Team:SydneyUni Australia/Project/Results
From 2013.igem.org
C.Squirrel (Talk | contribs) |
|||
| Line 88: | Line 88: | ||
<div class="pictext"> | <div class="pictext"> | ||
| - | <div class="pictextl" style="height: 100px; line-height: 100px">E. coli expressing ToMO converts indol to | + | <div class="pictextl" style="height: 100px; line-height: 100px">E. coli expressing ToMO converts indol to indigo:</div> |
<div class="pictextr" style="height: 100px;"> | <div class="pictextr" style="height: 100px;"> | ||
<center> | <center> | ||
Revision as of 03:52, 28 September 2013


Project Results
- Toluene-o-xylene monooxygenase (ToMO) from Pseuodomonas stutzeri OX1 has been shown to oxidise xylenes, toluene, benzene, styrene, napthalene (Bertoni et al, 1996) as well as tetrachloroethene, trichloroethene, dichloroethene and vinyl chloride (Shim et al, 2001). The enzyme was optimised for chlorinated ethene degradation (Varder & Wood, 2005), and gifted to our host lab in the plasmid pBS(Kan)ToMO.
- Bertoni, G., Bolognese, F., Galli, E., & Barbieri, P. (1996). Cloning of the genes for and characterization of the early stages of toluene and o-xylene catabolism in Pseudomonas stutzeri OX1. Applied and environmental microbiology, 62(10), 3704-3711.
- Shim, H., Ryoo, D., Barbieri, P., & Wood, T. (2001). Aerobic degradation of mixtures of tetrachloroethylene, trichloroethylene, dichloroethylenes, and vinyl chloride by toluene-o-xylene monooxygenase of Pseudomonas stutzeri OX1. Applied microbiology and biotechnology, 56(1-2), 265-269.
- Vardar, G., & Wood, T. K. (2005). Protein engineering of toluene-o-xylene monooxygenase from Pseudomonas stutzeri OX1 for enhanced chlorinated ethene degradation and o-xylene oxidation. Applied microbiology and biotechnology, 68(4), 510-517.
- We showed that ToMO can begin degradation of DCA through an assay for chloride ions (link to protocol) released as DCA is converted to chloroacetaldehyde. To our knowledge this has’t been shown by anyone else before.
- This is pretty cool, but during the middle of the year we decided to try synthesising the whole pathway rather than building it by conventional cloning. The length of the ToMO gene cluster meant it was too expensive for us to continue working with it.
- Progress
- Transformation
- We spent a week (12/9 to 16/9) optimising the transformation of our Gibson Assembly reaction product. We initially tried transformation into E. Coli EPI300 and yielded no transformants. We suspected that there may be a metabolic burden or harm to cells carrying our correctly assembled product, due to the strong constitutive expression of our designed promoter, Psyn. To account for this we tried transforming into E. Coli EPI400, which carries plasmids at low copy-number with an inducible increase in copy-number. We also tried incubating and growing cells at room temperature to lessen their growth rate, so that they might be able to better cope with any possible toxicity of the transformed Gibson Assembly reaction product. Neither of these were successful, however, we were able to screen 87 clones by transforming into a different strain, E. Coli TOP10.
- Screening
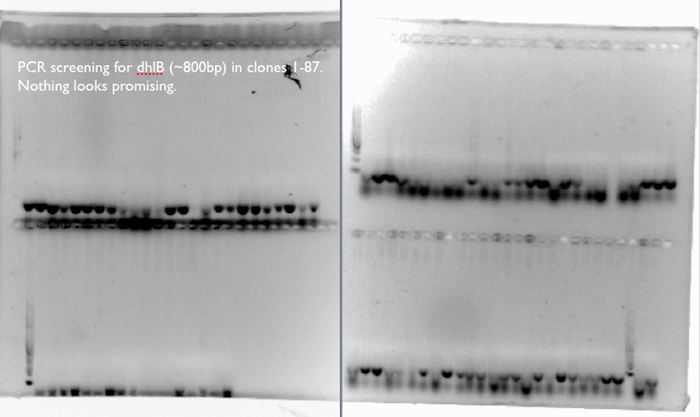
- We screened 87 clones for the presence of dhlB, a gene responsible for the breakdown of chloroacetate in our pathway, by incubating resting cells with chloroacetate and chloride assay (link to protocol). A few clones from each type of pathway looked promising, so we proceeded to extract plasmids from these for further investigation.
- Later on (link to calendar, it was about a week later, after a set of primers had arrived), we also tried screening for dhlB by PCR. We amplicon of interest would have spanned only a single overlap during Gibson Assembly, yet we failed to find a single clone containing the assembled (or misassembled) gene dhlB.
- GET PICTURES FOR ABOVE
- Plasmid Preps
- None of the clones from which we extracted plasmids contained the correctly assembled insert. By PCR and diagnostic restriction digests on these plasmids we were able to distinguish two different misassembled versions of our desired Gibson Assembly reaction product.
- PCR
- We thought it could have been possible to assemble our entire pathway from smaller fragments salvaged from our Gibson Assembly reaction product. This proved impossible, presumably due to the extent of heterogenous template including both correctly and incorrectly assembled gBlocks in the Gibson Assembly reaction product. It may have been possible to do something similar using IDT’s gBlocks as template, however, this was not possible as we’d used up all of one of gBlocks during a second Gibson Assembly.
- Lessons
- Constitutive Expression
- We suspect that some of the genes we tried to assemble (eg, p450, Nishino et al, 2013, dropboxed in ‘reading’) can harm the cells they're expressed in. If this is the case, then by a sort of natural screening we were only able to find colonies on plates that contain misassembled gBlocks that did not express these genes.
- Upon reflection, we approached the assembly of our pathway with an almost child-like ignorance and optimism. Our promoter was specifically designed to maximise expression of our construct, as if ‘the more pollutant degrading genes, the better’. If the hypothesis above is correct, then we might have had more success with an inducible promoter.
- Modularity
- Consider the parable of the watchmakers:
There once were two watchmakers, named Hora and Tempus, who manufactured very fine watches. Both of them were highly regarded, and the phones in their workshops rang frequently - new customers were constantly calling them. However, Hora prospered, while Tempus became poorer and poorer and ?nally lost his shop.What was the reason?
The watches the men made consisted of about 1,000 parts each. Tempus had so constructed his that if he had one partly assembled and had to put it down - to answer the phone say - it immediately fell to pieces and had to be reassembled from the elements. The better the customers liked his watches, the more they phoned him, the more difficult it became for him to find enough uninterrupted time to finish a watch.
The watches that Hora made were no less complex than those of Tempus. But he had designed them so that he could put together subassemblies of about ten elements each. Ten of these subassemblies, again, could be put together into a larger subassembly; and a system of ten of the latter sub-assemblies constituted the whole watch. Hence, when Hora had to put down a partly assembled watch in order to answer the phone, he lost only a small part of his work, and he assembled his watches in only a fraction of the man-hours it took Tempus.
H.A. Simon, The Architecture of Complexity, 1962. - The sequences we had synthesised as gBlocks by IDT were designed so that they could only be assembled in the whole DCA-degradation pathway, rather than as parts within pSB1C3 which could then be assembled piece-by-piece. This meant that our success relied on the correct assembly of the entire pathway, and when this failed, that we were unable to access parts of the pathway (without PCR assembly, or ordering new, complementary gBlocks).
- Plans
- Replacement of gBlocks
- By replacing a single gBlock (link to gBlock design, I’m talking about iGEMBLOCK1, which contains an overlap with pSB1C3, Psyn, an RBS and the beginning of the gene aldA) it might be possible to find correctly assembled Gibson products by substituting our strong constitutive promoter Psyn with a repressible promoter.
- With four new gBlocks, it might be possible to assemble some of the important genes in our pathway (aldA, p450, adh1b1) in a BioBrick vector, so that they could then be subsequently assembled piecewise.
- PCR Assembly
- It might be possible to PCR amplify genes directly from our gBlocks as template rather than our Gibson Assembly reaction product. Alternatively, with the design of new primers, the genes of interest might be amplified from gBlocks for cloning into a BioBrick vector. With either of these options, sequence fidelity may be an issue.
We expect a band at ~800bp for an amplification of dhlB, the gene in our pathway responsible for the degradation of chloroacetate. We don’t see it any of the 87 clones, which corresponds to our phenotypic assays of the clones, where few of the clones released many Cl- ions from the chloroacetate substrate.
We did a second plasmid prep of our clones because the first weren’t very clear. Desmond figured out that by digesting the plasmids with EcoRV we ought to see a single 2kb band if pSB has closed on itself. We don't see anything like this in our clones, but we know from the rfp that this works (pSB-rfp is about 3kb, and contains a single EcoRV site in the pSB backbone). If the plasmid contains our gBlocks (or at least iGEMBLOCK 1 with aldA), then we expect to see two bands - one at 1kb, a second at 5 or 6kb (depending whether we're looking at p450 or the adh plasmid). We don't see this either. We reckon that in most of the plasmids, some gBlocks have assembled in pSB1C3 but not iGEMBLOCK 1. In one of the clones (64) we see a band at 1kb but nothing else, so maybe iGEMBLOCK1 assembled in pSB, but not other gBlocks.
Interestingly, rfp looks spot-on while our results are similar to that in the last plasmid prep (everything slightly higher than 3kb, except 64). With rfp we expect a single band at 3kb (pSB is 2kb plus the rfp 'part' is 1 kb, and the whole construct contains a single EcoRV site in pSB). Notably, 64 was one of the small p450 colonies we patched from Gibson Assembly Product transformations.
- While struggling with Gibson Assembly of our whole pathway we turned to the extraction, cloning and characterisation of two parts in our pathway, [https://static.igem.org/mediawiki/2013/9/90/DCApathwaysHartman.jpg dhlB and dhlA ]. These two genes had been cloned previously into pUC19 by Jake Munro, a PhD student in our lab .
- Amplification
- We designed primers specifically for amplifying dhlB and dhlA, while removing a forbidden EcoRI site between them. Design with the primers allowed us to try cloning each gene by itself and also together into the shipping vector pSB1C3.
- Cloning
- After PCR screening of the junctions between our parts and pSB1C3, we extracted the plasmids from a few that looked OK for further confirmation and submission to the iGEM HQ.
- Characterisation of submitted parts with the constitutive promoter Pcat
- Estimated percentage degradation of chloroacetate and 1,2-DCA as determined by:

Where the Test cell supernatant is from BBa_K1115009 or BBa_K1115010, the Negative cell supernatant is from BBa_K1115008, and the substrate concentration is 2mM Chloroacetate or 1,2-DCA. Note that values of over 100% degradation are misleading and likely indicate endogenous Cl- production (i.e. Cl- production from other cellular processes unrelated to target substrate metaboloism.
- After sending dhlB and dhlA to iGEM HQ, we began characterisation by cloning a constitutive promoter (BBa_I14033) from the Distribution Kit in front of our parts.
- Amplification
- Pcat ([http://parts.igem.org/Part:BBa_I14033 BBa_I14033]) is 38bp, but with our primers produced a 280bp fragment, 4th well from left.
1.0% agarose Gel of PCR products from Distribution Kit: Loading order of was 1kb ladder (from the top: 10, 8, 6, 5, 4, 3, 2, 1.5, 1, 0.5kb), LacI generator PCR product (BBa_P0412 template), 100bp ladder (100, 200, 300, 400, 500, 6000, 700, 800, 900, 1200, 1500bp) PCat (BBa_I14033 template) and PLac. The band for PCat indicates the correct length including ends of pSB1C: 38bp + ~250bp=~290bp.
The PCat PCR products were combined (400uL) and column purified using the QiaQuick Kit (see Protocols tab). As we were attempting to construct an inducible system with PCat constitutively promoting the LacI generator and we had multiple LacI PCR products, we attempted a gel band extraction of the correct band but were unsuccessful.
Having lost our repressor part, we digested PCat with EcoRI and SpeI, and our dhlB-dhlA part BBa_K1115008 with EcoRI and XbaI at 37oC. Digested DNA was purified, mixed and ligated for one hour at room temperature to form a putative construct BBa_K1115009 which was transformed to chemically competent TOP10 cells.
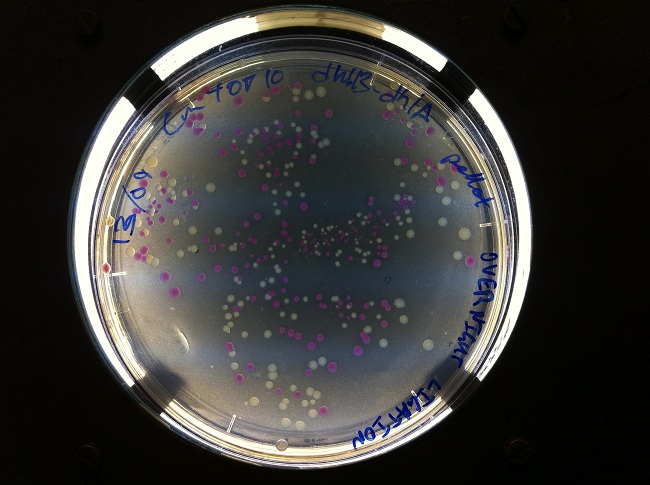
- After cloning into a plasmid containing dhlB and dhlA, we screened for clones expressing our construct. We made screening plates (contained LB-agar-chloramphenicol- 10mMchloroacetate-phenol red at pH 6.8) that allowed us to pick clones that looked like they were successfully expressing one of our genes of interest.
- We showed degradation of chloroacetate and DCA by chloride assay.
 "
"