Team:Yale/Project Bioassay
From 2013.igem.org
(Difference between revisions)
| Line 165: | Line 165: | ||
<div id="cssmenu1" style="width:730px; margin:0 auto;"> | <div id="cssmenu1" style="width:730px; margin:0 auto;"> | ||
<ul> | <ul> | ||
| - | <li><a href=' | + | <li><a href='https://2013.igem.org/Team:Yale/Project_Overview'><span>Project Overview</span></a></li> |
<li><a href='https://2013.igem.org/Team:Yale/Project_Validate'><span>Validate PLA Synthesis</span></a></li> | <li><a href='https://2013.igem.org/Team:Yale/Project_Validate'><span>Validate PLA Synthesis</span></a></li> | ||
<li class='active'><a href='https://2013.igem.org/Team:Yale/Project_Bioassay'><span>Develop Bioassay</span></a></li> | <li class='active'><a href='https://2013.igem.org/Team:Yale/Project_Bioassay'><span>Develop Bioassay</span></a></li> | ||
Revision as of 15:03, 23 September 2013
Contents |
Develop bioassay to screen PLA production
- We needed a way to detect the PLA once we produced it using the heterologous enzymes
- We decided to use the fluorescent dye Nile red, a intercellular lipid strain
- Nile red does not affect the growth of bacteria, and its fluorescence is quenched in water

This is a figure from Spiekermann et al. 1996 demonstrating Nile red staining of both PHB+ E. coli and PHB negative E. coli
Positive Control
- We used the 2012 Tokyo Tech Biobrick (BBa_K934001) as a positive control to attempt to detect Nile red fluorescence on our plate reader (ex. 530nm, em. 590nm)
- This plasmid had three enzymes which together give E. coli that ability to synthesize P(3HB) a similar plastic PLA
- Cells were grown for 24 hours in the presence of Nile red. The cells were washed and resuspended in PBS.
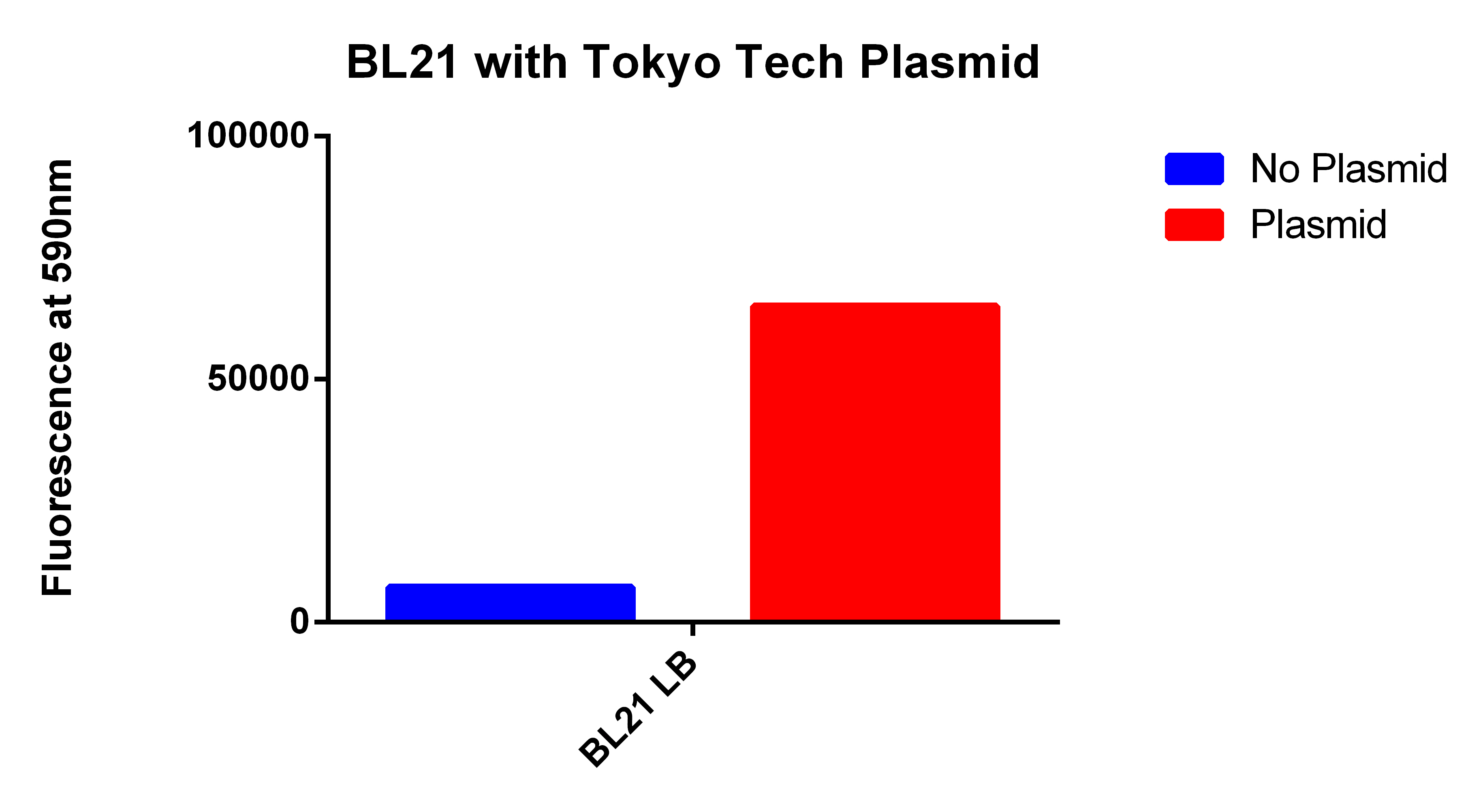
Our Construct
- We then proceeded to test out plasmid in the plate reader.
- Cells were grown for 24 hours with both enzymes induced and in the presence of Nile red. The cells were washed and resuspended in PBS. The readings were normalized for optical density.

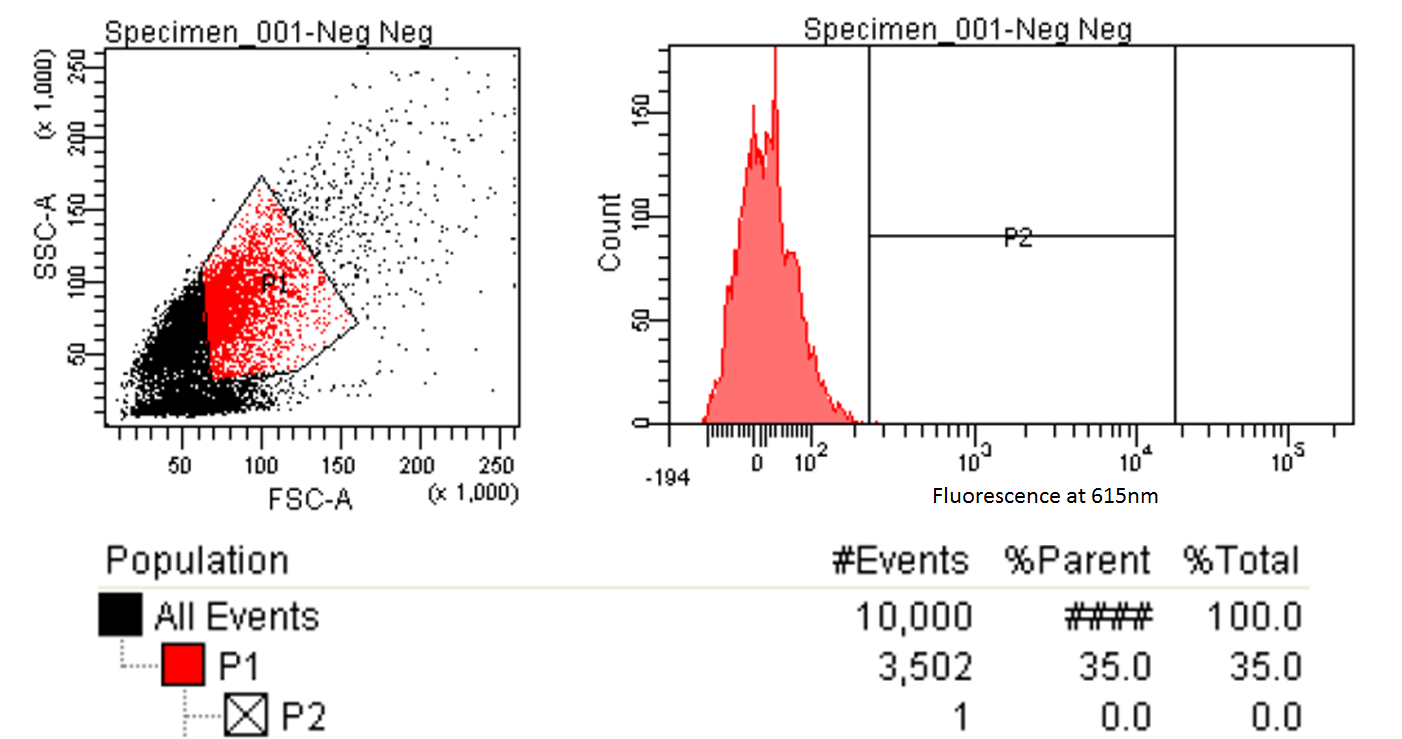
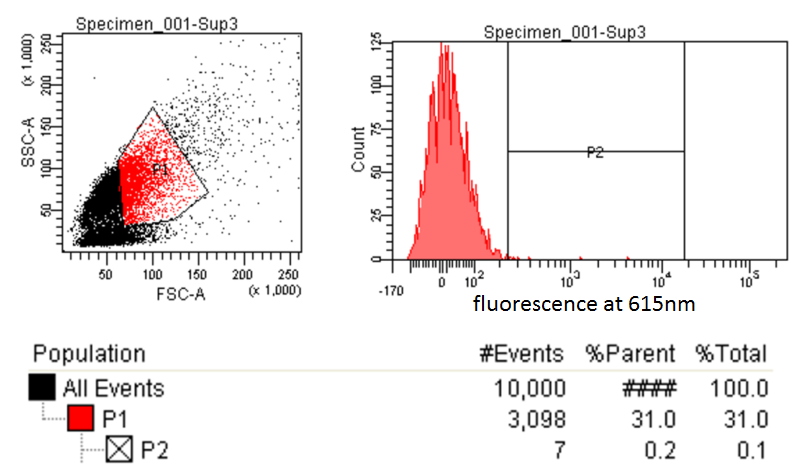
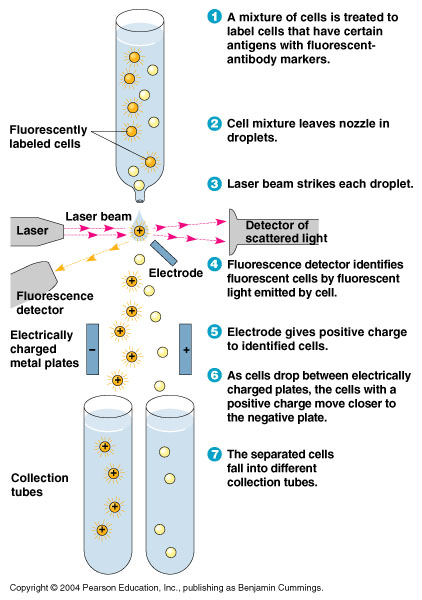
FACS Sorting
- In order to quickly screen the large diversity we planned to create using MAGE, we wanted to employ Fluorescence-activated cell sorting (FACS)
- FACS could sort cells based on the Nile red fluorescence, thus indicating those cells that have produced larger quantities of PLA
Testing for PLA
- Nile red has an emission maximum at 598nm when bound to p(3HB) granules according to Spiekermann et al. 1998
- Thus we decided to use PE-Texas Red which has an emission maximum at 615nm in order pick the best colonies.
- Next we tested these FACS sorted cells on the plate reader to see if we could detect a difference from the FACS sorted and non-FACS sorted cells

 "
"