brick. | ||
| + | |||
| + | '''Methods:''' | ||
| + | |||
| + | Please refer to <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#29/7/13" target="_blank">July 29th</a> </html> for the start of the protocol. Using the overnight ''B.subtilis'' strain.168 grown in MM media we carried out the [https://2013.igem.org/Team:Newcastle/Notebook/protocols#Bacillus_subtilis_168_Competent_cell_Prep_and_Transformation transformation protocol] and transformed them with our naked PCR switch brick product and pGFPrrnb (as a control). We then plated the ''B.subtilis'' transformed with pGFPrrnb onto LB agar + Chloramphenicol (5ug/ml) plates, and plated out the B.subtilis with the switch brick onto LB agar + Chloramphenicol (5ug/ml) + Xyl 0.8% plates. | ||
| + | |||
| + | |||
| + | '''Results:''' | ||
| + | Please refer to <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#31/7/13" target="_blank">July 31th</a> </html>. | ||
| + | |||
</div> | </div> | ||
| Line 908: | Line 1,393: | ||
==== Titles ==== | ==== Titles ==== | ||
<div> | <div> | ||
| - | * | + | *Transformation of ''B. subtilis'' with the switch biobrick |
*Visualising L-forms under Microfluidics | *Visualising L-forms under Microfluidics | ||
*Translating the cell wall model | *Translating the cell wall model | ||
| Line 915: | Line 1,400: | ||
<div> | <div> | ||
| - | ====Transformation | + | ====Transformation of ''B. subtilis'' with the switch biobrick==== |
| - | As it turns out the naked PCR product Switch BioBrick failed to be transformed, whilst the | + | |
| + | '''Aims and Methods:''' | ||
| + | |||
| + | Please refer to <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#29/7/13" target="_blank">July 29th</a> </html> and <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#30/7/13" target="_blank">July 30th</a> </html> | ||
| + | |||
| + | '''Results and Discussion:''' | ||
| + | |||
| + | As it turns out the naked PCR product Switch BioBrick failed to be transformed, whilst the positive control worked. We found out that the efficiency and probability of transformation to works with just naked PCR product is very low. So, we then planned to clone it into a plasmid first before transforming it into our ''Bacillus subtilis''. | ||
[[File:168%2BispA%3DLB%2BCm.jpg|frameless|'1000x900']] [[File:168%2Brrnb%3DLB%2BCM.jpg|frameless|'1000x900']] | [[File:168%2BispA%3DLB%2BCm.jpg|frameless|'1000x900']] [[File:168%2Brrnb%3DLB%2BCM.jpg|frameless|'1000x900']] | ||
| Line 932: | Line 1,426: | ||
<div> | <div> | ||
*Plasmid (pGFPrrnb) amplification | *Plasmid (pGFPrrnb) amplification | ||
| + | *L-Form simulation | ||
</div> | </div> | ||
==== Details ==== | ==== Details ==== | ||
<div> | <div> | ||
====Plasmid (pGFPrrnb) Amplification==== | ====Plasmid (pGFPrrnb) Amplification==== | ||
| + | |||
| + | '''Aims:''' | ||
| + | |||
| + | Amplify the pGFPrrnB. | ||
| + | |||
| + | '''Methods:''' | ||
| + | |||
We transformed the competent ''E.coli'' strain DH5alpha with plasmid pGFPrrnB taken from the spring 2013 BioBrick distribution kit using the [''E.coli'' transformation protocol]. We then innoculated ''B. subtilis'', from the yesterdays transformation plate, which contained pGFPrrnb into LB + Chloramphenicol (5ug/ml). | We transformed the competent ''E.coli'' strain DH5alpha with plasmid pGFPrrnB taken from the spring 2013 BioBrick distribution kit using the [''E.coli'' transformation protocol]. We then innoculated ''B. subtilis'', from the yesterdays transformation plate, which contained pGFPrrnb into LB + Chloramphenicol (5ug/ml). | ||
| + | |||
| + | ==== L-Form simulation ==== | ||
| + | Started working on Java based application to simulate L-form behavior. Current goal is to make something simple, maybe moving circles? | ||
</div> | </div> | ||
| Line 955: | Line 1,460: | ||
====Cloning switch brick==== | ====Cloning switch brick==== | ||
| + | |||
| + | '''Aim:''' | ||
| + | |||
| + | Cloning the switch brick into pSB1C3. | ||
| + | |||
| + | '''Methods:''' | ||
| + | |||
We extracted our pSB1C3 plasmid from the BioBrick distribution kit. Through the use of restriction enzymes (XbaI and SpeI) we cut out the RFP insert from the MCS of the plasmid. We ran the digested product on a 0.8% agarose gel and purified the DNA (plasmid backbone) through the use of [gel extraction purification]. | We extracted our pSB1C3 plasmid from the BioBrick distribution kit. Through the use of restriction enzymes (XbaI and SpeI) we cut out the RFP insert from the MCS of the plasmid. We ran the digested product on a 0.8% agarose gel and purified the DNA (plasmid backbone) through the use of [gel extraction purification]. | ||
| - | [[File:BarecillusgelelectrophoresisImage.jpeg| | + | [[File:BarecillusgelelectrophoresisImage.jpeg|300px]] |
| + | |||
| + | |||
| + | We checked the concentration of the purified DNA through nanodrop (spectrophotometer). | ||
| + | '''Results and Discussion:''' | ||
| - | + | It turned out that no DNA was present in the solution. This was due to too much elution buffer that was added to elute the DNA. This resulted in a very low concentration, and we will not be able to continue the [ligation protocol]. This protocol will need to be repeated the next day. Please refer to <html> | |
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#5/8/13" target="_blank">August 5th</a> </html> | ||
| - | [[File:BarecillusFail gel extraction pSB1C3.jpg]] | + | [[File:BarecillusFail gel extraction pSB1C3.jpg|500px]] |
</div> | </div> | ||
| Line 976: | Line 1,493: | ||
<div> | <div> | ||
====Cloning Switch Brick==== | ====Cloning Switch Brick==== | ||
| - | We started off by extracting the [http://parts.igem.org/wiki/index.php/Part:pSB1C3 pSB1C3] plasmid with RFP marker through [https://2013.igem.org/Team:Newcastle/Notebook/protocols#QIAprep_Spin_Miniprep_Kit Miniprep] from the ''E.coli'' liquid broth. We performed a double digest using XbaI and SpeI to isolate the insert from the plasmid backbone. We then ran the digested sample through gel electrophoresis to separate out the different parts. Through the use of gel extraction kit and protocol, we cut out the band of interest which was around 2000bp (the backbone). We then ligate the backbone with predigested switch brick through the use of T4 ligase via sticky end ligation in a 1:3 ratio. We left the ligated product in the 4 | + | |
| - | [[File:BareCillus Psb1c3 restrictyion purification.jpg| | + | '''Aim:''' |
| + | |||
| + | Cloning the switch brick into pSB1C3 | ||
| + | |||
| + | '''Methods:''' | ||
| + | |||
| + | We started off by extracting the [http://parts.igem.org/wiki/index.php/Part:pSB1C3 pSB1C3] plasmid with RFP marker through [https://2013.igem.org/Team:Newcastle/Notebook/protocols#QIAprep_Spin_Miniprep_Kit Miniprep] from the ''E.coli'' liquid broth. We performed a double digest using XbaI and SpeI to isolate the insert from the plasmid backbone. We then ran the digested sample through gel electrophoresis to separate out the different parts. Through the use of gel extraction kit and protocol, we cut out the band of interest which was around 2000bp (the backbone). We then ligate the backbone with predigested switch brick through the use of T4 ligase via sticky end ligation in a 1:3 ratio. We left the ligated product in the 4 ̊C fridge. | ||
| + | |||
| + | [[File:BareCillus Psb1c3 restrictyion purification.jpg|500px]] | ||
| + | |||
| + | Figure 1: Performed gel electrophoresis to confirm the size of the clone product. | ||
====Transforming [https://2013.igem.org/Team:Newcastle/Parts/HBsu-fp HBsu-(x)fp] BioBrick==== | ====Transforming [https://2013.igem.org/Team:Newcastle/Parts/HBsu-fp HBsu-(x)fp] BioBrick==== | ||
| Line 993: | Line 1,520: | ||
<div> | <div> | ||
*Amplifying Switch Brick | *Amplifying Switch Brick | ||
| - | * | + | *HBsu-(x)FP BioBrick Extraction |
*BioGame | *BioGame | ||
| + | *L-Form simulation | ||
</div> | </div> | ||
==== Details ==== | ==== Details ==== | ||
<div> | <div> | ||
====Amplifying Switch Brick==== | ====Amplifying Switch Brick==== | ||
| - | |||
| - | + | '''Aim:''' | |
| + | |||
| + | To amplify the switch brick. | ||
| + | |||
| + | '''Methods:'' | ||
| + | |||
| + | We transformed the pre-prepared competent ''E. coli'' DH5α cells with the Switch Brick that we have cloned in to the psB1C3 plasmid yesterday. These were then plated on to LB+chloramphenicol 5μm/ml and incubated at 37 ̊C; | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | Detail needed!! | ||
| + | |||
| + | ====HBsu-(x)FP BioBrick Extraction==== | ||
| + | |||
| + | '''Aim:''' | ||
| + | |||
| + | To extract the HBsu-RFP & HBsuGF BioBricks from the ''E.coli''. | ||
| + | |||
| + | |||
| + | '''Methods:''' | ||
| + | |||
We took ''E. coli'' DH5α that has been transformed with HBsu-GFP/HBsu-RFP BioBrick from the plate that was made yesterday and inoculated them onto LB to let them grow before extracting the plasmid out. | We took ''E. coli'' DH5α that has been transformed with HBsu-GFP/HBsu-RFP BioBrick from the plate that was made yesterday and inoculated them onto LB to let them grow before extracting the plasmid out. | ||
| + | |||
| + | Protocol to be continued on <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#7/8/13" target="_blank">August 7th</a> </html> | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | Please refer to <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#7/8/13" target="_blank">August 7th</a> </html> | ||
====BioGame==== | ====BioGame==== | ||
Further work has gone into developing the buttons, having now written the code for hit testing the clicks on the button. This code is a little buggy but will be improved as the program is progressed. Started to create the sub-menu that will appear when the "Install BioBrick" button is clicked. Currently this is just a blue box that will eventually contain a list of installable BioBricks. | Further work has gone into developing the buttons, having now written the code for hit testing the clicks on the button. This code is a little buggy but will be improved as the program is progressed. Started to create the sub-menu that will appear when the "Install BioBrick" button is clicked. Currently this is just a blue box that will eventually contain a list of installable BioBricks. | ||
| + | |||
| + | ==== L-Form simulation ==== | ||
| + | It's starting to look good, as a base for our L-Form simulation Ball game [http://www.ntu.edu.sg/home/ehchua/programming/java/J8a_GameIntro-BouncingBalls.html tutorial] was used. We managed to get couple balls moving around and colliding with each other. | ||
</div> | </div> | ||
| Line 1,014: | Line 1,572: | ||
<div> | <div> | ||
*HBsu-(x)FP BioBrick Extraction | *HBsu-(x)FP BioBrick Extraction | ||
| - | *Cloning HBsu-(x)FP BioBrick into | + | *Cloning HBsu-(x)FP BioBrick into pSB1C3 Plasmid |
*''B. subtilis'' str.168 Competent Cell Prep | *''B. subtilis'' str.168 Competent Cell Prep | ||
| + | *Inoculating Chinese Cabbage with L-forms | ||
*BioGame | *BioGame | ||
</div> | </div> | ||
| Line 1,021: | Line 1,580: | ||
<div> | <div> | ||
====HBsu-(x)FP BioBrick Extraction==== | ====HBsu-(x)FP BioBrick Extraction==== | ||
| - | Using the [https://2013.igem.org/Team:Newcastle/Notebook/protocols#QIAprep_Spin_Miniprep_Kit QIAgen Mini-Prep protocol] we extracted the HBsu-RFP & HBsu-GFP BioBricks from the ''E. coli''. We checked the concentration & quality of the plasmid using a [https://2013.igem.org/Team:Newcastle/Notebook/protocols#Nanodrop_quantification_of_DNA_extracts spectophotometer] | + | |
| + | '''Aims:''' | ||
| + | |||
| + | Please refer to <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#6/8/13" target="_blank">August 6th</a> </html> | ||
| + | |||
| + | '''Methods:''' | ||
| + | |||
| + | Continued protocol from <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#6/6/13" target="_blank">August 6th</a> </html> | ||
| + | |||
| + | Using the [https://2013.igem.org/Team:Newcastle/Notebook/protocols#QIAprep_Spin_Miniprep_Kit QIAgen Mini-Prep protocol] we extracted the HBsu-RFP & HBsu-GFP BioBricks from the ''E. coli''. We checked the concentration & quality of the plasmid using a [https://2013.igem.org/Team:Newcastle/Notebook/protocols#Nanodrop_quantification_of_DNA_extracts spectophotometer]. | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | The DNA concentration reading was the following | ||
| + | |||
| + | HBsu-GFP: 67.5 ng/μl | ||
| + | HBsu-RFP: 55.5 ng/μl | ||
| + | |||
| + | This is of good quality and will be used for cloning in the pSB1C3 plasmid on the same day. | ||
====Coloning HBsu-(x)FP BioBrick into psB1C3 Plasmid==== | ====Coloning HBsu-(x)FP BioBrick into psB1C3 Plasmid==== | ||
| - | + | ||
| + | '''Aim:''' | ||
| + | |||
| + | Clone the HBsu(x)FP BioBrick into pSB1C3 so it can be submitted to the repository. | ||
| + | |||
| + | '''Methods:''' | ||
| + | |||
| + | The HBsu-(x)FP BioBrick and the linearised pSB1C3 plasmid was cut using EcoRI and PstI. We then attempted to purify the BioBrick using the [https://2013.igem.org/Team:Newcastle/Notebook/protocols#QIAquick_Gel_Extraction_Protocol gel extraction protocol] | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | No bands were found after we have conducted the protocol. The protocol will be repeated on the next day. | ||
====''B. subtilis'' str.168 Competent Cell Prep==== | ====''B. subtilis'' str.168 Competent Cell Prep==== | ||
Detail needed!! | Detail needed!! | ||
| - | ==== | + | ====Inoculating Chinese Cabbage with L-forms==== |
| - | We washed and planted chinese cabbage seeds in preparation for the introduction of L-forms into seedlings following the [https://2013.igem.org/Team:Newcastle/Notebook/protocols#Production_of_L-form_containing_Plants Production of L-form containing Plants protocol] up to and including step four. The seeds were planted in Murishage and Skoog containing petri dishes and placed in a dark | + | |
| + | '''Aims:''' | ||
| + | |||
| + | To plant Chinese Cabbage seeds on petri dishes so it can germinate and can be inoculated with L-forms. | ||
| + | |||
| + | '''Methods:''' | ||
| + | |||
| + | We washed and planted chinese cabbage seeds in preparation for the introduction of L-forms into seedlings following the [https://2013.igem.org/Team:Newcastle/Notebook/protocols#Production_of_L-form_containing_Plants Production of L-form containing Plants protocol] up to and including step four. The seeds were planted in Murishage and Skoog containing petri dishes and placed in a dark cupboard to germinate. | ||
| + | |||
| + | The protocol was continued on <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#8/8/13" target="_blank">August 8th</a> </html> | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | Please refer to <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#9/8/13" target="_blank">August 9th</a> </html> | ||
====BioGame==== | ====BioGame==== | ||
| Line 1,041: | Line 1,646: | ||
<div> | <div> | ||
*Inoculating Plants with L-forms | *Inoculating Plants with L-forms | ||
| - | *B. subtilis Transformation of HBsu-(x)FP | + | *''B. subtilis'' Transformation of HBsu-(x)FP |
*Digest pMutin4 HBsu-GFP and HBsu-RFP | *Digest pMutin4 HBsu-GFP and HBsu-RFP | ||
</div> | </div> | ||
==== Details ==== | ==== Details ==== | ||
<div> | <div> | ||
| - | ====B. subtilis Transformation of HBsu-(x)FP==== | + | ====''B. subtilis'' Transformation of HBsu-(x)FP==== |
| - | We followed the [https://2013.igem.org/Team:Newcastle/Notebook/protocols#Bacillus_subtilis_168_Competent_cell_Prep_and_Transformation bacillus transformation protocol] | + | |
| + | '''Aim:''' | ||
| + | |||
| + | To transform ''B.subtilis'' with the HBsu-(x)FP biobrick into pMutin 4. | ||
| + | |||
| + | '''Methods:''' | ||
| + | |||
| + | We followed the [https://2013.igem.org/Team:Newcastle/Notebook/protocols#Bacillus_subtilis_168_Competent_cell_Prep_and_Transformation bacillus transformation protocol]. We then plated it out on LB + ery (5ug/ml) plates that were made earlier today. | ||
Plates: | Plates: | ||
| Line 1,060: | Line 1,672: | ||
Cell in LB x2 (to check viability of cells) | Cell in LB x2 (to check viability of cells) | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | Please refer to <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#9/8/13" target="_blank">August 9th</a> </html>. | ||
====Digest pMutin4 HBsu-GFP and HBsu-RFP==== | ====Digest pMutin4 HBsu-GFP and HBsu-RFP==== | ||
| - | + | ||
| + | '''Aim:''' | ||
| + | |||
| + | To digeste the pMutin4 HBsu-GFP and HBsu-RFP by fast digest from promega. | ||
| + | |||
| + | '''Methods:''' | ||
| + | |||
The reaction we ran was: | The reaction we ran was: | ||
| Line 1,087: | Line 1,710: | ||
After the digest we kept it in the -20 ̊C freezer. | After the digest we kept it in the -20 ̊C freezer. | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | Detail needed!! | ||
====Inoculating Plants with L-forms==== | ====Inoculating Plants with L-forms==== | ||
| + | |||
| + | '''Aim:''' | ||
| + | |||
| + | To plant Chinese Cabbage seeds on petri dishes so it can germinate and can be inoculated with L-forms. | ||
| + | |||
| + | '''Methods:''' | ||
| + | |||
| + | For the first part of the protocol please refer to <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#7/8/13" target="_blank">August 7th</a> </html>. | ||
We checked the seeds we planted yesterday and observed that the chinese cabbage seeds had begun to germinate. This meant that they were ready to absorb L-forms. We soaked 20 cabbage seeds for 3 hours in 5ml of L-form solution and another 20 seeds in 5%(w/v) mannitol solution to act as a control. The seeds were then washed with distilled water (to lyse extracellular L-forms) and replated on Murishage and Skoog agar plates. This was done following the [https://2013.igem.org/Team:Newcastle/Notebook/protocols#Production_of_L-form_containing_Plants Production of L-form containing Plants protocol] from stage five onwards. | We checked the seeds we planted yesterday and observed that the chinese cabbage seeds had begun to germinate. This meant that they were ready to absorb L-forms. We soaked 20 cabbage seeds for 3 hours in 5ml of L-form solution and another 20 seeds in 5%(w/v) mannitol solution to act as a control. The seeds were then washed with distilled water (to lyse extracellular L-forms) and replated on Murishage and Skoog agar plates. This was done following the [https://2013.igem.org/Team:Newcastle/Notebook/protocols#Production_of_L-form_containing_Plants Production of L-form containing Plants protocol] from stage five onwards. | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | Please refer to <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#9/8/13" target="_blank">August 9th</a> </html>. | ||
| + | |||
| + | |||
</div> | </div> | ||
| Line 1,098: | Line 1,741: | ||
<div> | <div> | ||
*L-form Protocol | *L-form Protocol | ||
| - | *Transformation | + | *''B. subtilis'' Transformation of HBsu-(x)FP |
*Plant Microscopy | *Plant Microscopy | ||
*<span style="color:red">Project Description Due</span> | *<span style="color:red">Project Description Due</span> | ||
| Line 1,108: | Line 1,751: | ||
====L-form Protocol==== | ====L-form Protocol==== | ||
We did the [https://2013.igem.org/Team:Newcastle/Notebook/protocols#Protoplasting_to_generate_L-form L-form protocol] to create more L-forms. Using this protocol, we created 4 flasks of L-forms which has HBsu-GFP tag on them. | We did the [https://2013.igem.org/Team:Newcastle/Notebook/protocols#Protoplasting_to_generate_L-form L-form protocol] to create more L-forms. Using this protocol, we created 4 flasks of L-forms which has HBsu-GFP tag on them. | ||
| - | ====Transformation==== | + | |
| + | ====''B. subtilis'' Transformation of HBsu-(x)FP==== | ||
| + | |||
| + | '''Aim and Methods:''' | ||
| + | |||
| + | Please refer to <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#8/8/13" target="_blank">August 8th</a> </html>. | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
The transformation conducted yesterday did not work after checking the plates. Only thing that grew was control in normal LB. | The transformation conducted yesterday did not work after checking the plates. Only thing that grew was control in normal LB. | ||
| + | |||
| + | [[File:LB ery 168H20 0908.jpg|frameless|300px]] | ||
| + | |||
| + | [[File:Barecillus LB ery 168rrnB 0908.jpg|frameless|300px]] | ||
| + | |||
| + | [[File:Barecillus LB ery HBsu-GFP 0908.jpg|frameless|300px]] | ||
| + | |||
| + | [[File:Barecillus LB ery HBsu-RFP 0908.jpg|frameless|300px]] | ||
====Plant microscopy==== | ====Plant microscopy==== | ||
| + | |||
| + | '''Aim and Methods:''' | ||
| + | |||
| + | Please refer to <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#7/8/13" target="_blank">August 7th</a> </html> and <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#8/8/13" target="_blank">August 8th</a> </html>. | ||
| + | |||
| + | '''Results and Discussion:''' | ||
| + | |||
Today we viewed the seedlings, that we had washed with L-forms yesterday,using bright field and fluorescence microscopy. When viewing an L-form washed seedling using fluorescence microscopy we noticed clearly defined green circles in several different areas of the plant. These were of the correct size (about 0.5um) to correspond to our GFP labelled L-forms. No such green circles were found in our control that had been washed in mannitol solution instead of L-form solution. These results are encouraging as they indicate we may have successfully inoculated plants with L-forms, although more evidence is needed. We also decided that when we next grow L-forms in plants, we should wash some seedlings in a solution of bacteria with cell walls to see if these can be taken up by plants. | Today we viewed the seedlings, that we had washed with L-forms yesterday,using bright field and fluorescence microscopy. When viewing an L-form washed seedling using fluorescence microscopy we noticed clearly defined green circles in several different areas of the plant. These were of the correct size (about 0.5um) to correspond to our GFP labelled L-forms. No such green circles were found in our control that had been washed in mannitol solution instead of L-form solution. These results are encouraging as they indicate we may have successfully inoculated plants with L-forms, although more evidence is needed. We also decided that when we next grow L-forms in plants, we should wash some seedlings in a solution of bacteria with cell walls to see if these can be taken up by plants. | ||
</div> | </div> | ||
| Line 1,119: | Line 1,788: | ||
==== Titles ==== | ==== Titles ==== | ||
<div> | <div> | ||
| - | * | + | *Transformation of HBsu-(x)FP into ''B. subtilis'' |
*L-form Growth Curve | *L-form Growth Curve | ||
| - | *Planting Seeds | + | *Planting Chinese Cabbage Seeds |
| + | *L-Form simulation | ||
</div> | </div> | ||
==== Details ==== | ==== Details ==== | ||
<div> | <div> | ||
| - | ==== | + | ====Transformation of HBsu-(x)FP into ''B. subtilis''==== |
| - | Prepared [https://2013.igem.org/Team:Newcastle/Notebook/protocols#Bacillus_subtilis_168_Competent_cell_Prep_and_Transformation competent cells] by putting B. subtilis 168 in 10ml MM Media and left it to grow overnight | + | |
| + | '''Aim:''' | ||
| + | |||
| + | To transform ''B. subtilis'' with HBsu-(x)FP. | ||
| + | |||
| + | '''Methods:''' | ||
| + | |||
| + | Prepared [https://2013.igem.org/Team:Newcastle/Notebook/protocols#Bacillus_subtilis_168_Competent_cell_Prep_and_Transformation competent cells] by putting ''B. subtilis'' 168 in 10ml MM Media and left it to grow overnight. For the next part of the protocol please refer to <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#13/8/13" target="_blank">August 13th</a> </html>. | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | Please refer to <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#13/8/13" target="_blank">August 13th</a> </html>. | ||
====L-form Growth Curve==== | ====L-form Growth Curve==== | ||
| - | |||
| - | ====Planting Seeds==== | + | '''Aim:''' |
| - | We washed and planted chinese cabbage seeds in preparation for the introduction of L-forms into seedlings following the [https://2013.igem.org/Team:Newcastle/Notebook/protocols#Production_of_L-form_containing_Plants Production of L-form containing Plants protocol] up to and including step four. The seeds were planted in Murishage and Skoog containing petri dishes and placed in a dark cuboard to germinate. | + | |
| + | To establish a growth curve for L-forms. | ||
| + | |||
| + | '''Methods:''' | ||
| + | |||
| + | We started doing the L-form growth curve. Initially we took OD reading of the L-form we grew over the weekend with a spectrophotometer. | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | The OD obtained was 0.246. We then diluted the solution down to get an OD of 0.05. That solution was then used to create the L-form growth curve. The initial readings of the two L-form samples were 0.05 and 0.046. | ||
| + | |||
| + | ====Planting Chinese Cabbage Seeds==== | ||
| + | |||
| + | '''Aim:''' | ||
| + | |||
| + | To plant seeds in order to wash them with L-forms and visualize them by confocal microscopy. | ||
| + | |||
| + | '''Methods:''' | ||
| + | |||
| + | We washed and planted chinese cabbage seeds in preparation for the introduction of L-forms into seedlings following the [https://2013.igem.org/Team:Newcastle/Notebook/protocols#Production_of_L-form_containing_Plants Production of L-form containing Plants protocol] up to and including step four. The seeds were planted in Murishage and Skoog containing petri dishes and placed in a dark cuboard to germinate. The washing part of the protocol will be done on <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#13/8/13" target="_blank">August 13th</a> </html>. | ||
| + | |||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | Please refer to <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#14/8/13" target="_blank">August 14th</a> </html>. | ||
| + | |||
| + | ==== L-Form simulation ==== | ||
| + | In last couple days we tried adding fusion, which was kind of hard to do, but in the end we succeeded doing that(although it still has couple of bugs, we will fix them asap). | ||
</div> | </div> | ||
| Line 1,139: | Line 1,850: | ||
==== Titles ==== | ==== Titles ==== | ||
<div> | <div> | ||
| - | *B. subtilis | + | *Transformation of HBsu-(x)FP into ''B. subtilis'' |
| - | * | + | *Cloning HBsu-GFP and HBsu-RFP into pSB1C3 |
| - | *Inoculating | + | *Inoculating L-forms into Chinese Cabbage |
| + | *L-form Growth Curve | ||
*Anything | *Anything | ||
</div> | </div> | ||
==== Details ==== | ==== Details ==== | ||
<div> | <div> | ||
| - | ====B. subtilis | + | ====Transformation of HBsu-(x)FP into ''B. subtilis''==== |
| - | We followed the [https://2013.igem.org/Team:Newcastle/Notebook/protocols#Bacillus_subtilis_168_Competent_cell_Prep_and_Transformation B. subtilis transformation protocol]. | + | |
| + | '''Aim:''' | ||
| + | |||
| + | To transform ''B. subtilis'' with HBsu-(x)FP. | ||
| + | |||
| + | '''Methods:''' | ||
| + | |||
| + | For the first part of the protocol please refer to <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#12/8/13" target="_blank">August 12th</a> </html>. We followed the [https://2013.igem.org/Team:Newcastle/Notebook/protocols#Bacillus_subtilis_168_Competent_cell_Prep_and_Transformation B. subtilis transformation protocol]. | ||
Plates: | Plates: | ||
| Line 1,162: | Line 1,882: | ||
Cell in LB x2 (to check viability of cells) | Cell in LB x2 (to check viability of cells) | ||
| + | |||
| + | [[File:Barecillus LB ery HBsu-GFP 1308.jpg|frameless|400px]] | ||
| + | |||
| + | Figure 1: HBsu-GFP on LB + ery (5ug/ml) plates | ||
| + | |||
| + | [[File:Barecillus LB ery HBsu-RFP 1308.jpg|frameless|400px]] | ||
| + | |||
| + | Figure 2: HBsu-RFP on LB + ery (5ug/ml) plates | ||
| + | |||
| + | [[File:Barecillus LB ery 168rrnB 1308.jpg|frameless|400px]] | ||
| + | |||
| + | Figure 3: rrnB on LB + ery (5ug/ml) plates x2 (positive control) | ||
| + | |||
| + | [[File:Barecillus LB ery 168H20 1308.jpg|frameless|400px]] | ||
| + | |||
| + | Figure 4: Water on LB + ery (5ug/ml) plates x2 (negative control) | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | Please refer to <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#14/8/13" target="_blank">August 14th</a> </html>. | ||
| + | |||
| + | |||
====Digest pMutin4 HBsu-GFP and HBsu-RFP==== | ====Digest pMutin4 HBsu-GFP and HBsu-RFP==== | ||
| - | + | ||
| + | '''Aim:''' | ||
| + | |||
| + | To digeste the pMutin4 HBsu-GFP and HBsu-RFP by fast digest from promega. | ||
| + | |||
| + | '''Methods:''' | ||
The reaction we ran was: | The reaction we ran was: | ||
| Line 1,182: | Line 1,930: | ||
We left this at 37 ̊C for 3 hours. | We left this at 37 ̊C for 3 hours. | ||
| - | After we linearized the plasmid, we ran a gel to confirm the size | + | After we linearized the plasmid, we ran 8ul digest reaction on a gel to confirm the size of the fragments (Figure 5). |
| - | + | '''Results and Discussion:''' | |
| - | + | ||
| - | + | ||
| + | [[File:Barecillus august22 cloning(pMutin4).jpg|frameless|800px]] | ||
| + | |||
| + | Figure 5: Gel to confirm size of HBsu-GFP and HBsu-RFP in pMutin4 | ||
| + | |||
| + | Lane 1: 10kB DNA Ladder | ||
| + | |||
| + | Lane 2-3: Empty | ||
| + | |||
| + | Lane 4: HBsu-RFP | ||
| + | |||
| + | Lane 5-6: Empty | ||
| + | |||
| + | Lane 7: Hbsu-GFP | ||
| + | |||
| + | Lane 8:Empty | ||
| + | |||
| + | Lane 9: 10kB DNA Ladder | ||
| + | |||
| + | Lane 10: Empty | ||
| + | |||
| + | Lane 11: 10kB DNA Ladder | ||
| + | |||
| + | |||
| + | After we have confirmed the size, we ran a 80ul digest and cut out the fragment of interest of HBsu-RFP and HBsu-GFP using the transilluminator and performed the [https://2013.igem.org/Team:Newcastle/Notebook/protocols#QIAquick_Gel_Extraction_Protocol gel extraction protocol]. | ||
| + | |||
| + | [[File:Barecillus august11 transilluminator(80).jpg|frameless|800px]] | ||
| + | |||
| + | Figure 6 | ||
| + | |||
| + | Lane 1: 10kB DNA Ladder | ||
| + | |||
| + | Lane 2-4: Empty | ||
| + | |||
| + | Lane 5: HBsu-RFP | ||
| + | |||
| + | Lane 6-7: Empty | ||
| + | |||
| + | Lane 8: 10kB DNA Ladder | ||
| + | |||
| + | Lane 9-11:Empty | ||
| + | |||
| + | Lane 12: Hbsu-GFP | ||
| + | |||
| + | After this was conducted we checked the nanodrop reading to find out the concentration of DNA which turned out to be for 20ng/ul HBsu-RFP and 17 ng/ul for HBsu-GFP). We then ligated them to the cut backbone that was done on the august 17th. Using the T4 ligase we ran a 1:1, 1:3, 1:5 and control for each sample. We then let it react in the thermocycler at 22 ̊C for an hour and stored at 4 ̊C overnight. | ||
| + | |||
| + | ====Inoculating L-forms into Chinese Cabbage==== | ||
| + | |||
| + | '''Aim and Methods:''' | ||
| + | |||
| + | We checked the seeds we planted on <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#12/8/13" target="_blank">August 12th</a> </html> and observed that the chinese cabbage seeds had begun to germinate. This meant that they were ready to absorb L-forms. We soaked 20 cabbage seeds for 3 hours in 5ml of L-form solution and another 20 seeds in 5%(w/v) mannitol solution to act as a control. We also created another control by soaking 20 seeds in ''B. subtilis'' rod cells. The seeds were then washed with distilled water (to lyse extracellular L-forms) and replated on Murishage and Skoog agar plates. This was done following the [https://2013.igem.org/Team:Newcastle/Notebook/protocols#Production_of_L-form_containing_Plants Production of L-form containing Plants protocol] from stage five onwards. | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | Please refer to <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#14/8/13" target="_blank">August 14th</a> </html>. | ||
| + | |||
| + | |||
| + | ====L-form Growth Curve==== | ||
| + | |||
| + | '''Aim and Methods:''' | ||
| + | |||
| + | Please refer to <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#12/8/13" target="_blank">August 12th</a> </html>. | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | An OD of 0.08 and 0.09 was obtained. | ||
| + | |||
| + | </div> | ||
== 8 == | == 8 == | ||
=== 14 === | === 14 === | ||
==== Titles ==== | ==== Titles ==== | ||
<div> | <div> | ||
| - | *Transformation | + | *Transformation of HBsu-(x)FP into ''B. subtilis'' |
*Competent Cells Preparation | *Competent Cells Preparation | ||
| + | *L-form Growth Curve | ||
*Plant Confocal Microscopy | *Plant Confocal Microscopy | ||
</div> | </div> | ||
==== Details ==== | ==== Details ==== | ||
<div> | <div> | ||
| - | ====Transformation==== | + | ====Transformation of HBsu-(x)FP into ''B. subtilis''==== |
| - | The transformation conducted on August 13th worked. We took some colonies and plated it on a [https://2013.igem.org/Team:Newcastle/Notebook/protocols#Making_Starch_Agar_Plates starch plate] we made earlier so the iodine test could be performed to confirm that HBsu-RFP and HBsu-GFP has integrated into the amyE region. | + | |
| + | '''Aim and Methods:''' | ||
| + | |||
| + | Please refer to <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#12/8/13" target="_blank">August 12th</a> </html> and <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#13/8/13" target="_blank">August 13th</a> </html>. | ||
| + | |||
| + | '''Results and Discussion:''' | ||
| + | |||
| + | The transformation conducted on August 13th worked. We took some colonies and plated it on a [https://2013.igem.org/Team:Newcastle/Notebook/protocols#Making_Starch_Agar_Plates starch plate] we made earlier so the iodine test could be performed to confirm that HBsu-RFP and HBsu-GFP has integrated into the amyE region (Figure 1-3). | ||
Plates: | Plates: | ||
| Line 1,210: | Line 2,036: | ||
pGFP-rrnB (positive control) | pGFP-rrnB (positive control) | ||
| + | |||
| + | [[File:Barecillus august14 frontview.jpg| 800px]] | ||
| + | |||
| + | Figure 1: Iodine Test (Front View of Plate 1) | ||
| + | |||
| + | [[File:Barecillus august14 backview.jpg| 800px]] | ||
| + | |||
| + | Figure 2: Iodine Test (Back View of Plate 1) | ||
| + | |||
| + | [[File:Barecillus august14 plate2.jpg| 800px]] | ||
| + | |||
| + | Figure 3: Iodine Test (Front View of Plate 2) | ||
====Competent Cells Preparation==== | ====Competent Cells Preparation==== | ||
| - | We inoculated some XL1-B in 10ml of LB overnight to use in the next day to prepare competent E.coli cells. | + | |
| + | '''Aim:''' | ||
| + | |||
| + | To make competent ''E.coli'' | ||
| + | |||
| + | '''Methods:''' | ||
| + | |||
| + | We inoculated some XL1-B in 10ml of LB overnight to use in the next day to prepare competent ''E.coli'' cells. Method continued on <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#15/8/13" target="_blank">August 15th</a> </html>. | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | Please refer to <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#15/8/13" target="_blank">August 15th</a> </html>. | ||
| + | |||
| + | ====L-form Growth Curve==== | ||
| + | |||
| + | '''Aim and Methods:''' | ||
| + | |||
| + | Please refer to <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#12/8/13" target="_blank">August 12th</a> </html>. | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | An OD of 0.1 and 0.12 was obtained | ||
====Plant Confocal microscopy==== | ====Plant Confocal microscopy==== | ||
| + | |||
| + | '''Aim and Methods:''' | ||
| + | |||
| + | Please refer to <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#12/8/13" target="_blank">August 12th</a> </html> and <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#13/8/13" target="_blank">August 13th</a> </html>. | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
Today we viewed our plants using confocal microscopy to try and detect any GFP-labelled bacteria. After viewing a few seeds washed in L-form solution under the microscope, we found a green fluorescing dot in the correct size range to be an L-form. No green dots were found in the rod cell or mannitol controls. Following these results we have decided to use confocal microscopy four days after washing our plants in L-form solution, rather than after one. As this co-insides with when [http://www.ncbi.nlm.nih.gov/pubmed/12859751 Tsomlexoglou et al.] used PCR to detect L-forms in their chinese cabbage. | Today we viewed our plants using confocal microscopy to try and detect any GFP-labelled bacteria. After viewing a few seeds washed in L-form solution under the microscope, we found a green fluorescing dot in the correct size range to be an L-form. No green dots were found in the rod cell or mannitol controls. Following these results we have decided to use confocal microscopy four days after washing our plants in L-form solution, rather than after one. As this co-insides with when [http://www.ncbi.nlm.nih.gov/pubmed/12859751 Tsomlexoglou et al.] used PCR to detect L-forms in their chinese cabbage. | ||
| Line 1,224: | Line 2,095: | ||
Figure 2. L-forms from the solution used to wash the Chinese cabbage. | Figure 2. L-forms from the solution used to wash the Chinese cabbage. | ||
| - | |||
</div> | </div> | ||
| Line 1,233: | Line 2,103: | ||
<div> | <div> | ||
*Iodine Test | *Iodine Test | ||
| - | *Making Competent E.coli | + | *Making Competent ''E.coli'' |
| - | *Preparation of IPTG reaction and B. subtilis Transformation | + | *Preparation of IPTG reaction and ''B. subtilis'' Transformation |
| + | *L-form Growth Curve | ||
</div> | </div> | ||
==== Details ==== | ==== Details ==== | ||
<div> | <div> | ||
====Iodine Test==== | ====Iodine Test==== | ||
| - | |||
| - | + | '''Aim:''' | |
| - | + | To carry out the iodine test to show... Detail needed!! | |
| - | B. Subtilis BSB1 (Negative control): Halo | + | '''Methods:''' |
| + | |||
| + | Detail needed!! | ||
| + | |||
| + | '''Results and Discussion:''' | ||
| + | |||
| + | ''B. Subtilis'' 168 (Negative control): Halo | ||
| + | |||
| + | ''B. Subtilis'' BSB1 (Negative control): Halo | ||
pGFP rrnB (Positive control): No Halo | pGFP rrnB (Positive control): No Halo | ||
| Line 1,255: | Line 2,133: | ||
When a halo is found, this shows that amyE is functional as it digests the starch around it. When a halo is not found, this means the amyE is non-functional. In this case, it means that the gene fragment has integrated into the amyE region causing it to be non-functional. | When a halo is found, this shows that amyE is functional as it digests the starch around it. When a halo is not found, this means the amyE is non-functional. In this case, it means that the gene fragment has integrated into the amyE region causing it to be non-functional. | ||
| - | ====Making Competent E.coli==== | + | ====Making Competent ''E.coli''==== |
| - | We followed the Competent E. coli protocol | + | |
| + | '''Aim:''' | ||
| + | |||
| + | To make competent ''E.coli'' for use on <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#21/8/13" target="_blank">August 21th</a> </html>. | ||
| + | |||
| + | '''Methods:''' | ||
| + | |||
| + | Please refer to <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#14/8/13" target="_blank">August 14th</a> </html> for the first part of the protocol. We followed the Competent ''E. coli'' protocol. | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | We yielded around 200 aliquots of E. coli competent cells. | ||
| + | |||
| + | ====Preparation of IPTG reaction and ''B. subtilis'' Transformation==== | ||
| + | We inoculated 2ml NB with ''B. subtilis'' 168 that is tagged with HBsu-GFP and another 2ml of NB with ''B. subtilis'' that is tagged with HBsu-RFP. This is then incubated at 37 ̊C to use for IPTG reaction. We also prepared 10ml of MM media and inoculated it with some ''B. subtilis'' 168. | ||
| + | |||
| + | ====L-form Growth Curve==== | ||
| + | |||
| + | '''Aim and Methods:''' | ||
| + | |||
| + | Please refer to <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#12/8/13" target="_blank">August 12th</a> </html>. | ||
| + | |||
| + | '''Results:''' | ||
| - | + | An OD of 0.06 and 0.07 was obtained. | |
| - | + | ||
</div> | </div> | ||
| Line 1,267: | Line 2,169: | ||
==== Titles ==== | ==== Titles ==== | ||
<div> | <div> | ||
| - | *Transformation of | + | *Transformation of Switch Brick into ''B. subtilis'' |
| + | *L-form Growth Curve | ||
| + | *L-Form simulation | ||
</div> | </div> | ||
==== Details ==== | ==== Details ==== | ||
<div> | <div> | ||
| - | ====Transformation of B. subtilis==== | + | ====Transformation of Switch Brick into ''B. subtilis''==== |
| - | Bacillus subtilis | + | |
| + | '''Aim:''' | ||
| + | |||
| + | To transform the switch biobrick into ''Bacillus subtilis'' 168 strain. | ||
| + | |||
| + | '''Methods:''' | ||
| + | |||
We followed the [https://2013.igem.org/Team:Newcastle/Notebook/protocols#Bacillus_subtilis_168_Competent_cell_Prep_and_Transformation B. subtilis transformation protocol] and plated it out on LB + Cm (5ug/ml) + Xyl (0.8%) plates that were made earlier today. | We followed the [https://2013.igem.org/Team:Newcastle/Notebook/protocols#Bacillus_subtilis_168_Competent_cell_Prep_and_Transformation B. subtilis transformation protocol] and plated it out on LB + Cm (5ug/ml) + Xyl (0.8%) plates that were made earlier today. | ||
| Line 1,281: | Line 2,191: | ||
rrnB on LB + Cm (5ug/ml)+ Xyl (0.8%) x1 (positive control) | rrnB on LB + Cm (5ug/ml)+ Xyl (0.8%) x1 (positive control) | ||
| - | 168 +Water on LB | + | ''B. subtilis'' 168 + Water on LB + Cm (5ug/ml)+ Xyl (0.8%) plates x1 (negative control) |
| - | + | ''B. subtilis'' 168 in LB x1 (to check viability of cells) | |
We left the plates to grow over the weekend. | We left the plates to grow over the weekend. | ||
| + | '''Results:''' | ||
| + | |||
| + | Please refer to <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#19/8/13" target="_blank">August 19th</a> </html>. | ||
| + | |||
| + | |||
| + | ====L-form Growth Curve==== | ||
| + | |||
| + | '''Aim and Methods:''' | ||
| + | |||
| + | Please refer to <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#12/8/13" target="_blank">August 12th</a> </html>. | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | An OD of 0.03 and 0.05 was obtained. | ||
| + | |||
| + | ==== L-Form simulation ==== | ||
| + | After fixing bugs with L-Form fusion, simulation is starting to look very good. Next step would be to add L-Form division, which might get complicated, because we will need to have additional objects which would serve as another L-Form. | ||
</div> | </div> | ||
| Line 1,294: | Line 2,223: | ||
==== Titles ==== | ==== Titles ==== | ||
<div> | <div> | ||
| + | *Transformation of Switch Brick into ''B. subtilis'' | ||
| + | *L-form Growth Curve | ||
*Lyon Presentation | *Lyon Presentation | ||
| - | *Planting Seeds | + | *Planting Chinese Cabbage Seeds |
</div> | </div> | ||
====Details==== | ====Details==== | ||
<div> | <div> | ||
| + | ====Transformation of Switch Brick into ''B. subtilis''==== | ||
| + | |||
| + | '''Aim and Methods:''' | ||
| + | |||
| + | Please refer to <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#16/8/13" target="_blank">August 16th</a> </html>. | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | We checked the transformation plate and it worked as expected. | ||
| + | |||
| + | Switch BioBrick on LB + Cm (5ug/ml)+ Xyl (0.8%) plates x1: Colonies Found | ||
| + | |||
| + | rrnB on LB + Cm (5ug/ml)+ Xyl (0.8%) x1 (positive control): Colonies Found | ||
| + | |||
| + | 168 +Water on LB + + Cm (5ug/ml)+ Xyl (0.8%) plates x1 (negative control): No Colonies Found | ||
| + | |||
| + | Cell in LB x1 (to check viability of cells): Colonies Found | ||
| + | |||
| + | This was done to confirm the characterization of our BioBrick. | ||
| + | |||
| + | [[File:Barecillus Switch brick 1908.jpg|frameless|300px]] | ||
| + | |||
| + | ====L-form Growth Curve==== | ||
| + | |||
| + | '''Aim and Methods:''' | ||
| + | |||
| + | Please refer to <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#12/8/13" target="_blank">August 12th</a> </html>. | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | An OD of 0.008 and 0.005 was obtained. | ||
| + | |||
====Lyon Presentation==== | ====Lyon Presentation==== | ||
Started working on the presentation layout for Lyon using [http://www.http://prezi.com/ Prezi]. The team feels this will enable us to create a much more visually appealing presentation, giving us not only the best chance of captivating our audience and conveying our findings, but also winning at the Jamboree. | Started working on the presentation layout for Lyon using [http://www.http://prezi.com/ Prezi]. The team feels this will enable us to create a much more visually appealing presentation, giving us not only the best chance of captivating our audience and conveying our findings, but also winning at the Jamboree. | ||
| Line 1,304: | Line 2,269: | ||
Initial design ideas have begun along the line of a microfluidics chip being used as a back-drop, with chambers used to contain relevant information (acting as "slides" as such). Along with the design ideas we have also started to compile a list of what information we want to have in the presentation, and how best to present this. | Initial design ideas have begun along the line of a microfluidics chip being used as a back-drop, with chambers used to contain relevant information (acting as "slides" as such). Along with the design ideas we have also started to compile a list of what information we want to have in the presentation, and how best to present this. | ||
| - | ====Planting Seeds==== | + | ====Planting Chinese Cabbage Seeds==== |
| - | We washed and planted chinese cabbage seeds in preparation for the introduction of L-forms into seedlings following the [https://2013.igem.org/Team:Newcastle/Notebook/protocols#Production_of_L-form_containing_Plants Production of L-form containing Plants protocol] up to and including step four. The seeds were planted in Murishage and Skoog containing petri dishes and placed in a dark | + | |
| + | '''Aim:''' | ||
| + | |||
| + | To plant seeds in order to wash them with L-forms and visualize them by confocal microscopy. | ||
| + | |||
| + | '''Methods:''' | ||
| + | |||
| + | We washed and planted chinese cabbage seeds in preparation for the introduction of L-forms into seedlings following the [https://2013.igem.org/Team:Newcastle/Notebook/protocols#Production_of_L-form_containing_Plants Production of L-form containing Plants protocol] up to and including step four. The seeds were planted in Murishage and Skoog containing petri dishes and placed in a dark cupboard to germinate. | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | Please refer to <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#23/8/13" target="_blank">August 23th</a> </html>. | ||
| + | |||
</div> | </div> | ||
| Line 1,313: | Line 2,291: | ||
<div> | <div> | ||
*Lyon Presentation | *Lyon Presentation | ||
| - | *Inoculating | + | *Inoculating L-forms into Chinese Cabbage |
| + | *L-form Growth Curve | ||
| + | *L-Form simulation | ||
</div> | </div> | ||
====Details==== | ====Details==== | ||
| Line 1,323: | Line 2,303: | ||
[[File:BareCillus_Pres1.jpg | 700px]] | [[File:BareCillus_Pres1.jpg | 700px]] | ||
| - | ====Inoculating | + | ====Inoculating L-forms into Chinese Cabbage==== |
| - | We checked the seeds we planted | + | |
| + | '''Aim and Methods:''' | ||
| + | |||
| + | We checked the seeds we planted on <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#19/8/13" target="_blank">August 19th</a> </html> and observed that the chinese cabbage seeds had begun to germinate. This meant that they were ready to absorb L-forms. We soaked 20 cabbage seeds for 3 hours in 5ml of L-form solution. We created 5%(w/v) mannitol, ''B. subtilis'' rod and ''E.coli'' rod controls. The seeds were then washed with distilled water (to lyse extracellular L-forms) and replated on Murishage and Skoog agar plates. This was done following the [https://2013.igem.org/Team:Newcastle/Notebook/protocols#Production_of_L-form_containing_Plants Production of L-form containing Plants protocol] from stage five onwards. | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | Please refer to <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#23/8/13" target="_blank">August 23th</a> </html>. | ||
| + | |||
| + | ====L-form Growth Curve==== | ||
| + | |||
| + | '''Aim and Methods:''' | ||
| + | |||
| + | Please refer to <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#12/8/13" target="_blank">August 12th</a> </html>. | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | An OD of 0.00 and 0.00 was obtained. For unknown circumstances, the L-form growth curve instead of increasing steadily throughout the week, it increased during the first 48hrs and then decreased throughout the rest of the week. We will reattempt to construct the L-form growth curve again at a later date. | ||
| + | |||
| + | ==== L-Form simulation ==== | ||
| + | Last couple days were quite busy in terms of L-Form simulation, was trying different ways to make L-Forms divide. Found a quite neat solution which required me to create another list, with empty objects (when L-Forms fuse, 1 of the previous L-Forms become redunandt), but using this method number of L-forms in simulation won't exceed initial number of balls. | ||
</div> | </div> | ||
| Line 1,331: | Line 2,334: | ||
==== Titles ==== | ==== Titles ==== | ||
<div> | <div> | ||
| + | *Characterisation of Switch Brick | ||
| + | *Cloning of HBsu-(x)fp pSB1C3 into ''E. coli'' XL1-B | ||
*Lyon Presentation | *Lyon Presentation | ||
</div> | </div> | ||
====Details==== | ====Details==== | ||
<div> | <div> | ||
| + | ====Characterization of switch brick==== | ||
| + | |||
| + | '''Aim:''' | ||
| + | |||
| + | To characterize the switch brick. | ||
| + | |||
| + | '''Methods:''' | ||
| + | |||
| + | We took some colonies that grew on <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#19/8/13" target="_blank">August 19th</a> </html> and put it in NB/MSM + xyl. We altered the xylose concentration between the range of 0-0.8% by increments of 0.2%. We then left it to grow for 12 hours before checking them under the microscope. | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | Please refer to <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#22/8/13" target="_blank">August 22nd</a> </html>. | ||
| + | |||
| + | ====Cloning of HBsu-(x)fp pSB1C3 into E.coli XL1-B==== | ||
| + | |||
| + | '''Aim:''' | ||
| + | |||
| + | To clone the ligation product that was obtained from <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#13/8/13" target="_blank">August 13th</a> </html> into competent E.coli XL1-B that was prepared on the <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#15/8/13" target="_blank">August 15th</a> </html>. | ||
| + | |||
| + | '''Methods:''' | ||
| + | |||
| + | Plates: | ||
| + | |||
| + | XL1B + 1:1 HBsu-GFP pSB1C3 on LB + Cm (5ug/ml) | ||
| + | |||
| + | XL1B + 1:3 HBsu-GFP pSB1C3 on LB + Cm (5ug/ml) | ||
| + | |||
| + | XL1B + 1:5 HBsu-GFP pSB1C3 on LB + Cm (5ug/ml) | ||
| + | |||
| + | XL1B + 1:1 HBsu-RFP pSB1C3 on LB + Cm (5ug/ml) | ||
| + | |||
| + | XL1B + 1:3 HBsu-RFP pSB1C3 on LB + Cm (5ug/ml) | ||
| + | |||
| + | XL1B + 1:5 HBsu-RFP pSB1C3 on LB + Cm (5ug/ml) | ||
| + | |||
| + | XL1B + No insert pSB1C3 on LB + Cm (5ug/ml) | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | Please refer to <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#22/8/13" target="_blank">August 22nd</a> </html>. | ||
| + | |||
====Lyon Presentation==== | ====Lyon Presentation==== | ||
The design idea created yesterday has been scrapped to further pursue the microfluidics theme less the abundance of animation & interactivity. Designs are currently being worked on. | The design idea created yesterday has been scrapped to further pursue the microfluidics theme less the abundance of animation & interactivity. Designs are currently being worked on. | ||
| Line 1,343: | Line 2,395: | ||
==== Titles ==== | ==== Titles ==== | ||
<div> | <div> | ||
| - | * | + | *Characterization of Switch Brick Results |
| - | * | + | *Cloning of HBsu-(x)fp pSB1C3 into E.coli XL1-B Results |
</div> | </div> | ||
==== Details ==== | ==== Details ==== | ||
<div> | <div> | ||
| - | + | ====Characterization of Switch Brick Results==== | |
| + | |||
| + | '''Aim and Methods:''' | ||
| + | |||
| + | Please refer to <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#21/8/13" target="_blank">August 21st</a> </html>. | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | The samples that had been growing for 12 hours were put under the microscope. | ||
| + | |||
| + | No xylose: All L-forms | ||
| + | |||
| + | 0.2 xylose: Most L-forms but a few rod cells | ||
| + | |||
| + | 0.4 xylose: Few L-forms more rod cells | ||
| + | |||
| + | 0.8 xylose: All rod cells | ||
| + | |||
| + | From this result, we can tell that our switch brick works as expected. When xylose is not present, almost all cells are L-forms. When more xylose is present, more rod cells survive. | ||
| + | |||
| + | ====Cloning of HBsu-(x)fp pSB1C3 into E.coli XL1-B Results==== | ||
| + | |||
| + | '''Aim and Methods:''' | ||
| + | |||
| + | Please refer to <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#21/8/13" target="_blank">August 21st</a> </html>. | ||
| + | |||
| + | |||
| + | '''Results and Discussion:''' | ||
| + | |||
| + | There were colonies for all of the plates except for the negative control. | ||
| + | |||
| + | [[File:Barecillus LB HBsuGFP 1,1 2108.jpg|frameless|400px]] | ||
| + | |||
| + | Figure 1: XL1B + 1:1 HBsu-GFP pSB1C3 on LB + Cm (5ug/ml) | ||
| + | |||
| + | [[File:Barecillus LB HBsuGFP 1,3 2108.jpg|frameless|400px]] | ||
| + | |||
| + | Figure 2: XL1B + 1:3 HBsu-GFP pSB1C3 on LB + Cm (5ug/ml) | ||
| + | |||
| + | [[File:Barecillus LB HBsuGFP 1,5 2108.jpg|frameless|400px]] | ||
| + | |||
| + | Figure 3: XL1B + 1:5 HBsu-GFP pSB1C3 on LB + Cm (5ug/ml) | ||
| + | |||
| + | [[File:Barecillus LB HBsuRFP 1,1 2108.jpg|frameless|400px]] | ||
| + | |||
| + | Figure 4: XL1B + 1:1 HBsu-RFP pSB1C3 on LB + Cm (5ug/ml) | ||
| + | |||
| + | [[File:Barecillus LB HBsuRFP 1,3 2108.jpg|frameless|400px]] | ||
| + | |||
| + | Figure 5: XL1B + 1:3 HBsu-RFP pSB1C3 on LB + Cm (5ug/ml) | ||
| + | |||
| + | [[File:Barecillus LB HBsuRFP 1,5 2108.jpg|frameless|400px]] | ||
| + | |||
| + | Figure 6: XL1B + 1:5 HBsu-RFP pSB1C3 on LB + Cm (5ug/ml) | ||
| + | |||
| + | [[File:Barecillus LB control 2108.jpg|frameless|400px]] | ||
| + | |||
| + | Figure 7: XL1B + No insert pSB1C3 on LB + Cm (5ug/ml) | ||
| + | |||
| + | We then inoculate one colony from each sample in to LB and let it grow overnight in order to harvest the plasmid. With the amplified plasmid we could then check that it is the correct size and confirm that cloning has worked (Figure 8). The HBsu-GFP and HBsu-RFP insert was confirmed to be the correct size of 1379bp and 1339bp respectively. However the switch brick has not been cut properly. Therefore, the protocol will be repeated. | ||
| + | |||
| + | [[File:Barecillus august22 cloning HBsu(pSB1C3).jpg|frameless|800px]] | ||
| + | |||
| + | Figure 8: Gel to Confirm Size of HBsu-GFP and HBsu-RFP insert size | ||
| + | |||
| + | Lane 1: 1kB Ladder | ||
| + | |||
| + | Lane 2: HBsu-GFP PstI | ||
| + | |||
| + | Lane 3: Hbsu-GFP EcoRI | ||
| + | |||
| + | Lane 4: HBsu-GFP PstI and EcoRI | ||
| + | |||
| + | Lane 5: HBsu-GFP Uncut | ||
| + | |||
| + | Lane 6: 1kB Ladder | ||
| + | |||
| + | Lane 7: HBsu-RFP PstI | ||
| + | |||
| + | Lane 8: Hbsu-RFP EcoRI | ||
| + | |||
| + | Lane 9: HBsu-RFP PstI and EcoRI | ||
| + | |||
| + | Lane 10: HBsu-RFP Uncut | ||
| + | |||
| + | Lane 11: 1kB Ladder | ||
| + | |||
| + | Lane 12: Switch Brick PstI | ||
| + | |||
| + | Lane 13: Switch Brick EcoRI | ||
| + | |||
| + | Lane 14: Switch Brick PstI and EcoRI | ||
| + | |||
| + | Lane 15: Switch Brick Uncut | ||
</div> | </div> | ||
| Line 1,360: | Line 2,507: | ||
==== Details ==== | ==== Details ==== | ||
<div> | <div> | ||
| + | ====Safety Form==== | ||
| + | We worked on the safety form that was due in for <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#30/8/13" target="_blank">August 30th</a> </html>. To complete the safety forms, we referred back to some COSHH forms and BioCOSHH forms of the lab for help. We also had to look up some of the bacterial strains we were using. This was to confirm the risk group and possible threats it may pose to the members of the lab, environment, general public and security through misuse. For more information on the safety form please refer to the following link. | ||
| + | |||
====Plant Confocal Microscopy==== | ====Plant Confocal Microscopy==== | ||
| + | |||
| + | '''Aim and Methods:''' | ||
| + | |||
| + | Please refer to <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#19/8/13" target="_blank">August 19th</a> </html> and <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#20/8/13" target="_blank">August 20th</a> </html> | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
We viewed 7 different Chinese cabbage seedlings washed with L-forms under the confocal microscope. In three of these plants we found clusters of green fluorescent dots about 0.5µm in diameter. We found no clusters of green fluorescent dots in three seedlings soaked in the mannitol control. | We viewed 7 different Chinese cabbage seedlings washed with L-forms under the confocal microscope. In three of these plants we found clusters of green fluorescent dots about 0.5µm in diameter. We found no clusters of green fluorescent dots in three seedlings soaked in the mannitol control. | ||
| Line 1,377: | Line 2,537: | ||
==== Titles ==== | ==== Titles ==== | ||
<div> | <div> | ||
| - | *Planting Seeds | + | *Miniprep on XL1-B cells with clone product |
| - | + | *Planting Chinese Cabbage Seeds | |
| + | |||
</div> | </div> | ||
==== Details ==== | ==== Details ==== | ||
<div> | <div> | ||
| - | ====Planting Seeds==== | + | ====Miniprep on XL1-B cells with clone product==== |
| + | |||
| + | '''Aim:''' | ||
| + | |||
| + | We conducted a miniprep on XL1-B cells containing the clone product from <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#22/8/13" target="_blank">August 22nd</a> </html>. | ||
| + | |||
| + | '''Methods:''' | ||
| + | |||
| + | *link to mini-prep protocol* | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | The spectrophotometer readings were: | ||
| + | |||
| + | HBsu-GFP pSB1C3: 80.7ng/ul | ||
| + | |||
| + | HBsu-RFP pSB1C3: 95.6ng/ul | ||
| + | |||
| + | We then digest the plasmid using EcoRI and PstI fast digest on a 10ul reaction. | ||
| + | |||
| + | 8ul of DNA | ||
| + | |||
| + | 0.5ul of PstI Fast Digest | ||
| + | |||
| + | 0.5ul of EcoRI Fast Digest | ||
| + | |||
| + | 1ul of 10x Fast Digest Buffer | ||
| + | |||
| + | Total: 10ul | ||
| + | |||
| + | We let the reaction run at 37 ̊C for 2 hours and ran the digest on a 0.7% agarose gel. After the gel has finished running, we viewed the gel under the transilluminator to check the size of the fragments. The fragments were all of the correct sizes so the cloning was successful. | ||
| + | |||
| + | ====Planting Chinese Cabbage Seeds==== | ||
| + | |||
| + | '''Aim:''' | ||
| + | |||
| + | To plant seeds in order to wash them with L-forms and visualize them by confocal microscopy. | ||
| + | |||
| + | '''Methods:''' | ||
| + | |||
We washed and planted chinese cabbage seeds in preparation for the introduction of L-forms into seedlings following the [https://2013.igem.org/Team:Newcastle/Notebook/protocols#Production_of_L-form_containing_Plants Production of L-form containing Plants protocol] up to and including step four. The seeds were planted in Murishage and Skoog containing petri dishes and placed in a dark cuboard to germinate. | We washed and planted chinese cabbage seeds in preparation for the introduction of L-forms into seedlings following the [https://2013.igem.org/Team:Newcastle/Notebook/protocols#Production_of_L-form_containing_Plants Production of L-form containing Plants protocol] up to and including step four. The seeds were planted in Murishage and Skoog containing petri dishes and placed in a dark cuboard to germinate. | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | Please refer to ?? | ||
| + | |||
</div> | </div> | ||
| Line 1,390: | Line 2,596: | ||
==== Titles ==== | ==== Titles ==== | ||
<div> | <div> | ||
| - | * | + | *L-Form simulation |
| - | * | + | *Characterisation of the HBsu-(x)fp |
| + | *Transformation of ''B. subtilis'' that contained HBsu-(x)FP with Switch Brick | ||
</div> | </div> | ||
==== Details ==== | ==== Details ==== | ||
<div> | <div> | ||
| - | + | ==== L-Form simulation ==== | |
| + | Simulation is nearly done, just giving finishing touches. One of them is improving movement of L-forms, so it's more realistic. Although it took a while to figure out the best movement algorithm, at first we tried doing it completely random, but it didn't work out quite well, so in the end we decided it to make not as random. So what we did is that let's say L-form moves 1px in x and y axis per second, so our algorithm would then generate random number from [-2;2] and then would add it up to the current speed. But, because of nature of randomness balls would move too fast or too slow, so we had to make "limits" so it doesn't get too fast or too slow. | ||
| + | |||
| + | ====Characterisation of the HBsu-(x)fp==== | ||
| + | |||
| + | '''Aim:''' | ||
| + | |||
| + | To characterize the HBsu-(x)FP BioBrick. From our cloning strategy, we planned to conduct a single cross over so the whole pMutin4 plasmid including the insert is integrated into the ''B. subtilis'' chromosomal DNA. After it has been integrated, our biobrick will be under the control of Pspac promoter. Pspac promoter is an IPTG inducible promoter. | ||
| + | |||
| + | '''Methods:''' | ||
| + | |||
| + | The ''B. subtilis'' 168 strain that has been transformed with HBsu-GFP and HBsu-RFP was inoculated on <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#15/8/13" target="_blank">August 15th</a> </html> to characterise the HBsu-(x)fp biobrick. | ||
| + | |||
| + | The concentration of IPTG was varied from a range of 0.05 – 0.8 mM to observe the conditions that yield the maximum concentration of product. We checked the fluorescence of this fusion protein at 10mins, 30mins, 60mins and 120mins after addition of IPTG to see how long it takes to get the maximum signal. | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | The results of this first trial were not very conclusive as there were cross contamination between samples with IPTG and the control. | ||
| + | |||
| + | ====Transformation of ''B. subtilis'' that contained HBsu-(x)FP with Switch Brick==== | ||
| + | |||
| + | '''Aim:''' | ||
| + | |||
| + | To transform ''B. subtilis'' that contained HBsu-(x)FP with Switch Brick as well. | ||
| + | |||
| + | '''Methods:''' | ||
| + | |||
| + | The the MM Media was inoculated with ''B. subtilis'' that was transformed with HBsu-GFP and HBsu-RFP and was left to grow overnight at 37 ̊C. This is to prepare for transformation with switch brick on <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#28/8/13" target="_blank">August 28th</a> </html>. | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | Please refer to <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#29/8/13" target="_blank">August 29th</a> </html> | ||
</div> | </div> | ||
| + | |||
== 8 == | == 8 == | ||
| Line 1,402: | Line 2,644: | ||
==== Titles ==== | ==== Titles ==== | ||
<div> | <div> | ||
| - | * | + | *Transformation of ''B. subtilis'' that contained HBsu-(x)FP with Switch Brick |
| - | * | + | *Safety Page |
</div> | </div> | ||
==== Details ==== | ==== Details ==== | ||
<div> | <div> | ||
| - | + | ==== Transforming Switch BioBrick into HBsu-(x)fp ''B. subtilis''==== | |
| + | |||
| + | '''Aim:''' | ||
| + | |||
| + | To transform ''B. subtilis'' that contained HBsu-(x)FP with Switch Brick as well. | ||
| + | |||
| + | '''Methods:''' | ||
| + | |||
| + | The protocol was continued from <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#27/8/13" target="_blank">August 27th</a> </html> | ||
| + | |||
| + | The switch biobrick was transformed into ''Bacillus subtilis'' 168 strain which already contained the HBsu-(x)fp. The [https://2013.igem.org/Team:Newcastle/Notebook/protocols#Bacillus_subtilis_168_Competent_cell_Prep_and_Transformation B. subtilis transformation protocol] was followed and samples were plated out on LB + Ery (5ug/ml)+ Cm (5ug/ml) + Xyl (0.8%) plates that were made earlier today. | ||
| + | |||
| + | Plates: | ||
| + | |||
| + | 168 HBsu-RFP + Switch BioBrick on LB + Ery (5ug/ml) + Cm (5ug/ml) + Xyl (0.8%) plates x1 | ||
| + | |||
| + | 168 HBsu-GFP + Switch BioBrick on LB + Ery (5ug/ml) + Cm (5ug/ml) + Xyl (0.8%) plates x1 | ||
| + | |||
| + | 168 HBsu-RFP + water on LB + Ery (5ug/ml) + Cm (5ug/ml) + Xyl (0.8%) plates x1 | ||
| + | |||
| + | 168 HBsu-GFP + water on LB + Ery (5ug/ml) + Cm (5ug/ml) + Xyl (0.8%) plates x1 | ||
| + | |||
| + | 168 HBsu-RFP + Switch BioBrick on LBx1 | ||
| + | |||
| + | 168 HBsu-GFP + Switch BioBrick on LB x1 | ||
| + | |||
| + | The plates were left to grow for the next two days. | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | Please refer to <html> | ||
| + | <a href="https://2013.igem.org/Team:Newcastle/Notebook/calendar#29/8/13" target="_blank">August 29th</a> </html>. | ||
| + | |||
| + | ====Safety Page==== | ||
| + | Having completed the safety form on August 23rd, we started to work on the safety page of the wiki. On the safety page, we included everything in the safety form but in more detail. We also included a detailed account of the lan training program and safety precautions. For more detail, please visit the [https://2013.igem.org/Team:Newcastle/HP/Safety safety page] | ||
| + | |||
</div> | </div> | ||
| Line 1,414: | Line 2,692: | ||
==== Titles ==== | ==== Titles ==== | ||
<div> | <div> | ||
| - | * | + | *Transformation of ''B. subtilis'' that contained HBsu-(x)FP with Switch Brick |
| - | * | + | *Part submission |
| + | |||
</div> | </div> | ||
==== Details ==== | ==== Details ==== | ||
<div> | <div> | ||
| - | + | ==== Transformation of ''B. subtilis'' that contained HBsu-(x)FP with Switch Brick==== | |
| + | |||
| + | We checked the transformation that we did previously and the results are the following: | ||
| + | |||
| + | Plates: | ||
| + | 168 HBsu-RFP + Switch BioBrick on LB + Ery (5ug/ml)+ Cm (5ug/ml) + Xyl (0.8%) plates x1: Colonies | ||
| + | |||
| + | 168 HBsu-GFP + Switch BioBrick on LB + Ery (5ug/ml)+ Cm (5ug/ml) + Xyl (0.8%) plates x1: No Colonies | ||
| + | |||
| + | 168 HBsu-RFP + water on LB + Ery (5ug/ml)+ Cm (5ug/ml) + Xyl (0.8%) plates x1: No Colonies | ||
| + | |||
| + | 168 HBsu-GFP + water BioBrick on LB + Ery (5ug/ml)+ Cm (5ug/ml) + Xyl (0.8%) plates x1: No Colonies | ||
| + | |||
| + | 168 HBsu-RFP + Switch BioBrick on LBx1: Colonies | ||
| + | |||
| + | 168 HBsu-GFP + Switch BioBrick on LB x1: Colonies | ||
| + | |||
| + | To check that the transformation with the switch brick works we inoculate some of the cells on the plate into NB/MSM + xyl (0.8%) and NB/MSM only. | ||
| + | |||
| + | ====Part submission==== | ||
| + | We worked on submitting three of our BioBrick to the iGEM registry. We put all the basic information about the part including the sequences. | ||
| + | |||
| + | Here are the parts: | ||
| + | |||
| + | [http://parts.igem.org/Part:BBa_K1185000 Switch BioBrick] | ||
| + | |||
| + | [http://parts.igem.org/Part:BBa_K1185001 HBsu-GFP] | ||
| + | |||
| + | [http://parts.igem.org/Part:BBa_K1185002 HBsu-RFP] | ||
| + | |||
</div> | </div> | ||
| Line 1,426: | Line 2,734: | ||
==== Titles ==== | ==== Titles ==== | ||
<div> | <div> | ||
| - | * | + | *Confirming Transforming Switch BioBrick into HBsu-(x)fp ''B. subtilis'' Has Worked |
| + | *L-form Protocol | ||
| + | *Preparing 168 with HBsu-GFP and HBsu-RFP for IPTG check | ||
| + | *L-form Conversion Time Lapse | ||
</div> | </div> | ||
| Line 1,432: | Line 2,743: | ||
<div> | <div> | ||
| - | + | ==== Confirming Transforming Switch BioBrick into HBsu-(x)fp ''B. subtilis'' has Worked==== | |
| + | |||
| + | '''Aim:''' | ||
| + | |||
| + | To confirm that the switch BioBrick has transformed into ''B. subtilis'' containing HBsu-(x)FP. | ||
| + | |||
| + | '''Methods:'' | ||
| + | |||
| + | Cells from the NB/MSM xyl (0.8%) and NB/MSM only plates were inoculated into a solution and the solution was checked. | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | NB/MSM + xyl: Stay as rod cells | ||
| + | |||
| + | NB/MSM only: L-forms | ||
| + | |||
| + | This suggests that the switch biobrick works even when we transform it into ''B. subtilis'' 168 with HBsu-(x)fp biobrick in it. | ||
| + | |||
| + | ====L-form Protocol==== | ||
| + | |||
| + | '''Aim:''' | ||
| + | |||
| + | To produce more L-form culture. | ||
| + | |||
| + | '''Methods:''' | ||
| + | |||
| + | The [https://2013.igem.org/Team:Newcastle/Notebook/protocols#Protoplasting_to_generate_L-form L-form Protocol] was followed to obtain more L-forms. | ||
| + | |||
| + | '''Results:''' | ||
| + | |||
| + | The L-form protocol was successful and new L-forms were obtained. | ||
| + | |||
| + | ====IPTG check for ''B. subtilis'' 168 containing HBsu-GFP and HBsu-RFP==== | ||
| + | |||
| + | '''Aim:''' | ||
| + | |||
| + | To check that HBsu-GFP and HBsu-RFP was controlled by IPTG. | ||
| + | |||
| + | Detail needed!! | ||
| + | |||
| + | '''Methods:''' | ||
| + | |||
| + | ''B. subtilis'' 168 with HBsu-GFP and HBsu-RFP was inoculated into LB+ery solution in preparation for IPTG check. | ||
| + | |||
| + | '''Results:''' | ||
| + | Details needed!! | ||
| + | |||
| + | ====L-form Conversion Time Lapse==== | ||
| + | Set up microscope to do time lapse photography to see the conversion of ''B. subtilis'' rod cell to L-forms. | ||
| + | |||
| + | </div> | ||
| + | |||
| + | == 9 == | ||
| + | === 2 === | ||
| + | ==== Titles ==== | ||
| + | <div> | ||
| + | *Wiki Design Plan | ||
| + | </div> | ||
| + | ==== Details ==== | ||
| + | <div> | ||
| + | ====Wiki Design Plan==== | ||
| + | We made rough sketches of adaptations to the front page of the wiki to make it more user friendly with less text and more symbols. This will make the wiki more visually appealing, directing readers to the overview page where the information will be found. There was also talk of redesigning the overview page to make it clearer where individual sub-project information could be found. | ||
| + | </div> | ||
| + | |||
| + | == 9 == | ||
| + | === 3 === | ||
| + | ==== Titles ==== | ||
| + | <div> | ||
| + | *Poster Text | ||
| + | </div> | ||
| + | ==== Details ==== | ||
| + | <div> | ||
| + | ==== Poster Text==== | ||
| + | We changed and adapted the poster text from the YSB1.0 conference according to feedback given, to make the format more visual and a clear. This included adding pictures, labels, captions and results for the Lyon Jamboree. | ||
| + | </div> | ||
| + | |||
| + | == 9 == | ||
| + | === 4 === | ||
| + | ==== Titles ==== | ||
| + | <div> | ||
| + | *Skype Conference with Edinburgh Team and Dr. Jane Calvert | ||
| + | </div> | ||
| + | ==== Details ==== | ||
| + | <div> | ||
| + | ====Skype Conference with Edinburgh Team and Jane Calvert==== | ||
| + | |||
| + | We had a conference call via skype to the Edinburgh iGEM team and Dr. Jane Calvert who works in the Science Technology and Innovation Studies, School of Social and Political Science at the University of Edinburgh. The aim of this conference was to gain ideas about how to approach the ethical concerns related to our project and possibly highlight opinions that haven't yet been considered. The call was very successful and there were multiple points made that we could consider and expand on for the ethics section of our project. | ||
| + | |||
| + | [[File:JaneCalvert_Edinburgh2.jpg|frameless|400px]] | ||
| + | |||
| + | [[File:JaneCalvert_Edinburgh.jpg|frameless|400px]] | ||
| + | |||
| + | </div> | ||
| + | == 9 == | ||
| + | === 6 === | ||
| + | ==== Titles ==== | ||
| + | <div> | ||
| + | *Switch BioBrick pre-submission check | ||
| + | *Creating L-form | ||
| + | </div> | ||
| + | ====Details==== | ||
| + | <div> | ||
| + | ====Switch BioBrick Digest==== | ||
| + | We have characterize the Switch BioBrick and shown that it works as expected. Now we are preparing sample for submission, just to make sure that the Switch BioBrick size is correct and has been cloned into pSB1C3 we digested the Switch BioBrick. | ||
| + | |||
| + | We performed 4 digest reactions, first is an uncut plasmid, second and third is a single digest with EcoRI, PstI in this order, and last one is the Double Digest with both enzymes. After leaving the reaction to run at 37oC for 3 hours, we then load them into 0.7% Agarose gel. | ||
| + | |||
| + | The results of the gel revealed that the Switch BioBrick is indeed the correct size and been cloned into pSB1C3 backbone. | ||
| + | |||
| + | [[File:BareCillus_Switch_BioBrick_Digest.jpg]] | ||
| + | |||
| + | Figure 1: Switch BioBrick digest product on a 0.7% agarose. | ||
| + | |||
| + | Lane 1: Uncut plasmid | ||
| + | |||
| + | Lane 2: Switch BioBrick Single Digest with EcoRI | ||
| + | |||
| + | Lane 3: Switch BioBrick Single Digest with PstI | ||
| + | |||
| + | Lane 4: Double Digest with EcoRI and PstI | ||
| + | |||
| + | We then picked the Switch BioBrick transformed ''E.coli'' XL1B colonies from the plate and inoculate 4 bottles of 10ml LB with it. This is to amplify the plasmid, and so will be able to do Plasmid Extraction MiniPrep to extract large quantities of plamid and ready for submission. | ||
| + | |||
| + | ====Making L-form==== | ||
| + | We picked ''B. subtilis'' str. 168 with HBsu-GFP + Switch BioBrick and HBsu-RFP + Switch BioBrick transformed into them from the LB+Cm+Ery+0.8%Xylose plate and innoculate them into 12 bottles of 10ml LB. Let them grew for 3 hours before applying lysosyme onto them to get L-form. These L-forms were then incubated at 30oC over the weekend, before being used on Monday to prove that they are unstable outside osmotically balance environment. | ||
</div> | </div> | ||
| Line 1,468: | Line 2,903: | ||
</div> | </div> | ||
| - | |||
{{Team:Newcastle/Sponsors}} | {{Team:Newcastle/Sponsors}} | ||
Latest revision as of 14:46, 30 September 2013

June
| Sun | Mon | Tue | Wed | Thu | Fri | Sat |
|---|---|---|---|---|---|---|
|
|
|
|
|
|
|
1
|
|
2
|
3
|
4
|
5
|
6
|
7
|
8
|
|
9
|
10
|
11
|
12
|
13
|
14
|
15
|
|
16
|
17
|
18
|
19
|
20
|
21
|
22
|
|
23
|
24
|
25
|
26
|
27
|
28
|
29
|
|
30
|
|
|
|
|
|
|
July
| Sun | Mon | Tue | Wed | Thu | Fri | Sat |
|---|---|---|---|---|---|---|
|
|
1
|
2
|
3
|
4
|
5
|
6
|
|
7
|
8
|
9
|
10
|
11
|
12
|
13
|
|
14
|
15
|
16
|
17
|
18
|
19
|
20
|
|
21
|
22
|
23
|
24
|
25
|
26
|
27
|
|
28
|
29
|
30
|
31
|
|
|
|
August
| Sun | Mon | Tue | Wed | Thu | Fri | Sat |
|---|---|---|---|---|---|---|
|
|
|
|
|
1
|
2
|
3
|
|
4
|
5
|
6
|
7
|
8
|
9
|
10
|
|
11
|
12
|
13
|
14
|
15
|
16
|
17
|
|
18
|
19
|
20
|
21
|
22
|
23
|
24
|
|
25
|
26
|
27
|
28
|
29
|
30
|
31
|
September
| Sun | Mon | Tue | Wed | Thu | Fri | Sat |
|---|---|---|---|---|---|---|
|
1
|
2
|
3
|
4
|
5
|
6
|
7
|
|
8
|
9
|
10
|
11
|
12
|
13
|
14
|
|
15
|
16
|
17
|
18
|
19
|
20
|
21
|
|
22
|
23
|
24
|
25
|
26
|
27
|
28
|
|
29
|
30
|
|
|
|
|
|
Contents
|
6
10
Titles
- Training
- Wiki
Details
Training
The teams split into small groups to discuss individual themes. They did some research into multiple aspects of the project including: wiki layout, t-shirt design and possible team logos. We also looked at past iGEM team wikis for inspirational ideas.
A talk on Computational Modelling and BioBrick concepts was given. This talk led the team to start building the biophysical model of the lipid membrane of Bacillus subtilis and modelling of lipid synthetic pathway. Later in the day, the team met to discuss initial aims and progress of our work so far.
Wiki
Initial designs for the wiki have been created and uploaded. Currently all style sheets and javascript pages are stored externally until the design has been ratified by the team. Front-page image carousel has been created and initialised with blank colours to demonstrate what it shall look like until actual photographs have been taken and edited.
6
11
Modelling Training
- Modelling Training
- Wiki
Details
Modelling Training
We started off working on a Modelling Exercise to identify components and species of the subtilin biosynthesis pathway. We also sketched down some potential logo ideas and created a shortlist of favorites.
We went on to learn about the differences and similarities between stochastic and deterministic modeling. We also used rule based modeling such as RuleBender and BioNetGen. Using the Michaelis-Menten kinetics model as an example, we alter the parameters and rates of reaction to observe the effects.
Wiki
The wiki design was agreed upon and further developed. A front-page image display has been created, developed and set up with dummy text and placeholder images. Discussions have been held over the layout for the calendar, the general consensus being that as few a clicks as possible should be needed to switch between and overall summary of what was done and the more detailed view. Designs are started with the idea being to create a large calendar holding daily headlines that spawn a non-obtrusive pop-up window containing more detail.
6
12
Modelling Training
- Modelling Training
- GUS cloning strategy
Details
Modelling Training
Today we continued learning about stochastic and deterministic modelling and looked at converting the rate parameters in RuleBender. We made the logo more detailed and narrowed it down to two ideas.
GUS cloning strategy
We began work on the GUS cloning strategy which we plan to use to create Beta-glucuronidase producing L-forms to grow and visualise L-forms in plants. The β-glucuronidase produced by the L-forms will cleave the colourless substrate 5-bromo-4-chloro-3-indolyl-β-D-glucuronide (X-Gluc) to produce a blue product allowing the L-form distribution in plant tissue to be observed. This would involve inserting [http://parts.igem.org/Part:BBa_K330002 part:BBa_K330002] into our bacteria. We also started a rough design of the BioBrick, selecting a promoter, Ribosome binding site and antibiotic marker.
6
13
Basic Lab Skills
- Basic Lab Skills
Details
Basic Lab Skills
Before we started work in the lab, we were briefed on various lab health and safety policies to ensure we maintained a safe working environment. We were then given a tour around the lab to inform us of the layout including waste disposal procedures, location of chemicals and use of equipment.
We were then taught how to calibrate pipettes and learned how to operate them. We are also shown how to inoculate bacteria to a liquid culture and how to prepare bacteria streak plates.
Transformation of GFP and RFP bricks into E. coli Competent Cells
Aim:
The aim of this experiment is to transform GFP and RFP BioBricks into Escherichia coli.
Methods:
Please refer to the Escherichia coli competent cells protocol and the E.coli Transformation Protocol .
We then plated those cells onto LB with 5ug/ml chloramphenicol plates.
Results:
Please see June 14th for results
6
14
Basic Lab Skills 2
- Transformation of GFP and RFP bricks into E. coli Competent Cells Results
- Wiki
Details
Transformation of GFP and RFP bricks into E. coli Competent Cells Results
Aim:
Please see June 14th for aim.
Methods:
Please see June 14th for methods.
Results:
We checked the plates to see if anything grew. The RFP plates showed a lot of colonies, while the GFP plates did not work as expected. The next thing we did was to view the plates under the fluorescent inverted microscope. In the afternoon,we had an introductory lecture on basic molecular biology and biobrick design.
Wiki
The calendar has now been created, with the emphasis on both user interaction (simple and obvious to use) as well as ease of editing (the members of the team not familiar with HTML and JQuery should easily be able and add to the calendar). A rough JQuery script has been written to convert standard wiki MediaWiki code into the format used by the Calendar. This will need extensive debugging before the Calendar can be put into use.
6
16
Titles
- Wiki
Details =
Wiki
The Calendar was fully debugged and deployed over the weekend whilst no one was in the lab. This means that as of tomorrow the entire team will be able to edit and add to the calendar with ease. A brief document has also been written that will provide details on how to edit some of the more complex sections of the wiki. This document will be added to as more and more functionality is added into the wiki.
6
17
Basic Lab Skills 3
- Basic Lab Skills
- Day Overview
- BioGame
- HBsu BioBrick clonning strategy
Details
Basic Lab Skills
Aim: To familiarize with miniprep, nanodrop, restriction digest and gel electrophoresis.
Methods:
We transfer colonies of bacteria into LB and extracted the plasmids via the miniprep protocol, and checked the concentration of plasmids through the use of nanodrop spectrophotometry. We then prepared a 1% Agarose gel and performed a single digest with XbaI and PstI and a double digest with both restriction enzymes. We loaded our samples to the gel with the DNA ladder and control (Figure 1).
Results:
Figure 1
Day Overview
The group started thinking about their cloning strategies for each theme of the project. We also discussed ideas for human practices and came up with the idea of making an educational game based on synthetic biology. At the end of the day we had a group meeting to discuss our progress and our ideas for the modelling workshop for the UCL meet up.
BioGame
Work began on the BioGame. Largely contained to simply choosing the best method of implementation for the game. The following options were considered:
- Android SDK (with no game engine/plug ins)
- LibGDX
- OpenGL
- LWJGL
Ultimately the choice was made to use LibGDX due to the documentation available on it, the ease at which games can be developed and the consistent structure it provides with standard gaming development. After this decision was made, a series of tutorials were found and followed to provide a base of knowledge going into developing the application itself.
HBsu- (x)fp BioBrick Cloning Strategy
The team has been looking in literature for a protein that binds non-specifically to DNA. This will aid the visualization of Genome Shuffling among L-forms by enabling the team to see sections of DNA that have come from two different Bacteria. The decision was made to use the HBsu protein which is encoded by the hbs gene in B.subtilis because it is well characterised and should remain associated with the DNA for a long period of time. The HBsu will be conjugated with a florescent protein such as GFP and RFP.
6
18
Basic Lab Skills 3
- Wiki work
- HBsu BioBrick Cloning Strategy
Details
Wiki work
We came up with a script, story board and a list of materials needed for the introductory video on our wiki. We had fun learning to crochet in order to create characters for the video. Furthermore, we updated the team profile page on the wiki by adding individual pictures of all team members.
HBsu BioBrick Cloning Strategy
An initial design for the BioBrick has been created. It includes Kanamycin resistance cassette [http://parts.igem.org/Part:BBa_P1003 BBa_P1003], a lacI Promoter under a strong RBS [http://partsregistry.org/Part:BBa_B0034 BBa_B0034], the BioBrick and a double terminator. Golden Braid assembly is planned to be used to create the HBsu BioBrick.
6
19
Cloning strategies 1
- Gus Reporter BioBrick
- BioGame
- HBsu BioBrick Cloning Strategy
Details
Gus Reporter BioBrick
We began working on the method for creating our construct. This would involve the use of restriction enzymes, primer design and gibson assembly. We also began creating a protocol for introducing our L-forms into plants.
BioGame
Today the game project was set up, created using the LibGDX-setup GUI in order to make projects for Android, Desktop and HTML deployment, all utilising LibGDX's built in features to run off a single "core" project. This was a big deciding factor in the choice to go with LIBGdx for developing the game as it allows us to quickly deploy across multiple platforms should we wish to do so. With the project set up, we have started to create assets such as textures and fonts to be loaded in and used within the game.
HBsu BioBrick Cloning Strategy
We plan to clone it into [http://parts.igem.org/wiki/index.php/Part:pSB1C3 pSB1C3] then PCR the insert before integrating it into the amyE integration plasmid (pSBBs1C). We discovered that the Kanamycin resistance is an E.coli version and wouldn't work in the B. subtilis. Different antibiotic markers that would work in B.subtilis are trying to be found.
6
20
Cloning strategies 2
- GUS Cloning Strategy
- BioGame
- HBsu BioBrick Cloning Strategy
Details
GUS Cloning Strategy
We continued work on the cloning strategies as well as starting modelling the gus reporter system using BioNetGen. We produced a preliminary model of the gus reporter system. However, this model needs to be modified by adding parameters. We have also looked through some papers to find parameters required for the model.
Figure 1: The framework of the GUS model. The red line indicates the blue product in the GUS reporter system. The blue line represents substrate 5-bromo-4-chloro-3indolyl glucuronide (X-Gluc). The green line represents the beta B-glucuronidase.
BioGame
The Asset class has been created and all currently designed assets loaded into the game. A very simple (and temporary) cartoon bacteria has been loaded in and displayed on the screen along with the team logo. The pre-loading screen itself has not yet been developed but all constructs to create it are now in place. The percentage of loading done is simply outputted to the Java console for debugging purposes..
HBsu BioBrick Cloning Strategy
Designed Primers to be used in the Golden Braid assembly, and checked the melting temperature and self-annealing to make sure they work perfectly. Uses RFP (BBa_E1010) and GFP (BBa_E0040) to conjugate it to the HBsu protein in order to observe the recombination.
6
21
Cloning strategies 3
- GUS Cloning strategy
- HBsu BioBrick Cloning Strategy
Details
GUS Reporter Gene Cloning Strategy
We further developed our cloning strategies and decided on the basic design of our construct. We made a preliminary design of the construct using Gene Designer to give a visual representation of how our construct should look like. From this image (Figure 1), we could see the positioning of all the parts, which will be more convenient for us when we want to devise a method to produce this.
We also improved the logo so it's ready for T-shirt printing. (picture from Alina)
HBsu BioBrick Cloning Strategy
A discussion with our supervisor revealed that the assembly system might be very time consuming. So it was decided that DNA 2.0 would be used to synthesis the HBsu BioBrick.
6
24
Progress
- Groningen plasmid Mini-prep
- BioGame
- HBsu BioBrick Cloning Strategy
Details
Groningen plasmid Mini-prep
Aim:
To isolate the groningen plasmid.
Methods:
We performed a mini prep of the Groningen 2012 integration plasmid DNA grown in E.coli overnight at 37 ̊C. The purity was tested using NanoDrop.
We confirmed the insert size by first cut with fast digest XbaI and BamHI and then running the product on a gel to confirm the presence of the plasmid in the purified pDNA.
Results:
Figure 1: Gel electrophoresis of Groningen plasmid.
Lane 1: DNA Ladder
Lane 2: Uncut plasmid
Lane 3: XbaI
Lane 4: BamHI
Lane 5: XbaI and BamHI
Discussion:
The gel shows that in lane 5 where the plasmid is cut with both XbaI and BamHI, two fragments are present. This is what we expected to have happened as the plasmid is cut at two places, isolating an insert of interest. The smaller fragment is the insert (RFP reporter gene) while the bigger fragment is the plasmid backbone.
BioGame
Further work on the game architecture has been done today. The main BioBrick class been created to provide common methods that all BioBricks will use. A very basic BioBrick has been created, and can be added upon touch to the Bacteria. This is simply to demonstrate what will happen when a BioBrick is added and the possibilities therein.
HBsu BioBrick Cloning Strategy
Due to the high cost of sequencing, the integration method of the HBsu BioBrick is now to clone the gene construct into the pMutin4 plasmid and use a single cross-over via the amyE gene to integrate it into the bacteria genome. Also the antibiotic marker and promoter is no longer being used because after SCO the whole plasmid will be integrated and as such will contain the promoter Pspac and Kanamycin cassette.
6
25
Modelling
- Preparation for UCL
- B.subtilis Transformation
- Microfliuidics Introduction
- HBsu BioBrick Cloning Strategy
Details
Preparation for UCL
We started preparing for our project presentation for UCL, and have started to prepare a presentation for our workshop about BioNetGen modelling.
B.subtilis Transformation
Aim:
To transform the bacteria with Groningen plasmid.
Methods:
A single colony of B.subtilis strain 168 was picked and transferred into MM media and left to grow at 37 ̊C overnight. We continued the protocol on June 26th .
Microfluidics Introduction
Our iGEM team attended a talk about microfluidics given by Sunny Park. Microfluidics is vital to some of our sub-projects (Genome Shuffling and Shape Shifting), therefore we looked into the mechanism of operation, possible uses and creation of design for master moulds on the silicon wafer.
HBsu BioBrick Cloning Strategy
The HBsu-GFP and HBsu-RFP BioBricks were sent off for synthesis. We then created an overnight liquid culture of LR2 rod B.subtilis cells in NB/MSM media, ready to be used to make L-form plates to obtain microscopic pictures.
6
26
Modelling
- Preparation for UCL
- B.subtilis Transformation
Details
Preparation for UCL
The team continued work on our modelling presentation for the UCL meetup. We added information about ordinary differential equation vs stochastic modelling and about Markov chain (Figure 1).
Figure 1: Markov Chain
B.subtilis Transformation
Aim:
To transform the bacteria with Groningen plasmid.
Methods:
Please refer to June 25th for the first part of the protocol.
We transferred 1ml of the prepared B.subtilis str. 168 into fresh MM media. We then tranformed the bacteria with the Groningen plasmid following the B.subtilis str. 168 transformation protocol. We also made some frozen competent B.subtilis str. 168 in glycerol.
Results:
Please refer to June 27th .
6
27
Modelling
- Preparation for UCL
- Transformation Prep
Details
Preparation for UCL
We began brainstorming what we should cover in our modelling workshop presentation for the YSB 1.0. We decided to first describe biological modelling and talk about stochastic vs differential models. Before introducing BioNetGen,talking about it's benefits and covering the basics of this language.
B.subtilis Transformation
Aim and Methods:
For Aim and Protocol, please refer to June 25th and June 26th .
Results:
No growth has been observed on the chloramphenicol plates apart from a single colony which might be just background growth.
6
28
Titles
- Transform B.subtilis 168 with Groningen 2012 plasmid and RFP
Details
Groningen plasmid characterisation
Aim:
To characterise the [http://parts.igem.org/wiki/index.php?title=Part:BBa_K818000 Groningen 2012 integration plasmid].
Methods:
The transformation was done using the protocol for competent Bacillus subtilis.
Positive control: pGFPrrnB plasmid
Negative control: water
We then plated the transformants into LB + 5ug/ml Chloramphenicol plates.
Results and Discussion
Please refer July 1st .
7
1
Titles
- Grow B.subtilis 168 for transformation
- Sucrose test
Details
Results from Transformation
Aim and Methods:
Please refer June 28th .
Results:
We checked the transformation plates from the June 28th and found colonies have grown. We decided to conduct the sucrose test to check whether the cell contained actual Groningen plasmid. (see below)
Sucrose test
Aim:
To check whether the colonies on the Cm plate with B.subtilis 168 containing Groningen 2012 integration plasmid actually contained transformant cells rather than a random mutants/contaminant.
Methods:
Transfer the culture of into LB + 1% sucrose media and leaving it to grow over night. Protocol to be continued on July 2nd .
Results and Discussion:
Please refer July 2nd .
Transform B.subtilis 168 with Groningen 2012 plasmid and RFP
Aim:
In case if the transformation of Groningen plasmid into B. subtilis did not work, we decided to inoculated 5 ml of freshly prepared MM media with B.subtilis str.168 colonies to grow overnight in preparation to repeat the protocol.
Methods
A single colony of B.subtilis strain 168 was picked and transferred into MM media and left to grow at 37 ̊C overnight. We continued the protocol on July 2nd .
Results and Discussion:
Please refer July 3rd .
7
2
Titles
- Transforming B.subtilis 168
- Results of sucrose test
Details
Sucrose Test
Aim:
To check whether the colonies on the Cm plate with B.subtilis 168 containing Groningen 2012 integration plasmid actually contained transformant cells rather than a random mutants/contaminant.
Methods:
Following from the protocol indicated on July 1st , we checked the pH of the solution of cells from the Cm plate with Groningen 2012 integration plasmid incubated in sucrose rich media overnight to check for presence of acidic metabolites of sucrose and hence the activity of AmyE.
Results
The results were as hoped, as the pH of both the samples we prepared was the same as the pH of the +ve control (Bacillus 168 cells):4.7, 4.8, 5.04.
Transformation
Aim:
To transform B. subtilis with Groningen 2012 integration plasmid.
Methods:
We transformed B.subtilis 168 cells with Groningen 2012 integration plasmid and with pGMP (+ve control) and no plasmid at all (-ve control). Plated the colonies out according to the protocol and left to grow over night.
Results and Discussion:
Please see July 3rd for results and discussion.
7
3
Titles
- Check the results of transformation
- Set up the test for Amylase
Details
Groningen Plasmid Transformation Results
Aim and Methods:
Please refer to July 2nd
Results:
No colonies were found on the chloramphenicol plates B.subtilis 168 with Groningen 2012 plasmid, however there were a few colonies on the positive control, B.subtilis 168 with pGMP (Figure 1) integration plasmid plates and no colonies on B.subtilis 168 LB + chloramphenicol plates.
Figure 1: B. subtilis 168 + pGMP on LB + Cm plate. Although a few colonies are found, it is not confirmed whether the pGMP insert is present.
Amylase Test
Aim:
To confirm that the pGMP plasmid was integrated in all of the colonies.
Methods:
Transfer a few colonies onto a freshly made LB + Starch agar plates. Check the amylase function using the iodine test for starch on July 4th .
Results and Discussion
Please refer to July 4th
7
4
Titles
- Modelling Presentation
- Checking starch plates
Details
Modelling Presentation
We made a few changes to our modelling presentation and practised our delivery in preparation for the presentation to members of the school of bioinformatics.
Amylase Test
Aim and Methods:
Please refer to July 3rd
Results:
The starch plates from yesterday were checked and it was found that they have overgrown.
Figure 1: Overgrown starch plates.
Discussion:
The experiment will be repeated.
Repeating Amylase Test
Aim:
Please refer to July 3rd
Methods:
Starch agar plates were made including the antibiotic chloramphenicol (Cm) in the media. After these plates were made, colonies were transferred directly onto the plates and were left overnight.
Results:
Please refer to July 5th
7
5
Titles
- Workshop Practice Presentation
- Starch test
Details
Workshop Practice Presentation
We gave a workshop practice presentation to members of the school of bioinformatics. It was decided that we shouldn't talk about the subtilin two-components system model, because it might be too complex for our workshop. It was suggested that our workshop needed to be a little less like a presentation, with more interaction for the participants.
Starch test
Aim and Methods:
Please refer to July 3rd
Results: The starch plates were checked and a lawn of bacteria was found. The starch test was done anyway using vaporised iodine. The test showed a positive result for pGFP as it turned black (Figure 1: left plate) showing no amylase was present and there for it had transformed. The B.subtilis (positive control) did not go black because the starch was consumed by the bacteria as expected (Figure 1: white plate). Also E.coli was left with the Groningen plasmid in 10ml of LB over the weekend for purification. This was done because stocks of the Groningen plasmid were running low.
Figure 1: Results from Iodine Test. Black plate contains pGFP, white plate contains B. subtilis and crystalized plate is just iodine crystals
Discussion
From the results, we can confirm that pGFP rrnB has an unfunctional amyE and hence it is unable to digest the starch, resulting in a black plate. The B. subtilis plate is white because there is a functional amyE and therefore the starch has been digested. From this experiment, we can tell that pGFP rrnB has been integrated into the correct region while there is no insert in that region in the B.subtilis plate.
7
8
Titles
- Updated SGM tutorial
- Finalized workshop presentation
Details
SGM tutorial
We added a modelling section to the SGM tutorial for potential funding. Deciding what information to include was a challenge due to the time constraint. We considered some of the exercises in the workshop for the tutorial.
UCL Presentation Practice
We practiced the UCL presentation to everyone on the team before everyone on the team gave feedback and constructive criticism which was recorded and addressed.
Workshop presentation
The slides for the workshop were edited according to criticism from the feedback session on the 5th. We also began producing a handout with exercises to complete during the workshop.
7
9
Titles
- Modelling Workshop
- Groningen plasmid Mini-prep
- B.subtilis Transformation
Details
Modelling Workshop Feedback Form
We produced a feedback form for attendees of our Workshop. This will be useful to access our workshops success and to improve its content for any future occasions we deliver a modelling workshop.
Groningen plasmid Mini-prep
Aim and Methods:
Please refer to June 24th
Results:
The Groningen plasmid was purified using plasmid Miniprep of previously prepared E.coli. The purity was checked using the NanoDrop and a result of 183.7 was obtained meaning the DNA was usable in subsequent protocols.
B.subtilis Transformation
Aim:
To transform the bacteria with Groningen plasmid.
Methods:
A single colony of B.subtilis strain 168 was picked and transferred into MM media and left to grow at 37 ̊C overnight. We continued the protocol on July 10th .
7
10
Titles
- B.subtilis transformation
- Receiving L-form Strain
Details
B.subtilis transformation
Aim:
To transform the bacteria with Groningen plasmid.
Methods:
Please refer to July 9th for the first part of the protocol.
The B.subtilis transformation was carried out again to see if it would successfully integrate the Groningen plasmid when a lower concentration of Cm antibiotic is used in the plates. The plates used include:
- B.subtilis transformed with 10µl Groningen plasmid on 5µg/ml Cm, 12.5µg/ml Cm, Amp and LB
- B.subtilis transformed with 20µl Groningen plasmid on 5µg/ml Cm and 12.5µg/ml Cm
- B.subtilis transformed with 10µl pGFP (positive control) on 5µg/ml Cm, 12.5µg/ml Cm
- B.subtilis with water (negative control) on 5µg/ml Cm, 12.5µg/ml Cm and Amp
Results:
Please refer to July 11th .
Receiving B.subtilis L-Form
Today we received two plates of B.subtilis L-Form on a LB + 0.8% Xylose plate, courtesy of Dr. Ling Juan Wu, which contains the original switch brick(BioBrick incompatable). One of the L-form has a GFP marker on its genome, and the other contain RFP. We also received the sequence of this strains genome.
7
11
Titles
- B.subtilis transformation
- Sucrose test
- Chromosomal Extraction
Details
B.subtilis transformation
Aim and Methods:
Please refer to July 9th and July 10th .
Results: The following are pictures of the plates:
Figure 1: B.subtilis transformed with 10µl Groningen plasmid on 5µg/ml Cm, 12.5µg/ml Cm, Amp and LB
Figure 2: B.subtilis transformed with 10µl pGFP (positive control) on 5µg/ml Cm, 12.5µg/ml Cm
Sucrose test
Aim:
To check whether the colonies on the Cm plate with B.subtilis 168 containing Groningen 2012 integration plasmid actually contained transformant cells rather than a random mutants/contaminant.
Methods:
Both sucrose and glucose agar was made, left to cool, plated and left to set. Then the culture was transfered into LB + 1% sucrose media and leaving it to grow over night.
Results and Discussion:
Please refer July 16th .
Extracting Chromosome from the B. subtilis received
Using the protocol and kit supplied by Qiagen, we extracted the chromosome out of the B. subtilis that contain the Switch Brick in it. The spec reading showed that the DNA is actually present and to prevent the chromosome to be degraded it was stored in -20oC.
7
12
Titles
- YSB 1.0 Conference
- Modelling Workshop
Details
YSB 1.0 Conference
The team travelled down to London for the Young Synthetic Biologists (YSB1.0) conference with the other UK iGEM teams. On arrival at the conference, the team put up their poster for display and proceeded to listen to other teams giving presentations about their projects. After lunch Yana, Geoff, Justas and James gave a modelling workshop on Rule Based Modelling with BioNetGen. During this time other team members attended various workshops about BioArt and public engagement. Discussions were ongoing throughout the day regarding teams posters and our own project.
More details can be found on the YSB1.0 page.
7
13
Titles
- YSB 1.0 Conference
- Presentation
Details
YSB 1.0 Conference
On arrival at the second day of the conference the team had the opportunity to view the posters and projects of other teams. A 10 minute presentation about our project was also given, followed by 5 minutes of questions. The presentation was very well received, resulting in many interesting questions and generating a lot of interest about the project. The entire conference split into groups after the presentations to discuss topics including modelling, wiki design, parts characterisation, human practices and SynBioSoc.
More details can be found on the YSB1.0 page.
7
15
Titles
- Sequence Analysis
- Primer design
Details
Switch Brick Sequence Analysis
We looked through the chromosomal sequence of the L-form B.subtilis we acquired from Dr. Ling Juan Wu, and located the region (ranging from the pbpb up to the murE gene) and sequence of the Switch Brick. Inside the strain that we received, the Pxyl and the Chloramphenicol resistance gene were already present in the region so it saved us a bit of work as we didn't have to clone it in. However, the sequence contains multiple illegal restriction sites for the BioBrick (EcoRI and PstI). To fix this issue, we decided to make a to synonymous change (no AA change) to the sequence where the restriction sites are located. The Switch Brick sequence is then sent off to DNA 2.0 for synthesis. Whilst waiting for the synthesised BioBrick to arrive, we will be using the current Switch BioBrick which contain EcoRI and PstI for our experiments.
Primer design for switch BioBrick
We designed primers to clone out and amplify the switch BioBrick from the strain that we received.
The primers were as follows:
BBa_K1185000_1_Forward Primer (20bp + XbaI tag + 'AAA')
5’ AAATTCTAGAGCAAAAGAAAGCAAAAGAGGA 3’
BBa_K1185000_1_Reverse Primer (19bp + SpeI tag + 'AAA')
5’ AAATACTAGTATTGTCCCTGTTATCCCAAT 3’
7
16
Titles
- Making Plant Agar Plates
- Sucrose test
Details
Making Agar Plates for our Plants
Aim:
Make some M&S Agar Plates for growing plants.
Methods:
Some Murashige and Skoog Medium (M&S Medium) was made. This medium was ordered in powder form and water had to be added to it to be made into a solution. The solutions were then autoclaved to be used to make the agar for growing plants.
After the solutions have been autoclaved, we made some agar with the M&S Medium. We weighed out 3.0g of agar no.2 into 2x500mL bottles. 250mL of M&S Medium was then added and mixed. The agar was then autoclaved and then cooled at 50 ̊C. When it is ready to be poured, we poured the agar onto 25 plates and left it to set.
Sucrose test
Aim and Methods:
Please refer to July 11th .
Results and Discussion:
Inconclusive results were found as the agar was cloudy and individual colonies were not clear. For this reason a sucrose test was set up to see if the cells containing the Groningen plasmid from the plates had integrated the plasmid, consequently interrupting the sacA gene and so failing to metabolise sucrose and showing as more basic. For a positive control we used B.subtilis and sucrose (this was expected to be acidic) and our negative control was LB and sucrose (this was expected to be more basic as no metabolism). These cultures were left overnight. To see results please refer to July 17th .
7
17
Titles
- Sucrose test
- Transforming B.subtilis 168
- Planting Seeds
- Shape-shifting modelling
Details
Sucrose test
Aim and Method: Please refer to July 11th .
Results and Discussion: The sucrose test was carried out on the cultures using a PH meter. The results observed were as follows:
- LB + sucrose + B.subtilis = 5.15
- LB + B.subtilis = 6.77
- LB + Groningen plasmid + sucrose = 4.23
- LB + Groningen plasmid = 6.75
- LB + sucrose = 7.07
These results show that cells with the Groningen plasmid had functional sucrase showing the plasmid has not been integrated so colonies must have been contaminants.
Transforming B.subtilis 168
Aim:
To transform B. subtilis with Groningen 2012 integration plasmid.
Methods:
Preparation for B.subtilis transformation was carried out; 5 ml of freshly prepared MM media was inoculated with B.subtilis 168 colonies to grow overnight. Also E.coli with Groningen plasmid was put in LB media and left to grow overnight. Protocol continued on July 18th .
Results and Discussion:
Please see July 19th for results and discussion.
Planting Chinese Cabbage Seeds
Aim:
Plant some Chinese Cabbage seeds to prepare for washing with L-forms.
Methods:
We washed and planted cabbage and french dwarf bean seeds in preparation for the introduction of L-forms into seedlings following the Production of L-form containing Plants protocol up to and including step four. The seeds were planted in Murishage and Skoog containing petri dishes and placed in a dark cupboard to germinate.
The washing of plant seeds with L-forms will be done on July 18th .
Shape-Shifting modelling
Modelling tasks for the shape-shifting team were identified:
- Model cell shape using cytoskeleton structure
- Model cell growth
- Limit the growth of the cell by a certain shape (star or triangle)
- OPTIONAL Change the membrane rigidity parameters and run the simulation again.
The shape of the cell is influenced by membrane fluidity and structure of the cytoskeleton. As cytoskeleton is present in both eukaryotic and prokaryotic cells. The former cells have no cell wall, so it is reasonable to assume that the shape of the L-form cell would obey similar rules as eukaryotic cell would. It has been determined that cytoskeleton behaves as an isotropic fluid. Therefore it's realsonable to rely on two-phase flow modelling frame (Holmes, Edelstein-Keshet, 2012).
7
18
Titles
- Innoculating Plants with L-forms
- Groningen Plasmid Mini-prep
- B.subtilis Transformation
Details
Innoculating Plants with L-forms
Aim and Methods:
The seeds that were planted on July 17th were checked and it was observed that the cabbage seeds had begun to germinate. This meant that they were ready to absorb L-forms. We soaked 5 cabbage seeds for 3 hours in 5ml of L-form solution and another 5 seeds in 5%(w/v) mannitol solution to act as a control. The seeds were then washed with distilled water (to lyse extracellular L-forms) and replated on Murishage and Skoog agar plates. This was done following the Production of L-form containing Plants protocol from stage five onwards.
Groningen Plasmid Mini-prep
Aim:
To isolate the groningen plasmid.
Methods:
We performed a mini prep of the Groningen 2012 integration plasmid DNA grown in E.coli overnight at 37 ̊C. The purity was tested using NanoDrop. We confirmed the insert size by first cut with fast digest XbaI and BamHI and then running the product on a gel to confirm the presence of the plasmid in the purified pDNA.
Results
This was done in 8 micro centrifuge tubes. The purity of each was tested using a NanoDrop.The average purity of the 8 tubes was 324ng/µl.
B.subtilis Transformation
Aim:
To transform B. subtilis with Groningen 2012 integration plasmid.
Methods:
Please refer to July 17th for the first part of the protocol. The B.subtilis transformation was carried out again to see if it would successfully integrate the Groningen plasmid when a higher concentration of the plasmid was used. The plates used include:
- pGFP (positive control)on 5µg/ml Cm and 12.5µg/ml Cm
- Groningen plasmid 30 µl on 5µg/ml Cm and 12.5µg/ml Cm
- Groningen plasmid 45 µl on 5µg/ml Cm and 12.5µg/ml Cm
- Groningen plasmid 60 µl on 5µg/ml Cm and 12.5µg/ml Cm
- Water (negative control) on 5µg/ml Cm and 12.5µg/ml Cm
Results and Discussion:
Please see July 19th for results and discussion.
7
19
Titles
- B.subtilis Transformation
- Modelling
Details
B.subtilis Transformation
Aim and Methods: Please refer to July 17th .
Results and Discussion:
The results of the last transformation of B.subtilis with Groningen 2012 integration plasmid look promising. Everything has worked as expected. The insignificant difference in the number of colonies grown on plate with cells inoculated with different amount of plasmid indicates that the transformation occurred as efficiently as it could at around 9ug of DNA per reaction tube. The reason why nothing grew on the plates with 12.5ug/ml of chloramphenicol may be the inefficiency of the promoter used. The colony counts are recorded in a table below.
| Agar/Transformants | pGFP (positive control) | Groningen plasmid 30 µl | Groningen plasmid 45 µl | Groningen plasmid 60 µl | Water |
|---|---|---|---|---|---|
| Nutrient Agar + 5µg/ml Cm | 134 Colonies | 9 Colonies | 12 Colonies | 7 Colonies | 0 Colonies |
| LB Agar = 12.5µg/ml Cm | 142 Colonies | 1 Colonies | 2 Colonies | 3 Colonies | 0 Colonies |
To confirm the integration of the plasmid the test for sucrose metabolism will be carried out on Monday.
Modelling
Preparation work for modelling, learning to use VCell software. We started working our way through simple tutorials to get more familiar with the program before doing "real work".
- Cell Shape model
- The structure and function of cytoskeleton were studied. The cell behaves like an isotropic fluid in terms of shape and movement, therefore some of the factors which influence the shape could be identified: Cell Volume, surface area, surface tension, hydrostatic pressure.
- It turns out that eukaryotic cytoskeleton is characterised as a two-phase viscous, responsive and contractile liquid. If bacterial cytoskeleton is similar, it would complicate the modelling by a fair bit. More insight into the structure and physical properties of bacterial cytoskeleton is needed. In terms of cell growth and division it will be useful to look into the properties of FtsZ and MreB as well as properties of the vesicular membrane and physical triggers of the process of 'blebbing'. So that's a job for Monday.
7
22
Titles
- Made SMM media
- Made sucrose and glucose agar
- Creating L-forms
- BioGame
Details
Made SMM media
More SMM media was made, in order to make the sucrose and glucose plates which will be used to test if the Groningen plasmid has been integrated.
First trial on creating B.subtilis L-form
Aim:
To create L-forms.
Methods:
We used the plate of B.subtilis rod cells (containg the switch BioBrick) that Dr. Ling Juan Wu provided us on the 15th July. This was carried out using both; Direct Method and Protoplasting to generate L-form.
Results:
Both protocols were successful in creating L-forms.
Made sucrose and glucose agar
Both sucrose and glucose agar was made, and sent for auto claving
BioGame
Work restarted on the BioGame. The Sprite class has been extended to cope with both static and animated Sprites utilising Sprite Sheets. An animated background has been created for the game and further work is planned on the user interface in the coming days.
7
23
Titles
- Tested the Lysozyme Protoplast Formation Protocol
- Sucrose and Glucose test
- Improving the poster design
Details
Tested the Lysozyme Protoplast Formation Protocol
The rest of the team members tested the protocol and further suggestions for improvement were made. Despite the suggestion, all teams were successful in creating L-forms. (microscope picture)
Sucrose and Glucose test
Aim:
To check whether the colonies on the Cm plate with B.subtilis 168 containing Groningen 2012 integration plasmid actually contained transformed cells rather than a random mutants/contaminant.
Methods:
The agar was melted on heat plates for half an hour, left to cool, plated and left to set in the fume cupboard. Then the colonies from the transformation plates were plated out onto the agar. Both B.subtilis with the Groningen plasmid at concentrations of 30, 45 and 60 µl and B.subtilis alone (negative control) were put onto both sucrose and glucose plates.
Results:
Please refer to July 23th .
Improving the poster design
Progress on the poster design was made by thinking of different alterations to the layout to make the poster more readable. The favorite was selected as a starting point for the final design.
7
24
Titles
- Sucrose and Glucose plates
- Poster design
- Wiki
- BioGame
Details
Sucrose and Glucose plates
Aim and Methods:
Please refer to July 23th .
Results:
Both the Sucrose and Glucose plates overgrew which means that the Groningen plasmid is not integrated. our next step is to consider whether to sequence the plasmid using the BioBrick primers.
Poster Design
Today, the layout was developed further and shown to the group. The next steps would be to introduce the text, logos and illustrations.
BioGame
A loading bar has now been added for the BioGame, providing the user graphical representation of how long they will be waiting for the game to load. This then opens into the main game screen where your Bacterium can be seen. The next stage is to add the User Interface allowing the user to "take care" of their Bacterium and install BioBricks.
7
25
Titles
- PCR Switch BioBrick
- Planting Seeds
Details
PCR Switch BioBrick
The Primers ordered to clone the Switch BioBrick arrived. Using the chromosome extracted from the B. subtilis which contain the Switch BioBrick we perform PCR to amplify the BioBrick. The spectophotometer reading for the PCR product showed good results (Figure 1) , however when running it on agarose gel to check for the size it didn't show (expected product around 2,666bp)(Figure 2).
Figure 1: DNA concentration reading by conducting Nanodrop spectrophotometry
Figure 2:Gel to check the size of the BioBrick (To label)
Planting Seeds
Aim:
To plant seeds in order to wash them with L-forms and visualized by confocal microscopy.
Methods:
We washed and planted chinese cabbage seeds in preparation for the introduction of L-forms into seedlings following the Production of L-form containing Plants protocol up to and including step four. The seeds were planted in Murishage and Skoog containing petri dishes and placed in a dark cupboard to germinate.
Results: Due to contamination did not get results. Please refer to July 26th for more detail
7
26
Titles
- Cloning Switch Brick via PCR
- Travel Arrangements
- L-form culture contaminate
- Treating Plants with L-forms
Details
PCR Switch Brick
'Aim:
To confirm the size of the switch brick.
Methods:
We performed another round of PCR to amplify the Switch BioBrick from the extracted chromosome. The spec reading show good results (Figure 1), and we run an agarose gel to confirm its present. From the gel we can see a ban around 2,000-3,000bp region (Figure 2), and our BioBrick is around 2,600bp this conclude that we have successfully amplified our BioBrick.
Figure 1: Switch brick spectrophotometer reading with Nanodrop
Figure 2: Gel electrophoresis to confirm size of switch brick size(To label)
Travel Arrangements
Research was done into the most convenient way to travel to the regional jamboree in Lyon in October. A spread sheet was made of the different travel options so a balance could be found between cost and time efficiency. This included timings, prices and duration of trains, flights and buses.
Contamination by Staphylococcus aureus
We checked our stocks L-form culture under the inverted microscopes and found out that all of the culture have been contaminated by S. aureus.
Treating Plants with L-forms
The above mentioned S. aureus contamination means we do not have any L-forms to wash our seedlings in and so cannot continue with the Production of L-form containing Plants protocol. We will have to grow new seedlings once we have some L-forms free from contamination.
7
29
Titles
- Creating L-form
- Transformation of B. subtilis with the switch biobrick
- MM media innoculation
- Cell wall synthesis modelling
Details
Creating L-form culture
Aim:
To make an L-form culture.
Methods:
Using the Protoplasting protocol, we converted both of the L-form strains we possess (GFP and RFP marker) into L-form. We left them to grow for 2 days in 30 ̊C incubator.
Transformation of B. subtilis with the switch biobrick
Aim:
To transform B.subtilis str.168 with the switch brick.
Methods:
Inoculate 10ml of MM medium with B.subtilis str.168 and left to grow overnight in 37 ̊C for preparation of Transformation.
The protocol to be continued on July 30th
Results:
Cell wall synthesis modelling
Found a model of Peptidoglycan biosynthesis in B.subtilis on the BioModels Database. Generated automatically from the Kegg pathway, so the model is quite complicated and unclear. We plan on converting the SBML to bngl (i.e. useable in BioNetGen and adding the ability to switch the cell wall on and off- simulating our main BioBrick
7
30
Titles
- Cell Shape Model
- Transformation of B. subtilis with the switch biobrick
Details
Cell Shape Model
Substantial progress was made towards the understanding of fundamental approaches to mathematical modelling. Dr. David Swailes from the Department of Mechanical Engineering has kindly agreed to help us with our task. It was thought sensible to divide the project into three parts. First model the growth of the vesicle which is not limited by anything, then model the process of growth in a confined environment from the time just after the vesicle grows to touch the sides of the chamber, followed by a model of the transition between the two phases. Tasks for Wednesday: research more on the physiology of the vesicle growth.
Transformation of B. subtilis with the switch biobrick
Aim:
To transform B.subtilis str.168 with the switch brick.
Methods:
Please refer to July 29th for the start of the protocol. Using the overnight B.subtilis strain.168 grown in MM media we carried out the transformation protocol and transformed them with our naked PCR switch brick product and pGFPrrnb (as a control). We then plated the B.subtilis transformed with pGFPrrnb onto LB agar + Chloramphenicol (5ug/ml) plates, and plated out the B.subtilis with the switch brick onto LB agar + Chloramphenicol (5ug/ml) + Xyl 0.8% plates.
Results:
Please refer to
July 31th .
7
31
Titles
- Transformation of B. subtilis with the switch biobrick
- Visualising L-forms under Microfluidics
- Translating the cell wall model
Details
Transformation of B. subtilis with the switch biobrick
Aims and Methods:
Please refer to July 29th and July 30th
Results and Discussion:
As it turns out the naked PCR product Switch BioBrick failed to be transformed, whilst the positive control worked. We found out that the efficiency and probability of transformation to works with just naked PCR product is very low. So, we then planned to clone it into a plasmid first before transforming it into our Bacillus subtilis.
Visualising L-forms under Microfluidics
We created a silicon wafer of the mother machine using the master mold courtesy of Sunny Park. Once the wafer set we removed any impurities using a combination of dd.H2O, acetone, and isopropanol. In order to let the silicon wafer hold together we heated with plasma under vacuum condition. We then load L-forms with two different tags on them (GFP and mCherry expression). The results showed potential endosymbiosis or cell fusion between the L-forms.
Cell wall modelling
Opened the peptidoglycan biosynthesis model in copasi, to try and understand what was happening. The species names were particularly complicated so we attempted to simplify them, whilst making clear what was being referred to. There are 54 species so this took some time.
8
1
Titles
- Plasmid (pGFPrrnb) amplification
- L-Form simulation
Details
Plasmid (pGFPrrnb) Amplification
Aims:
Amplify the pGFPrrnB.
Methods:
We transformed the competent E.coli strain DH5alpha with plasmid pGFPrrnB taken from the spring 2013 BioBrick distribution kit using the [E.coli transformation protocol]. We then innoculated B. subtilis, from the yesterdays transformation plate, which contained pGFPrrnb into LB + Chloramphenicol (5ug/ml).
L-Form simulation
Started working on Java based application to simulate L-form behavior. Current goal is to make something simple, maybe moving circles?
8
2
Titles
- L-Form simulation
- E.coli Innoculation
- Cloning switch brick
Details
L-Form simulation
Started programming L-form genomic fusion simulation using Java. So far everything went smoothly, we have some bacteria basic movement mechanics, and soon will be looking into how to make two bacteria fuse when they meet together.
E.coli Innoculation
We innoculated the transformed E.coli (pGFPrrnb) that we did yesterday into LB.
Cloning switch brick
Aim:
Cloning the switch brick into pSB1C3.
Methods:
We extracted our pSB1C3 plasmid from the BioBrick distribution kit. Through the use of restriction enzymes (XbaI and SpeI) we cut out the RFP insert from the MCS of the plasmid. We ran the digested product on a 0.8% agarose gel and purified the DNA (plasmid backbone) through the use of [gel extraction purification].
We checked the concentration of the purified DNA through nanodrop (spectrophotometer).
Results and Discussion:
It turned out that no DNA was present in the solution. This was due to too much elution buffer that was added to elute the DNA. This resulted in a very low concentration, and we will not be able to continue the [ligation protocol]. This protocol will need to be repeated the next day. Please refer to August 5th
8
5
Titles
- Cloning Switch Brick
- Transforming HBsu-(x)fp BioBrick
- BioGame
Details
Cloning Switch Brick
Aim:
Cloning the switch brick into pSB1C3
Methods:
We started off by extracting the [http://parts.igem.org/wiki/index.php/Part:pSB1C3 pSB1C3] plasmid with RFP marker through Miniprep from the E.coli liquid broth. We performed a double digest using XbaI and SpeI to isolate the insert from the plasmid backbone. We then ran the digested sample through gel electrophoresis to separate out the different parts. Through the use of gel extraction kit and protocol, we cut out the band of interest which was around 2000bp (the backbone). We then ligate the backbone with predigested switch brick through the use of T4 ligase via sticky end ligation in a 1:3 ratio. We left the ligated product in the 4 ̊C fridge.
Figure 1: Performed gel electrophoresis to confirm the size of the clone product.
Transforming HBsu-(x)fp BioBrick
The synthesised construct arrived fully cloned into the pMutin4 plasmid from DNA2.0 and we rehydrated the DNA by adding 10ul of elution buffer. We transformed the plasmid into competent E.coli DH5-alpha and plated it out on LB + Amp plate.
BioGame
Work recommenced on the BioGame with the beginning of the User Interface creation for the main game screen. This began by simply designing some basic sprite sheets for the button states, and then moved on to creating the button class. The button class started off by extending my own Sprite class, choosing to not use LibGDX's built in sprite or button class. I feel this would give me more control over the functionality of the objects, also as some of the documentation on the built in button class is a little light, it means I know exactly what it is doing.
8
6
Titles
- Amplifying Switch Brick
- HBsu-(x)FP BioBrick Extraction
- BioGame
- L-Form simulation
Details
Amplifying Switch Brick
Aim:
To amplify the switch brick.
'Methods:
We transformed the pre-prepared competent E. coli DH5α cells with the Switch Brick that we have cloned in to the psB1C3 plasmid yesterday. These were then plated on to LB+chloramphenicol 5μm/ml and incubated at 37 ̊C;
Results:
Detail needed!!
HBsu-(x)FP BioBrick Extraction
Aim:
To extract the HBsu-RFP & HBsuGF BioBricks from the E.coli.
Methods:
We took E. coli DH5α that has been transformed with HBsu-GFP/HBsu-RFP BioBrick from the plate that was made yesterday and inoculated them onto LB to let them grow before extracting the plasmid out.
Protocol to be continued on August 7th
Results:
Please refer to August 7th
BioGame
Further work has gone into developing the buttons, having now written the code for hit testing the clicks on the button. This code is a little buggy but will be improved as the program is progressed. Started to create the sub-menu that will appear when the "Install BioBrick" button is clicked. Currently this is just a blue box that will eventually contain a list of installable BioBricks.
L-Form simulation
It's starting to look good, as a base for our L-Form simulation Ball game [http://www.ntu.edu.sg/home/ehchua/programming/java/J8a_GameIntro-BouncingBalls.html tutorial] was used. We managed to get couple balls moving around and colliding with each other.
8
7
Titles
- HBsu-(x)FP BioBrick Extraction
- Cloning HBsu-(x)FP BioBrick into pSB1C3 Plasmid
- B. subtilis str.168 Competent Cell Prep
- Inoculating Chinese Cabbage with L-forms
- BioGame
Details
HBsu-(x)FP BioBrick Extraction
Aims:
Please refer to August 6th
Methods:
Continued protocol from August 6th
Using the QIAgen Mini-Prep protocol we extracted the HBsu-RFP & HBsu-GFP BioBricks from the E. coli. We checked the concentration & quality of the plasmid using a spectophotometer.
Results:
The DNA concentration reading was the following
HBsu-GFP: 67.5 ng/μl HBsu-RFP: 55.5 ng/μl
This is of good quality and will be used for cloning in the pSB1C3 plasmid on the same day.
Coloning HBsu-(x)FP BioBrick into psB1C3 Plasmid
Aim:
Clone the HBsu(x)FP BioBrick into pSB1C3 so it can be submitted to the repository.
Methods:
The HBsu-(x)FP BioBrick and the linearised pSB1C3 plasmid was cut using EcoRI and PstI. We then attempted to purify the BioBrick using the gel extraction protocol
Results:
No bands were found after we have conducted the protocol. The protocol will be repeated on the next day.
B. subtilis str.168 Competent Cell Prep
Detail needed!!
Inoculating Chinese Cabbage with L-forms
Aims:
To plant Chinese Cabbage seeds on petri dishes so it can germinate and can be inoculated with L-forms.
Methods:
We washed and planted chinese cabbage seeds in preparation for the introduction of L-forms into seedlings following the Production of L-form containing Plants protocol up to and including step four. The seeds were planted in Murishage and Skoog containing petri dishes and placed in a dark cupboard to germinate.
The protocol was continued on August 8th
Results:
Please refer to August 9th
BioGame
Upon adding buttons into the Install BioBrick sub-menu, it was noted that any button placed over the top of the clicked button on the parent menu is started off in the "over" position and as such is automatically registering a hit. To solve this the hit test code as been completely refactored for a more accurate hit test. To aid this, ActionListeners have been added into the Button class so as to handle the desire events upon click. This will enable coders to write anonymous classes for attaching to individual buttons.
8
8
Titles
- Inoculating Plants with L-forms
- B. subtilis Transformation of HBsu-(x)FP
- Digest pMutin4 HBsu-GFP and HBsu-RFP
Details
B. subtilis Transformation of HBsu-(x)FP
Aim:
To transform B.subtilis with the HBsu-(x)FP biobrick into pMutin 4.
Methods:
We followed the bacillus transformation protocol. We then plated it out on LB + ery (5ug/ml) plates that were made earlier today.
Plates:
HBsu-GFP on LB + ery (5ug/ml) plates x2
HBsu-RFP on LB + ery (5ug/ml) plates x2
rrnB on LB + ery (5ug/ml) plates x2 (positive control)
Water on LB + ery (5ug/ml) plates x2 (negative control)
Cell in LB x2 (to check viability of cells)
Results:
Please refer to August 9th .
Digest pMutin4 HBsu-GFP and HBsu-RFP
Aim:
To digeste the pMutin4 HBsu-GFP and HBsu-RFP by fast digest from promega.
Methods:
The reaction we ran was:
45ul DNA
1ul 10x fast digest buffer
1ul PstI
1ul EcoRI
2ul Water
Total: 50ul
We left this at 37 ̊C for 3 hours.
Then we also cut the linearised pSB1C3 backbone that was sent from iGEM using the same restriction enzyme. The reaction used is:
4ul of DNA (25ng/ul)
4ul of master mix (Contains restriction enzyme EcoRI and PstI)
After the digest we kept it in the -20 ̊C freezer.
Results:
Detail needed!!
Inoculating Plants with L-forms
Aim:
To plant Chinese Cabbage seeds on petri dishes so it can germinate and can be inoculated with L-forms.
Methods:
For the first part of the protocol please refer to August 7th . We checked the seeds we planted yesterday and observed that the chinese cabbage seeds had begun to germinate. This meant that they were ready to absorb L-forms. We soaked 20 cabbage seeds for 3 hours in 5ml of L-form solution and another 20 seeds in 5%(w/v) mannitol solution to act as a control. The seeds were then washed with distilled water (to lyse extracellular L-forms) and replated on Murishage and Skoog agar plates. This was done following the Production of L-form containing Plants protocol from stage five onwards.
Results:
Please refer to August 9th .
8
9
Titles
- L-form Protocol
- B. subtilis Transformation of HBsu-(x)FP
- Plant Microscopy
- Project Description Due
Details
Wiki
We finalized the project description and put it onto the wiki page.
L-form Protocol
We did the L-form protocol to create more L-forms. Using this protocol, we created 4 flasks of L-forms which has HBsu-GFP tag on them.
B. subtilis Transformation of HBsu-(x)FP
Aim and Methods:
Please refer to August 8th .
Results:
The transformation conducted yesterday did not work after checking the plates. Only thing that grew was control in normal LB.
Plant microscopy
Aim and Methods:
Please refer to August 7th and August 8th .
Results and Discussion:
Today we viewed the seedlings, that we had washed with L-forms yesterday,using bright field and fluorescence microscopy. When viewing an L-form washed seedling using fluorescence microscopy we noticed clearly defined green circles in several different areas of the plant. These were of the correct size (about 0.5um) to correspond to our GFP labelled L-forms. No such green circles were found in our control that had been washed in mannitol solution instead of L-form solution. These results are encouraging as they indicate we may have successfully inoculated plants with L-forms, although more evidence is needed. We also decided that when we next grow L-forms in plants, we should wash some seedlings in a solution of bacteria with cell walls to see if these can be taken up by plants.
8
12
Titles
- Transformation of HBsu-(x)FP into B. subtilis
- L-form Growth Curve
- Planting Chinese Cabbage Seeds
- L-Form simulation
Details
Transformation of HBsu-(x)FP into B. subtilis
Aim:
To transform B. subtilis with HBsu-(x)FP.
Methods:
Prepared competent cells by putting B. subtilis 168 in 10ml MM Media and left it to grow overnight. For the next part of the protocol please refer to August 13th .
Results:
Please refer to August 13th .
L-form Growth Curve
Aim:
To establish a growth curve for L-forms.
Methods:
We started doing the L-form growth curve. Initially we took OD reading of the L-form we grew over the weekend with a spectrophotometer.
Results:
The OD obtained was 0.246. We then diluted the solution down to get an OD of 0.05. That solution was then used to create the L-form growth curve. The initial readings of the two L-form samples were 0.05 and 0.046.
Planting Chinese Cabbage Seeds
Aim:
To plant seeds in order to wash them with L-forms and visualize them by confocal microscopy.
Methods:
We washed and planted chinese cabbage seeds in preparation for the introduction of L-forms into seedlings following the Production of L-form containing Plants protocol up to and including step four. The seeds were planted in Murishage and Skoog containing petri dishes and placed in a dark cuboard to germinate. The washing part of the protocol will be done on August 13th .
Results:
Please refer to August 14th .
L-Form simulation
In last couple days we tried adding fusion, which was kind of hard to do, but in the end we succeeded doing that(although it still has couple of bugs, we will fix them asap).
8
13
Titles
- Transformation of HBsu-(x)FP into B. subtilis
- Cloning HBsu-GFP and HBsu-RFP into pSB1C3
- Inoculating L-forms into Chinese Cabbage
- L-form Growth Curve
- Anything
Details
Transformation of HBsu-(x)FP into B. subtilis
Aim:
To transform B. subtilis with HBsu-(x)FP.
Methods:
For the first part of the protocol please refer to August 12th . We followed the B. subtilis transformation protocol.
Plates:
HBsu-GFP on LB + ery (5ug/ml) plates x2
HBsu-RFP on LB + ery (5ug/ml) plates x2
rrnB on LB + ery (5ug/ml) plates x2 (positive control)
Water on LB + ery (5ug/ml) plates x2 (negative control)
Water on LB + Cm (5ug/ml) plates x2 (negative control)
Cell in LB x2 (to check viability of cells)
Figure 1: HBsu-GFP on LB + ery (5ug/ml) plates
Figure 2: HBsu-RFP on LB + ery (5ug/ml) plates
Figure 3: rrnB on LB + ery (5ug/ml) plates x2 (positive control)
Figure 4: Water on LB + ery (5ug/ml) plates x2 (negative control)
Results:
Please refer to August 14th .
Digest pMutin4 HBsu-GFP and HBsu-RFP
Aim:
To digeste the pMutin4 HBsu-GFP and HBsu-RFP by fast digest from promega.
Methods:
The reaction we ran was:
45ul DNA
1ul 10x fast digest buffer
1ul PstI
1ul EcoRI
2ul Water
Total: 50ul
We left this at 37 ̊C for 3 hours.
After we linearized the plasmid, we ran 8ul digest reaction on a gel to confirm the size of the fragments (Figure 5).
Results and Discussion:
Figure 5: Gel to confirm size of HBsu-GFP and HBsu-RFP in pMutin4
Lane 1: 10kB DNA Ladder
Lane 2-3: Empty
Lane 4: HBsu-RFP
Lane 5-6: Empty
Lane 7: Hbsu-GFP
Lane 8:Empty
Lane 9: 10kB DNA Ladder
Lane 10: Empty
Lane 11: 10kB DNA Ladder
After we have confirmed the size, we ran a 80ul digest and cut out the fragment of interest of HBsu-RFP and HBsu-GFP using the transilluminator and performed the gel extraction protocol.
Figure 6
Lane 1: 10kB DNA Ladder
Lane 2-4: Empty
Lane 5: HBsu-RFP
Lane 6-7: Empty
Lane 8: 10kB DNA Ladder
Lane 9-11:Empty
Lane 12: Hbsu-GFP
After this was conducted we checked the nanodrop reading to find out the concentration of DNA which turned out to be for 20ng/ul HBsu-RFP and 17 ng/ul for HBsu-GFP). We then ligated them to the cut backbone that was done on the august 17th. Using the T4 ligase we ran a 1:1, 1:3, 1:5 and control for each sample. We then let it react in the thermocycler at 22 ̊C for an hour and stored at 4 ̊C overnight.
Inoculating L-forms into Chinese Cabbage
Aim and Methods:
We checked the seeds we planted on August 12th and observed that the chinese cabbage seeds had begun to germinate. This meant that they were ready to absorb L-forms. We soaked 20 cabbage seeds for 3 hours in 5ml of L-form solution and another 20 seeds in 5%(w/v) mannitol solution to act as a control. We also created another control by soaking 20 seeds in B. subtilis rod cells. The seeds were then washed with distilled water (to lyse extracellular L-forms) and replated on Murishage and Skoog agar plates. This was done following the Production of L-form containing Plants protocol from stage five onwards.
Results:
Please refer to August 14th .
L-form Growth Curve
Aim and Methods:
Please refer to August 12th .
Results:
An OD of 0.08 and 0.09 was obtained.
8
14
Titles
- Transformation of HBsu-(x)FP into B. subtilis
- Competent Cells Preparation
- L-form Growth Curve
- Plant Confocal Microscopy
Details
Transformation of HBsu-(x)FP into B. subtilis
Aim and Methods:
Please refer to August 12th and August 13th .
Results and Discussion:
The transformation conducted on August 13th worked. We took some colonies and plated it on a starch plate we made earlier so the iodine test could be performed to confirm that HBsu-RFP and HBsu-GFP has integrated into the amyE region (Figure 1-3).
Plates:
HBsu-RFP
HBsu-GFP
168 BSB1 (negative control)
pGFP-rrnB (positive control)
Figure 1: Iodine Test (Front View of Plate 1)
Figure 2: Iodine Test (Back View of Plate 1)
Figure 3: Iodine Test (Front View of Plate 2)
Competent Cells Preparation
Aim:
To make competent E.coli
Methods:
We inoculated some XL1-B in 10ml of LB overnight to use in the next day to prepare competent E.coli cells. Method continued on August 15th .
Results:
Please refer to August 15th .
L-form Growth Curve
Aim and Methods:
Please refer to August 12th .
Results:
An OD of 0.1 and 0.12 was obtained
Plant Confocal microscopy
Aim and Methods:
Please refer to August 12th and August 13th .
Results:
Today we viewed our plants using confocal microscopy to try and detect any GFP-labelled bacteria. After viewing a few seeds washed in L-form solution under the microscope, we found a green fluorescing dot in the correct size range to be an L-form. No green dots were found in the rod cell or mannitol controls. Following these results we have decided to use confocal microscopy four days after washing our plants in L-form solution, rather than after one. As this co-insides with when [http://www.ncbi.nlm.nih.gov/pubmed/12859751 Tsomlexoglou et al.] used PCR to detect L-forms in their chinese cabbage.
Figure 1. Possible L-form inside Chinese cabbage. The bright green dot is the correct size to be an L-form. The green fluorescence image used to detect the L-form is superimposed onto a bright-field image of plant matter.
Figure 2. L-forms from the solution used to wash the Chinese cabbage.
8
15
Titles
- Iodine Test
- Making Competent E.coli
- Preparation of IPTG reaction and B. subtilis Transformation
- L-form Growth Curve
Details
Iodine Test
Aim:
To carry out the iodine test to show... Detail needed!!
Methods:
Detail needed!!
Results and Discussion:
B. Subtilis 168 (Negative control): Halo
B. Subtilis BSB1 (Negative control): Halo
pGFP rrnB (Positive control): No Halo
HBsu-RFP: No Halo
HBsu-GFP: No Halo
When a halo is found, this shows that amyE is functional as it digests the starch around it. When a halo is not found, this means the amyE is non-functional. In this case, it means that the gene fragment has integrated into the amyE region causing it to be non-functional.
Making Competent E.coli
Aim:
To make competent E.coli for use on August 21th .
Methods:
Please refer to August 14th for the first part of the protocol. We followed the Competent E. coli protocol.
Results:
We yielded around 200 aliquots of E. coli competent cells.
Preparation of IPTG reaction and B. subtilis Transformation
We inoculated 2ml NB with B. subtilis 168 that is tagged with HBsu-GFP and another 2ml of NB with B. subtilis that is tagged with HBsu-RFP. This is then incubated at 37 ̊C to use for IPTG reaction. We also prepared 10ml of MM media and inoculated it with some B. subtilis 168.
L-form Growth Curve
Aim and Methods:
Please refer to August 12th .
Results:
An OD of 0.06 and 0.07 was obtained.
8
16
Titles
- Transformation of Switch Brick into B. subtilis
- L-form Growth Curve
- L-Form simulation
Details
Transformation of Switch Brick into B. subtilis
Aim:
To transform the switch biobrick into Bacillus subtilis 168 strain.
Methods:
We followed the B. subtilis transformation protocol and plated it out on LB + Cm (5ug/ml) + Xyl (0.8%) plates that were made earlier today.
Plates:
Switch BioBrick on LB + Cm (5ug/ml)+ Xyl (0.8%) plates x1
rrnB on LB + Cm (5ug/ml)+ Xyl (0.8%) x1 (positive control)
B. subtilis 168 + Water on LB + Cm (5ug/ml)+ Xyl (0.8%) plates x1 (negative control)
B. subtilis 168 in LB x1 (to check viability of cells)
We left the plates to grow over the weekend.
Results:
Please refer to August 19th .
L-form Growth Curve
Aim and Methods:
Please refer to August 12th .
Results:
An OD of 0.03 and 0.05 was obtained.
L-Form simulation
After fixing bugs with L-Form fusion, simulation is starting to look very good. Next step would be to add L-Form division, which might get complicated, because we will need to have additional objects which would serve as another L-Form.
8
19
Titles
- Transformation of Switch Brick into B. subtilis
- L-form Growth Curve
- Lyon Presentation
- Planting Chinese Cabbage Seeds
Details
Transformation of Switch Brick into B. subtilis
Aim and Methods:
Please refer to August 16th .
Results:
We checked the transformation plate and it worked as expected.
Switch BioBrick on LB + Cm (5ug/ml)+ Xyl (0.8%) plates x1: Colonies Found
rrnB on LB + Cm (5ug/ml)+ Xyl (0.8%) x1 (positive control): Colonies Found
168 +Water on LB + + Cm (5ug/ml)+ Xyl (0.8%) plates x1 (negative control): No Colonies Found
Cell in LB x1 (to check viability of cells): Colonies Found
This was done to confirm the characterization of our BioBrick.
L-form Growth Curve
Aim and Methods:
Please refer to August 12th .
Results:
An OD of 0.008 and 0.005 was obtained.
Lyon Presentation
Started working on the presentation layout for Lyon using [http://www.http://prezi.com/ Prezi]. The team feels this will enable us to create a much more visually appealing presentation, giving us not only the best chance of captivating our audience and conveying our findings, but also winning at the Jamboree.
Initial design ideas have begun along the line of a microfluidics chip being used as a back-drop, with chambers used to contain relevant information (acting as "slides" as such). Along with the design ideas we have also started to compile a list of what information we want to have in the presentation, and how best to present this.
Planting Chinese Cabbage Seeds
Aim:
To plant seeds in order to wash them with L-forms and visualize them by confocal microscopy.
Methods:
We washed and planted chinese cabbage seeds in preparation for the introduction of L-forms into seedlings following the Production of L-form containing Plants protocol up to and including step four. The seeds were planted in Murishage and Skoog containing petri dishes and placed in a dark cupboard to germinate.
Results:
Please refer to August 23th .
8
20
Titles
- Lyon Presentation
- Inoculating L-forms into Chinese Cabbage
- L-form Growth Curve
- L-Form simulation
Details
Lyon Presentation
The presentation team has started getting to grips with Prezi and trying to find out exactly what it is capable of. We have discovered that the easiest way of creating animations is to create .swf files using Adobe Flash which, luckily, one of our members has a decent amount of experience with. Unfortunately it has also come to light that Prezi does not handle interactive animations well, meaning that animations will work much better if they loop endlessly. Whilst it is possibly to build simply interactivity into the animation, it requires and ActionScript 3.0 "hack" that could result in some unpredictable behavior. In the interest of reliability and owing to not wanting any unexpected events whilst giving the presentation, the interactive elements have been dropped out of the design.
Due to this the microfluidics theme has come under question, as the complexity of it may outweigh the overall result. One team member has, as such, come up with a preliminary design along a different theme. This can be seen in the image below and also by clicking [http://prezi.com/4jcdd8as8qrm/?utm_campaign=share&utm_medium=copy this link].
Inoculating L-forms into Chinese Cabbage
Aim and Methods:
We checked the seeds we planted on August 19th and observed that the chinese cabbage seeds had begun to germinate. This meant that they were ready to absorb L-forms. We soaked 20 cabbage seeds for 3 hours in 5ml of L-form solution. We created 5%(w/v) mannitol, B. subtilis rod and E.coli rod controls. The seeds were then washed with distilled water (to lyse extracellular L-forms) and replated on Murishage and Skoog agar plates. This was done following the Production of L-form containing Plants protocol from stage five onwards.
Results:
Please refer to August 23th .
L-form Growth Curve
Aim and Methods:
Please refer to August 12th .
Results:
An OD of 0.00 and 0.00 was obtained. For unknown circumstances, the L-form growth curve instead of increasing steadily throughout the week, it increased during the first 48hrs and then decreased throughout the rest of the week. We will reattempt to construct the L-form growth curve again at a later date.
L-Form simulation
Last couple days were quite busy in terms of L-Form simulation, was trying different ways to make L-Forms divide. Found a quite neat solution which required me to create another list, with empty objects (when L-Forms fuse, 1 of the previous L-Forms become redunandt), but using this method number of L-forms in simulation won't exceed initial number of balls.
8
21
Titles
- Characterisation of Switch Brick
- Cloning of HBsu-(x)fp pSB1C3 into E. coli XL1-B
- Lyon Presentation
Details
Characterization of switch brick
Aim:
To characterize the switch brick.
Methods:
We took some colonies that grew on August 19th and put it in NB/MSM + xyl. We altered the xylose concentration between the range of 0-0.8% by increments of 0.2%. We then left it to grow for 12 hours before checking them under the microscope.
Results:
Please refer to August 22nd .
Cloning of HBsu-(x)fp pSB1C3 into E.coli XL1-B
Aim:
To clone the ligation product that was obtained from August 13th into competent E.coli XL1-B that was prepared on the August 15th .
Methods:
Plates:
XL1B + 1:1 HBsu-GFP pSB1C3 on LB + Cm (5ug/ml)
XL1B + 1:3 HBsu-GFP pSB1C3 on LB + Cm (5ug/ml)
XL1B + 1:5 HBsu-GFP pSB1C3 on LB + Cm (5ug/ml)
XL1B + 1:1 HBsu-RFP pSB1C3 on LB + Cm (5ug/ml)
XL1B + 1:3 HBsu-RFP pSB1C3 on LB + Cm (5ug/ml)
XL1B + 1:5 HBsu-RFP pSB1C3 on LB + Cm (5ug/ml)
XL1B + No insert pSB1C3 on LB + Cm (5ug/ml)
Results:
Please refer to August 22nd .
Lyon Presentation
The design idea created yesterday has been scrapped to further pursue the microfluidics theme less the abundance of animation & interactivity. Designs are currently being worked on.
8
22
Titles
- Characterization of Switch Brick Results
- Cloning of HBsu-(x)fp pSB1C3 into E.coli XL1-B Results
Details
Characterization of Switch Brick Results
Aim and Methods:
Please refer to August 21st .
Results:
The samples that had been growing for 12 hours were put under the microscope.
No xylose: All L-forms
0.2 xylose: Most L-forms but a few rod cells
0.4 xylose: Few L-forms more rod cells
0.8 xylose: All rod cells
From this result, we can tell that our switch brick works as expected. When xylose is not present, almost all cells are L-forms. When more xylose is present, more rod cells survive.
Cloning of HBsu-(x)fp pSB1C3 into E.coli XL1-B Results
Aim and Methods:
Please refer to August 21st .
Results and Discussion:
There were colonies for all of the plates except for the negative control.
Figure 1: XL1B + 1:1 HBsu-GFP pSB1C3 on LB + Cm (5ug/ml)
Figure 2: XL1B + 1:3 HBsu-GFP pSB1C3 on LB + Cm (5ug/ml)
Figure 3: XL1B + 1:5 HBsu-GFP pSB1C3 on LB + Cm (5ug/ml)
Figure 4: XL1B + 1:1 HBsu-RFP pSB1C3 on LB + Cm (5ug/ml)
Figure 5: XL1B + 1:3 HBsu-RFP pSB1C3 on LB + Cm (5ug/ml)
Figure 6: XL1B + 1:5 HBsu-RFP pSB1C3 on LB + Cm (5ug/ml)
Figure 7: XL1B + No insert pSB1C3 on LB + Cm (5ug/ml)
We then inoculate one colony from each sample in to LB and let it grow overnight in order to harvest the plasmid. With the amplified plasmid we could then check that it is the correct size and confirm that cloning has worked (Figure 8). The HBsu-GFP and HBsu-RFP insert was confirmed to be the correct size of 1379bp and 1339bp respectively. However the switch brick has not been cut properly. Therefore, the protocol will be repeated.
Figure 8: Gel to Confirm Size of HBsu-GFP and HBsu-RFP insert size
Lane 1: 1kB Ladder
Lane 2: HBsu-GFP PstI
Lane 3: Hbsu-GFP EcoRI
Lane 4: HBsu-GFP PstI and EcoRI
Lane 5: HBsu-GFP Uncut
Lane 6: 1kB Ladder
Lane 7: HBsu-RFP PstI
Lane 8: Hbsu-RFP EcoRI
Lane 9: HBsu-RFP PstI and EcoRI
Lane 10: HBsu-RFP Uncut
Lane 11: 1kB Ladder
Lane 12: Switch Brick PstI
Lane 13: Switch Brick EcoRI
Lane 14: Switch Brick PstI and EcoRI
Lane 15: Switch Brick Uncut
8
23
Titles
- Lyon Jamboree Attendance Fee Due
- Plant Confocal Microscopy
Details
Safety Form
We worked on the safety form that was due in for August 30th . To complete the safety forms, we referred back to some COSHH forms and BioCOSHH forms of the lab for help. We also had to look up some of the bacterial strains we were using. This was to confirm the risk group and possible threats it may pose to the members of the lab, environment, general public and security through misuse. For more information on the safety form please refer to the following link.
Plant Confocal Microscopy
Aim and Methods:
Please refer to August 19th and August 20th
Results:
We viewed 7 different Chinese cabbage seedlings washed with L-forms under the confocal microscope. In three of these plants we found clusters of green fluorescent dots about 0.5µm in diameter. We found no clusters of green fluorescent dots in three seedlings soaked in the mannitol control.
Figure 1. Cluster of L-forms inside a Chinese cabbage seedling.
Figure 2. Image of mannitol-soaked control exhibiting faint green auto-fluorescence, allowing us to see the outline of plant cells.
8
26
Titles
- Miniprep on XL1-B cells with clone product
- Planting Chinese Cabbage Seeds
Details
Miniprep on XL1-B cells with clone product
Aim:
We conducted a miniprep on XL1-B cells containing the clone product from August 22nd .
Methods:
- link to mini-prep protocol*
Results:
The spectrophotometer readings were:
HBsu-GFP pSB1C3: 80.7ng/ul
HBsu-RFP pSB1C3: 95.6ng/ul
We then digest the plasmid using EcoRI and PstI fast digest on a 10ul reaction.
8ul of DNA
0.5ul of PstI Fast Digest
0.5ul of EcoRI Fast Digest
1ul of 10x Fast Digest Buffer
Total: 10ul
We let the reaction run at 37 ̊C for 2 hours and ran the digest on a 0.7% agarose gel. After the gel has finished running, we viewed the gel under the transilluminator to check the size of the fragments. The fragments were all of the correct sizes so the cloning was successful.
Planting Chinese Cabbage Seeds
Aim:
To plant seeds in order to wash them with L-forms and visualize them by confocal microscopy.
Methods:
We washed and planted chinese cabbage seeds in preparation for the introduction of L-forms into seedlings following the Production of L-form containing Plants protocol up to and including step four. The seeds were planted in Murishage and Skoog containing petri dishes and placed in a dark cuboard to germinate.
Results:
Please refer to ??
8
27
Titles
- L-Form simulation
- Characterisation of the HBsu-(x)fp
- Transformation of B. subtilis that contained HBsu-(x)FP with Switch Brick
Details
L-Form simulation
Simulation is nearly done, just giving finishing touches. One of them is improving movement of L-forms, so it's more realistic. Although it took a while to figure out the best movement algorithm, at first we tried doing it completely random, but it didn't work out quite well, so in the end we decided it to make not as random. So what we did is that let's say L-form moves 1px in x and y axis per second, so our algorithm would then generate random number from [-2;2] and then would add it up to the current speed. But, because of nature of randomness balls would move too fast or too slow, so we had to make "limits" so it doesn't get too fast or too slow.
Characterisation of the HBsu-(x)fp
Aim:
To characterize the HBsu-(x)FP BioBrick. From our cloning strategy, we planned to conduct a single cross over so the whole pMutin4 plasmid including the insert is integrated into the B. subtilis chromosomal DNA. After it has been integrated, our biobrick will be under the control of Pspac promoter. Pspac promoter is an IPTG inducible promoter.
Methods:
The B. subtilis 168 strain that has been transformed with HBsu-GFP and HBsu-RFP was inoculated on August 15th to characterise the HBsu-(x)fp biobrick.
The concentration of IPTG was varied from a range of 0.05 – 0.8 mM to observe the conditions that yield the maximum concentration of product. We checked the fluorescence of this fusion protein at 10mins, 30mins, 60mins and 120mins after addition of IPTG to see how long it takes to get the maximum signal.
Results:
The results of this first trial were not very conclusive as there were cross contamination between samples with IPTG and the control.
Transformation of B. subtilis that contained HBsu-(x)FP with Switch Brick
Aim:
To transform B. subtilis that contained HBsu-(x)FP with Switch Brick as well.
Methods:
The the MM Media was inoculated with B. subtilis that was transformed with HBsu-GFP and HBsu-RFP and was left to grow overnight at 37 ̊C. This is to prepare for transformation with switch brick on August 28th .
Results:
Please refer to August 29th
8
28
Titles
- Transformation of B. subtilis that contained HBsu-(x)FP with Switch Brick
- Safety Page
Details
Transforming Switch BioBrick into HBsu-(x)fp B. subtilis
Aim:
To transform B. subtilis that contained HBsu-(x)FP with Switch Brick as well.
Methods:
The protocol was continued from August 27th
The switch biobrick was transformed into Bacillus subtilis 168 strain which already contained the HBsu-(x)fp. The B. subtilis transformation protocol was followed and samples were plated out on LB + Ery (5ug/ml)+ Cm (5ug/ml) + Xyl (0.8%) plates that were made earlier today.
Plates:
168 HBsu-RFP + Switch BioBrick on LB + Ery (5ug/ml) + Cm (5ug/ml) + Xyl (0.8%) plates x1
168 HBsu-GFP + Switch BioBrick on LB + Ery (5ug/ml) + Cm (5ug/ml) + Xyl (0.8%) plates x1
168 HBsu-RFP + water on LB + Ery (5ug/ml) + Cm (5ug/ml) + Xyl (0.8%) plates x1
168 HBsu-GFP + water on LB + Ery (5ug/ml) + Cm (5ug/ml) + Xyl (0.8%) plates x1
168 HBsu-RFP + Switch BioBrick on LBx1
168 HBsu-GFP + Switch BioBrick on LB x1
The plates were left to grow for the next two days.
Results:
Please refer to August 29th .
Safety Page
Having completed the safety form on August 23rd, we started to work on the safety page of the wiki. On the safety page, we included everything in the safety form but in more detail. We also included a detailed account of the lan training program and safety precautions. For more detail, please visit the safety page
8
29
Titles
- Transformation of B. subtilis that contained HBsu-(x)FP with Switch Brick
- Part submission
Details
Transformation of B. subtilis that contained HBsu-(x)FP with Switch Brick
We checked the transformation that we did previously and the results are the following:
Plates: 168 HBsu-RFP + Switch BioBrick on LB + Ery (5ug/ml)+ Cm (5ug/ml) + Xyl (0.8%) plates x1: Colonies
168 HBsu-GFP + Switch BioBrick on LB + Ery (5ug/ml)+ Cm (5ug/ml) + Xyl (0.8%) plates x1: No Colonies
168 HBsu-RFP + water on LB + Ery (5ug/ml)+ Cm (5ug/ml) + Xyl (0.8%) plates x1: No Colonies
168 HBsu-GFP + water BioBrick on LB + Ery (5ug/ml)+ Cm (5ug/ml) + Xyl (0.8%) plates x1: No Colonies
168 HBsu-RFP + Switch BioBrick on LBx1: Colonies
168 HBsu-GFP + Switch BioBrick on LB x1: Colonies
To check that the transformation with the switch brick works we inoculate some of the cells on the plate into NB/MSM + xyl (0.8%) and NB/MSM only.
Part submission
We worked on submitting three of our BioBrick to the iGEM registry. We put all the basic information about the part including the sequences.
Here are the parts:
[http://parts.igem.org/Part:BBa_K1185000 Switch BioBrick]
[http://parts.igem.org/Part:BBa_K1185001 HBsu-GFP]
[http://parts.igem.org/Part:BBa_K1185002 HBsu-RFP]
8
30
Titles
- Confirming Transforming Switch BioBrick into HBsu-(x)fp B. subtilis Has Worked
- L-form Protocol
- Preparing 168 with HBsu-GFP and HBsu-RFP for IPTG check
- L-form Conversion Time Lapse
Details
Confirming Transforming Switch BioBrick into HBsu-(x)fp B. subtilis has Worked
Aim:
To confirm that the switch BioBrick has transformed into B. subtilis containing HBsu-(x)FP.
'Methods:
Cells from the NB/MSM xyl (0.8%) and NB/MSM only plates were inoculated into a solution and the solution was checked.
Results:
NB/MSM + xyl: Stay as rod cells
NB/MSM only: L-forms
This suggests that the switch biobrick works even when we transform it into B. subtilis 168 with HBsu-(x)fp biobrick in it.
L-form Protocol
Aim:
To produce more L-form culture.
Methods:
The L-form Protocol was followed to obtain more L-forms.
Results:
The L-form protocol was successful and new L-forms were obtained.
IPTG check for B. subtilis 168 containing HBsu-GFP and HBsu-RFP
Aim:
To check that HBsu-GFP and HBsu-RFP was controlled by IPTG.
Detail needed!!
Methods:
B. subtilis 168 with HBsu-GFP and HBsu-RFP was inoculated into LB+ery solution in preparation for IPTG check.
Results:
Details needed!!
L-form Conversion Time Lapse
Set up microscope to do time lapse photography to see the conversion of B. subtilis rod cell to L-forms.
9
2
Titles
- Wiki Design Plan
Details
Wiki Design Plan
We made rough sketches of adaptations to the front page of the wiki to make it more user friendly with less text and more symbols. This will make the wiki more visually appealing, directing readers to the overview page where the information will be found. There was also talk of redesigning the overview page to make it clearer where individual sub-project information could be found.
9
3
Titles
- Poster Text
Details
Poster Text
We changed and adapted the poster text from the YSB1.0 conference according to feedback given, to make the format more visual and a clear. This included adding pictures, labels, captions and results for the Lyon Jamboree.
9
4
Titles
- Skype Conference with Edinburgh Team and Dr. Jane Calvert
Details
Skype Conference with Edinburgh Team and Jane Calvert
We had a conference call via skype to the Edinburgh iGEM team and Dr. Jane Calvert who works in the Science Technology and Innovation Studies, School of Social and Political Science at the University of Edinburgh. The aim of this conference was to gain ideas about how to approach the ethical concerns related to our project and possibly highlight opinions that haven't yet been considered. The call was very successful and there were multiple points made that we could consider and expand on for the ethics section of our project.
9
6
Titles
- Switch BioBrick pre-submission check
- Creating L-form
Details
Switch BioBrick Digest
We have characterize the Switch BioBrick and shown that it works as expected. Now we are preparing sample for submission, just to make sure that the Switch BioBrick size is correct and has been cloned into pSB1C3 we digested the Switch BioBrick.
We performed 4 digest reactions, first is an uncut plasmid, second and third is a single digest with EcoRI, PstI in this order, and last one is the Double Digest with both enzymes. After leaving the reaction to run at 37oC for 3 hours, we then load them into 0.7% Agarose gel.
The results of the gel revealed that the Switch BioBrick is indeed the correct size and been cloned into pSB1C3 backbone.
Figure 1: Switch BioBrick digest product on a 0.7% agarose.
Lane 1: Uncut plasmid
Lane 2: Switch BioBrick Single Digest with EcoRI
Lane 3: Switch BioBrick Single Digest with PstI
Lane 4: Double Digest with EcoRI and PstI
We then picked the Switch BioBrick transformed E.coli XL1B colonies from the plate and inoculate 4 bottles of 10ml LB with it. This is to amplify the plasmid, and so will be able to do Plasmid Extraction MiniPrep to extract large quantities of plamid and ready for submission.
Making L-form
We picked B. subtilis str. 168 with HBsu-GFP + Switch BioBrick and HBsu-RFP + Switch BioBrick transformed into them from the LB+Cm+Ery+0.8%Xylose plate and innoculate them into 12 bottles of 10ml LB. Let them grew for 3 hours before applying lysosyme onto them to get L-form. These L-forms were then incubated at 30oC over the weekend, before being used on Monday to prove that they are unstable outside osmotically balance environment.
9
18
Titles
- BioBrick DNA Due
Details
The DNA of our BioBricks needs to be submitted to the registry by this date.
 "
"