Team:SydneyUni Australia/Project/Background
From 2013.igem.org
| Line 1: | Line 1: | ||
{{Team:SydneyUni_Australia/Style}} | {{Team:SydneyUni_Australia/Style}} | ||
{{Team:SydneyUni_Australia/Header}} | {{Team:SydneyUni_Australia/Header}} | ||
| + | |||
| + | __NOTOC__ | ||
== '''Botany Bay'''== | == '''Botany Bay'''== | ||
| Line 25: | Line 27: | ||
== '''Our Project''' == | == '''Our Project''' == | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
'''Goals''' | '''Goals''' | ||
| Line 40: | Line 36: | ||
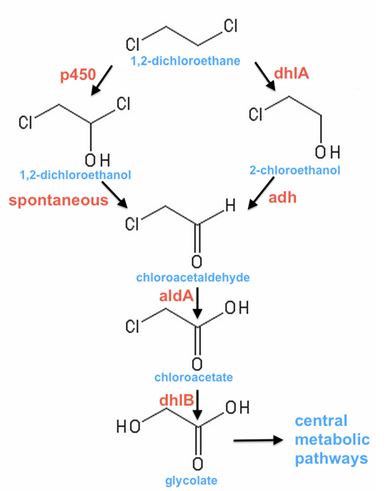
[[File:DCApathwaysHartman.jpg|center]] | [[File:DCApathwaysHartman.jpg|center]] | ||
| + | |||
| + | |||
'''As far as we are aware, there are only two paths of DCA degradation, as in the picture above.''' | '''As far as we are aware, there are only two paths of DCA degradation, as in the picture above.''' | ||
*One begins with oxidation, and has been documented in Pseudomonas spp. [strains DE2 (Stucki et al. 1983) and DCA1 (Hage and Hartmans, 1999)], as well as toluene degraders [Pseudomonas mendocina KR1 (McClay, 1996)] and also a DCA-degrader that our supervisor Nick Coleman,isolated a decade ago [Polaromonas JS666 (Nishino et al, 2013)]. | *One begins with oxidation, and has been documented in Pseudomonas spp. [strains DE2 (Stucki et al. 1983) and DCA1 (Hage and Hartmans, 1999)], as well as toluene degraders [Pseudomonas mendocina KR1 (McClay, 1996)] and also a DCA-degrader that our supervisor Nick Coleman,isolated a decade ago [Polaromonas JS666 (Nishino et al, 2013)]. | ||
*The other pathway begins by removing a Cl- ion, eg, Xanthobacter autotrophicus GJ10 (Janssen, 1994), A. aquaticus strains AD25 and AD27 (van den Wijngaard et al., 1992) and some South African strains isolated from contaminated waste-water (Govender, 2011). I’m pretty sure Jake Munro (PhD kid) from Coleman’s lab has also characterised some of the second type from Botany Bay. | *The other pathway begins by removing a Cl- ion, eg, Xanthobacter autotrophicus GJ10 (Janssen, 1994), A. aquaticus strains AD25 and AD27 (van den Wijngaard et al., 1992) and some South African strains isolated from contaminated waste-water (Govender, 2011). I’m pretty sure Jake Munro (PhD kid) from Coleman’s lab has also characterised some of the second type from Botany Bay. | ||
| + | |||
| + | |||
'''Neither of these pathways have been built from scratch.''' | '''Neither of these pathways have been built from scratch.''' | ||
Revision as of 15:02, 27 September 2013


Botany Bay
There is evidence of settlement by the Eora people (Indigenous Australians) in Sydney for many thousands of years. Many household English words are inherited from their language, including dingo, wombat, waratah, wallaby, etc. The best explanation for this is that when Captain Cook initially landed on the continent of Australia, way back in 1770 at Botany Bay, the Eora people were first he met. Thus Botany Bay is not only a significant historical landmark, but represents a collision of worlds, languages, cultures, and relationships to the environment.
Today, Botany Bay is part of the modern city of Sydney - it’s a suburb with schools, supermarkets, churches, and so on. It’s also been the site of an Orica (previously ICI Australia) manufacturing plant since the 1940’s. During this period of industrial activity in a period of relatively absent environmental awareness and regulation, there has been extensive chemical pollution of the Orica site, leading to contamination of the groundwater at Botany Bay. In 1998, a plume of DCA (1,2-dichloroethane) was discovered in the groundwater. Since then the contamination has been tracked, treated, modelled, debated, and more. Residents are banned from using the water for drinking, washing, watering the garden - anything.
The pollution at Botany Bay is not only a hassle for local residents or a justification for Orica’s new Groundwater Treatment Plant. It is a symbol of a people out of touch with their environment, who forget their own national story, who solve the problems they cause by looking elsewhere - bringing in water for domestic use and attempting to export chlorinated hydrocarbons overseas.
References
- http://www.oricabotanytransformation.com/?page=25
- http://en.wikipedia.org/wiki/Eora
- http://www.water.nsw.gov.au/Water-management/Water-quality/Groundwater/Botany-Sand-Beds-aquifer/Botany-Sands-Aquifer/default.aspx
Chlorinated Hydrocarbons
Conventional Treatment
Our Project
Goals
- Construct and compare two of the proposed pathways of DCA biodegradation.
- Characterise the components of the DCA-degradation pathway for admission into the Registry of Standard Parts.
- (If we get time!) Demonstrate the integration of our the pathway in the chromosome of Pseudomonas stutzeri via natural transformation and site-specific recombination.
As far as we are aware, there are only two paths of DCA degradation, as in the picture above.
- One begins with oxidation, and has been documented in Pseudomonas spp. [strains DE2 (Stucki et al. 1983) and DCA1 (Hage and Hartmans, 1999)], as well as toluene degraders [Pseudomonas mendocina KR1 (McClay, 1996)] and also a DCA-degrader that our supervisor Nick Coleman,isolated a decade ago [Polaromonas JS666 (Nishino et al, 2013)].
- The other pathway begins by removing a Cl- ion, eg, Xanthobacter autotrophicus GJ10 (Janssen, 1994), A. aquaticus strains AD25 and AD27 (van den Wijngaard et al., 1992) and some South African strains isolated from contaminated waste-water (Govender, 2011). I’m pretty sure Jake Munro (PhD kid) from Coleman’s lab has also characterised some of the second type from Botany Bay.
Neither of these pathways have been built from scratch.
- This is true in the sense that people have learned about DCA-degradation by taking the pathway apart, ie, by cloning a single gene and testing it out in a foreign host (usually E. Coli) and seeing whether it degrades a substrate or set of substrates. Part of the philosophy behing Synthetic Biology is about learning by building, which makes our project a neat way to build on extensive research characterising individual genes.
- It is also true in the sense that pollutant-degrading bacteria of interest can learn to degrade xenobiotics by piecing together genes from vastly different origins, and enzymes with activity on very different substrates, often by horizontal gene transfer (Jim Spain (http://www.ce.gatech.edu/people/faculty/671/overview) gave a talk at our school describing this process). Our project has involved a similar process of appropriating genes from different origins, but using online databases of sequences rather than the genepool of a particular microcosm.
 "
"