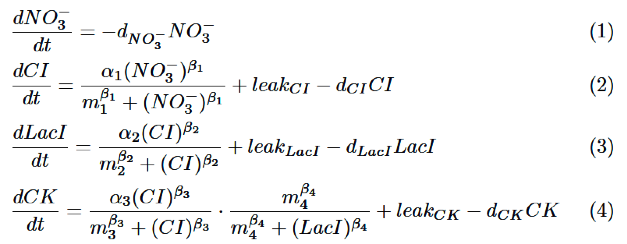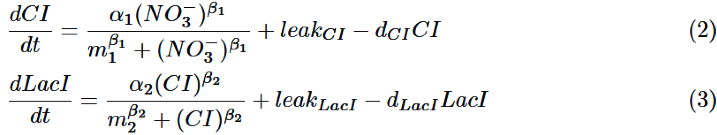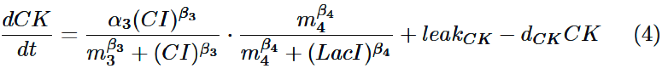Team:Tokyo Tech/Modeling/Incoherent Feed Forward Loop
From 2013.igem.org
| Line 7: | Line 7: | ||
<h2><p>In order to achieve the natural plants’ temporal pattern for producing plant hormones in E. coli, we introduced an incoherent feed forward loop, including new hybrid promoter part ([http://parts.igem.org/Part:BBa_K1139150 BBa_K1139150]). | <h2><p>In order to achieve the natural plants’ temporal pattern for producing plant hormones in E. coli, we introduced an incoherent feed forward loop, including new hybrid promoter part ([http://parts.igem.org/Part:BBa_K1139150 BBa_K1139150]). | ||
</p> | </p> | ||
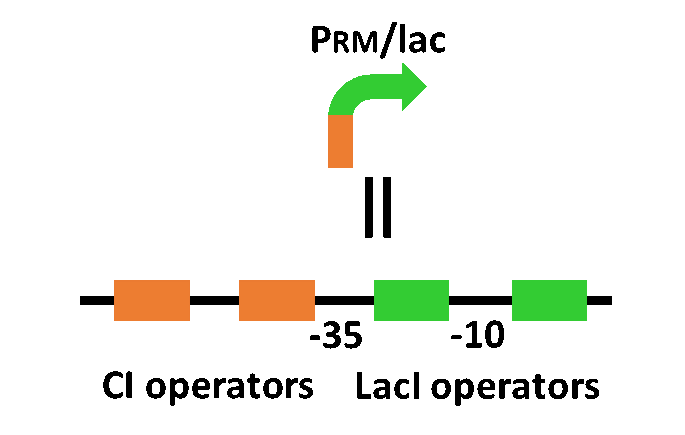
| - | <p>The system of our designed incoherent feed forward loop is shown in Fig. 4-2-1 | + | <p>The system of our designed incoherent feed forward loop is shown in Fig. 4-2-1. We newly developed PRM/lac hybrid promoter, which is activated by CI and repressed by LacI (Fig. 4-2-2.). We plan to ligate a hormone synthase part downstream of this hybrid promoter. While PRM/lac activation by CI is a single-step reaction, repression by LacI is a two-step reaction. Thus, the activation of PRM/lac is faster than its repression. This time lag between activation and repression is important for generating a temporal pattern of plant hormone production. |
</p></h2> | </p></h2> | ||
| - | [[Image:Titech2013_IFFL_Fig_4-2-1_Our_designed_incoherent_feed_forward_loop.png|400px|thumb|left|Fig. 4-2-1 | + | [[Image:Titech2013_IFFL_Fig_4-2-1_Our_designed_incoherent_feed_forward_loop.png|400px|thumb|left|Fig. 4-2-1. Our designed incoherent feed forward loop]] |
| - | [[Image:Titech2013_IFFL_Fig_4-2-2_Our new_designed_hybrid_promoter.png|400px|thumb|left|Fig. 4-2-2 | + | [[Image:Titech2013_IFFL_Fig_4-2-2_Our new_designed_hybrid_promoter.png|400px|thumb|left|Fig. 4-2-2. Our new designed hybrid promoter]] |
<h2> | <h2> | ||
<p> | <p> | ||
| Line 21: | Line 21: | ||
Our model is described by the following differential equations based on Hill equation. | Our model is described by the following differential equations based on Hill equation. | ||
</p> | </p> | ||
| - | [[Image:Titech2013_IFFL_Fig_4-2-3_Equations_for_our_modeling.png|500px|thumb|center|Fig. 4-2-3 | + | [[Image:Titech2013_IFFL_Fig_4-2-3_Equations_for_our_modeling.png|500px|thumb|center|Fig. 4-2-3. Our Equations for our modeling]] |
<p> | <p> | ||
We will explain each equation. | We will explain each equation. | ||
| Line 28: | Line 28: | ||
As for NO3-, we only take the degradation into consideration, assuming that NO3- act directly on nitrate promoter. | As for NO3-, we only take the degradation into consideration, assuming that NO3- act directly on nitrate promoter. | ||
</p> | </p> | ||
| - | [[Image:Titech2013_IFFL_Fig_4-2-4_Equation_of_NO3.png|500px|thumb|center|Fig. 4-2-4 | + | [[Image:Titech2013_IFFL_Fig_4-2-4_Equation_of_NO3.png|500px|thumb|center|Fig. 4-2-4. Equation of NO3-]] |
<p> | <p> | ||
CI is activated by NO3-, LacI is activated by CI. Behaviors of CI and LacI are described by equation (2) and (3). | CI is activated by NO3-, LacI is activated by CI. Behaviors of CI and LacI are described by equation (2) and (3). | ||
</p> | </p> | ||
| - | [[Image:Titech2013_IFFL_Fig_4-2-5_Equation_of_CI_and_LacI.png|500px|thumb|center|Fig. 4-2-5 | + | [[Image:Titech2013_IFFL_Fig_4-2-5_Equation_of_CI_and_LacI.png|500px|thumb|center|Fig. 4-2-5. Equations of CI and LacI]] |
<p> | <p> | ||
Finally, behavior of PRM/lac hybrid promoter is shown as multiplication of activation by CI and repression by LacI. Therefore, the dynamics of cytokinin is described by equation (4). | Finally, behavior of PRM/lac hybrid promoter is shown as multiplication of activation by CI and repression by LacI. Therefore, the dynamics of cytokinin is described by equation (4). | ||
</p> | </p> | ||
| - | [[Image:Titech2013_IFFL_Fig_4-2-6_Equation_of_cytokinin.png|500px|thumb|center|Fig. 4-2-6 | + | [[Image:Titech2013_IFFL_Fig_4-2-6_Equation_of_cytokinin.png|500px|thumb|center|Fig. 4-2-6. Equation of cytokinin]] |
<p> | <p> | ||
We set the parameters as follows: | We set the parameters as follows: | ||
</p> | </p> | ||
| - | [[Image:Titech2013_IFFL_Fig_4-2-7_Descriptions_of_parameters.png|700px|thumb|center|Fig. 4-2-7 | + | [[Image:Titech2013_IFFL_Fig_4-2-7_Descriptions_of_parameters.png|700px|thumb|center|Fig. 4-2-7. Descriptions of the parameters]] |
<p> | <p> | ||
The values of parameters are set as follows: | The values of parameters are set as follows: | ||
</p> | </p> | ||
| - | [[Image:Titech2013_IFFL_Fig_4-2-8_Values_of_the_parameters.png|500px|thumb|center|Fig. 4-2-8 | + | [[Image:Titech2013_IFFL_Fig_4-2-8_Values_of_the_parameters.png|500px|thumb|center|Fig. 4-2-8. Values of the parameters]] |
</h2> | </h2> | ||
<h3>2-2. Simulation results of incoherent feed forward loop</h3> | <h3>2-2. Simulation results of incoherent feed forward loop</h3> | ||
| Line 50: | Line 50: | ||
Fig. 4-2-9. shows the simulation results about the relation between input (NO3-) and output (cytokinin) in our circuit. The red line represent the concentration of NO3-, the green line represents the concentration of cytokinin. | Fig. 4-2-9. shows the simulation results about the relation between input (NO3-) and output (cytokinin) in our circuit. The red line represent the concentration of NO3-, the green line represents the concentration of cytokinin. | ||
</p> | </p> | ||
| - | [[Image:Titech2013_IFFL_Fig_4-2-9_Time-dependent_change_of_the_concentrations_of_NO3-_and_cytokinin.png|500px|thumb|center|Fig. 4-2-9 | + | [[Image:Titech2013_IFFL_Fig_4-2-9_Time-dependent_change_of_the_concentrations_of_NO3-_and_cytokinin.png|500px|thumb|center|Fig. 4-2-9. Time-dependent change of the concentrations of NO3- and cytokinin]] |
<p> | <p> | ||
As shown in Fig. 4-2-9. we verified the pulse-generation of cytokinin as the output with an input of nitric acid NO3-. We will discuss the process of the pulse-generation by looking at the expression level of each substance. | As shown in Fig. 4-2-9. we verified the pulse-generation of cytokinin as the output with an input of nitric acid NO3-. We will discuss the process of the pulse-generation by looking at the expression level of each substance. | ||
| Line 57: | Line 57: | ||
First, NO3- as an input activates CI. CI continues to increase until the input is completely degraded because the expression level of CI depends on the volume of the input (Fig. 4-2-10.). | First, NO3- as an input activates CI. CI continues to increase until the input is completely degraded because the expression level of CI depends on the volume of the input (Fig. 4-2-10.). | ||
</p> | </p> | ||
| - | [[Image:Titech2013_IFFL_Fig_4-2-10_Time-dependent_change_of_the_concentrations_of_NO3-_and_CI.png|500px|thumb|center|Fig. 4-2-10 | + | [[Image:Titech2013_IFFL_Fig_4-2-10_Time-dependent_change_of_the_concentrations_of_NO3-_and_CI.png|500px|thumb|center|Fig. 4-2-10. Time-dependent change of the concentrations of NO3- and CI]] |
<p> | <p> | ||
Next, Fig. 4-2-11. shows that LacI and cytokinin are activated by CI. Also, in Fig. 4-2-12., which shows the enlarged view of Fig. 4-2-11. around 0min, LacI is expressed after the expression of CI. | Next, Fig. 4-2-11. shows that LacI and cytokinin are activated by CI. Also, in Fig. 4-2-12., which shows the enlarged view of Fig. 4-2-11. around 0min, LacI is expressed after the expression of CI. | ||
</p> | </p> | ||
| - | [[Image:Titech2013_IFFL_Fig_4-2-11_Time-dependent_change_of_the_concentrations_of_CI,_LacI,_cytokinin.png|500px|thumb|center|Fig. 4-2-11 | + | [[Image:Titech2013_IFFL_Fig_4-2-11_Time-dependent_change_of_the_concentrations_of_CI,_LacI,_cytokinin.png|500px|thumb|center|Fig. 4-2-11. Time-dependent change of the concentrations of CI, LacI, cytokinin]] |
<br> | <br> | ||
| - | [[Image:Titech2013_IFFL_Fig_4-2-12_Enlarged_view_of_Fig. 4-2-11 | + | [[Image:Titech2013_IFFL_Fig_4-2-12_Enlarged_view_of_Fig. 4-2-11.png|500px|thumb|center|Fig. 4-2-12. Enlarged view of Fig. 4-2-11.]] |
<p> | <p> | ||
| Line 76: | Line 76: | ||
Fig. 4-2-13. shows the change in the waveforms of cytokinin depending on the maximum level of LacI expression. | Fig. 4-2-13. shows the change in the waveforms of cytokinin depending on the maximum level of LacI expression. | ||
</p> | </p> | ||
| - | [[Image:Titech2013_IFFL_Fig_4-2-13_Waveforms_of_cytokinin_depending_on_the_maximum_level_of_LacI_expression.png|500px|thumb|center|Fig. 4-2-13 | + | [[Image:Titech2013_IFFL_Fig_4-2-13_Waveforms_of_cytokinin_depending_on_the_maximum_level_of_LacI_expression.png|500px|thumb|center|Fig. 4-2-13. Waveforms of cytokinin depending on the maximum level of LacI expression]] |
<p> | <p> | ||
| Line 89: | Line 89: | ||
Here, we will predict the activity of LacI by considering IPTG, which represses LacI. Decreasing of LacI activity is described by equation (5). | Here, we will predict the activity of LacI by considering IPTG, which represses LacI. Decreasing of LacI activity is described by equation (5). | ||
</p> | </p> | ||
| - | [[Image:Titech2013_IFFL_Fig_4-2-14_Equation_of_decreasing_LacI_activity.png|500px|thumb|center|Fig. 4-2-14 | + | [[Image:Titech2013_IFFL_Fig_4-2-14_Equation_of_decreasing_LacI_activity.png|500px|thumb|center|Fig. 4-2-14. Equation of decreasing LacI activity]] |
<p> | <p> | ||
Substituting equation (5) for equation (4), we obtain equation (6). | Substituting equation (5) for equation (4), we obtain equation (6). | ||
</p> | </p> | ||
| - | [[Image:Titech2013_IFFL_Fig_4-2-15_Equation_of_cytokinin_led_from_equation_(4)_and_(5).png|500px|thumb|center|Fig. 4-2-15 | + | [[Image:Titech2013_IFFL_Fig_4-2-15_Equation_of_cytokinin_led_from_equation_(4)_and_(5).png|500px|thumb|center|Fig. 4-2-15. Equation of cytokinin led from equation (4) and (5)]] |
<p> | <p> | ||
| - | Shown in equation (6), pulse width of the cytokinin expression can be controlled by external IPTG. In simulation by changing the value of γ, we obtained the result shown in Fig. 4-2-16 | + | Shown in equation (6), pulse width of the cytokinin expression can be controlled by external IPTG. In simulation by changing the value of γ, we obtained the result shown in Fig. 4-2-16. |
</p> | </p> | ||
| - | [[Image:Titech2013_IFFL_Fig_4-2-16_Changes_of_cytokinin_expression_time_controlled_by_IPTG.png|500px|thumb|center|Fig. 4-2-16 | + | [[Image:Titech2013_IFFL_Fig_4-2-16_Changes_of_cytokinin_expression_time_controlled_by_IPTG.png|500px|thumb|center|Fig. 4-2-16. Changes of cytokinin expression time controlled by IPTG]] |
<p> | <p> | ||
Therefore, we confirmed that the pulse width of the cytokinin expression can be controlled by the volume of IPTG. | Therefore, we confirmed that the pulse width of the cytokinin expression can be controlled by the volume of IPTG. | ||
Revision as of 02:45, 27 September 2013
Contents |
Modeling for Farming
Modeling for the incoherent feed forward loop
1. Introduction
In order to achieve the natural plants’ temporal pattern for producing plant hormones in E. coli, we introduced an incoherent feed forward loop, including new hybrid promoter part ([http://parts.igem.org/Part:BBa_K1139150 BBa_K1139150]).
The system of our designed incoherent feed forward loop is shown in Fig. 4-2-1. We newly developed PRM/lac hybrid promoter, which is activated by CI and repressed by LacI (Fig. 4-2-2.). We plan to ligate a hormone synthase part downstream of this hybrid promoter. While PRM/lac activation by CI is a single-step reaction, repression by LacI is a two-step reaction. Thus, the activation of PRM/lac is faster than its repression. This time lag between activation and repression is important for generating a temporal pattern of plant hormone production.
By following mathematical model, we predicted that we can achieve pulse-generation of cytokinin as output, using nitric acid (NO3-) as input. In addition, we analyzed the relation between the waveform and the expression level of LacI.
2. Dynamic behavior of our circuit
2-1. Building a mathematical model
Our model is described by the following differential equations based on Hill equation.
We will explain each equation.
As for NO3-, we only take the degradation into consideration, assuming that NO3- act directly on nitrate promoter.
CI is activated by NO3-, LacI is activated by CI. Behaviors of CI and LacI are described by equation (2) and (3).
Finally, behavior of PRM/lac hybrid promoter is shown as multiplication of activation by CI and repression by LacI. Therefore, the dynamics of cytokinin is described by equation (4).
We set the parameters as follows:
The values of parameters are set as follows:
2-2. Simulation results of incoherent feed forward loop
Fig. 4-2-9. shows the simulation results about the relation between input (NO3-) and output (cytokinin) in our circuit. The red line represent the concentration of NO3-, the green line represents the concentration of cytokinin.
As shown in Fig. 4-2-9. we verified the pulse-generation of cytokinin as the output with an input of nitric acid NO3-. We will discuss the process of the pulse-generation by looking at the expression level of each substance.
First, NO3- as an input activates CI. CI continues to increase until the input is completely degraded because the expression level of CI depends on the volume of the input (Fig. 4-2-10.).
Next, Fig. 4-2-11. shows that LacI and cytokinin are activated by CI. Also, in Fig. 4-2-12., which shows the enlarged view of Fig. 4-2-11. around 0min, LacI is expressed after the expression of CI.
Finally, Fig. 4-2-11. also shows that cytokinin is repressed by LacI after it is activated by CI. Cytokinin is expressed during this time lag between the activation and the repression.
2-3. Relation between the waveform and the maximum level of LacI expression
In 2-2, we confirmed that the expression of LacI is deeply concerned with the expression of cytokinin. Here, we analyze the waveform of cytokinin output by changing the maximum level of LacI expression (α2). Fig. 4-2-13. shows the change in the waveforms of cytokinin depending on the maximum level of LacI expression.
We see that the waveform of cytokinin output changes depending on the maximum level of LacI expression. In considering the application of this system, we predict that we can adapt the output waveforms to the circumstances of plants.
2-4. Relation between the output waveform and IPTG
Here, we will predict the activity of LacI by considering IPTG, which represses LacI. Decreasing of LacI activity is described by equation (5).
Substituting equation (5) for equation (4), we obtain equation (6).
Shown in equation (6), pulse width of the cytokinin expression can be controlled by external IPTG. In simulation by changing the value of γ, we obtained the result shown in Fig. 4-2-16.
Therefore, we confirmed that the pulse width of the cytokinin expression can be controlled by the volume of IPTG.
 "
"