Team:Goettingen/Team/DAC
From 2013.igem.org
(→Results and Discussion) |
FMCommmichau (Talk | contribs) |
||
| Line 55: | Line 55: | ||
By conducting several experiments, we proved that the truncated DacA protein (BBa_K1045003) was not only active ''in vivo'', but also ''in vitro''. Moreover, we were able to purify the diadenylate cyclase in large scale for determining its 3D structure! Using the structure data, one can now search for chemical compounds that interfere with the activity of the cyclase either by testing with available chemical libraries or by computational modeling. In the following text, the experiments will be explained in more detail. However, if you wish to get even more details, please visit the [[Team:Goettingen/Parts|Parts Registry]] or [[Team:Goettingen/NoteBook|our LabBook.]] | By conducting several experiments, we proved that the truncated DacA protein (BBa_K1045003) was not only active ''in vivo'', but also ''in vitro''. Moreover, we were able to purify the diadenylate cyclase in large scale for determining its 3D structure! Using the structure data, one can now search for chemical compounds that interfere with the activity of the cyclase either by testing with available chemical libraries or by computational modeling. In the following text, the experiments will be explained in more detail. However, if you wish to get even more details, please visit the [[Team:Goettingen/Parts|Parts Registry]] or [[Team:Goettingen/NoteBook|our LabBook.]] | ||
| - | The truncated DacA protein ([http://parts.igem.org/Part:BBa_K1045003 BBa_K1045003]) was extended with an N-terminal Step-tag allowing easy purification steps. The expression of our protein was brought under the control of a T7 | + | The truncated DacA protein ([http://parts.igem.org/Part:BBa_K1045003 BBa_K1045003]) was extended with an N-terminal Step-tag allowing easy purification steps. The expression of our protein was brought under the control of a T7 promoter, enabling us to induce the expression by addition of Isopropyl-β-D-thiogalactopyranoside (IPTG). The plasmid was then cloned into the ''E. coli'' strain BL21, which is a protein expression strain. The Gram-negative bacterium ''E. coli'' does not produce c-di-AMP and is not severely affected by the signaling molecule in contrast to Gram-positive bacteria. |
In order to analyze the cyclase activity ''in vivo'', the ''E. coli'' clones were induced to express the protein by the addition of IPTG. The cells were then lysed to extract c-di-AMP from the cells. | In order to analyze the cyclase activity ''in vivo'', the ''E. coli'' clones were induced to express the protein by the addition of IPTG. The cells were then lysed to extract c-di-AMP from the cells. | ||
| Line 83: | Line 83: | ||
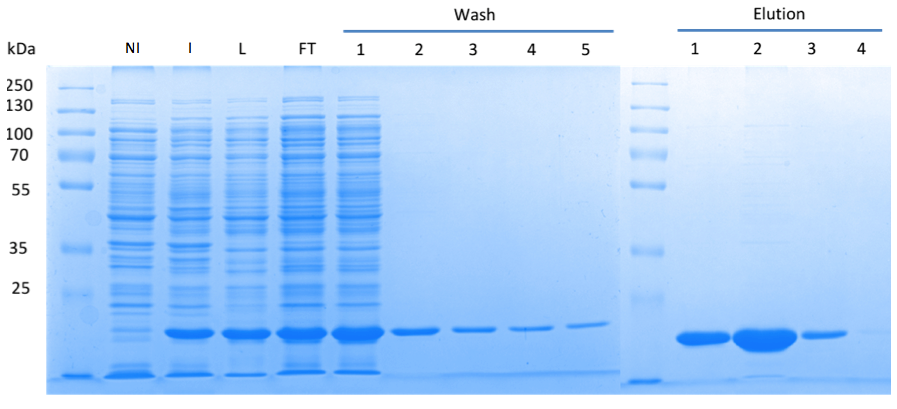
Fig. 3. '''SDS gel showing overexpression and purification of DacA.''' (A) Lane 1: Thermo Scientific PageRuler Plus Prestained Protein Ladder; NI: Non-induced, Clone contained the cyclase domain of which the activity was not induced; I: Induced, Clone contained the cyclase domain of which the activity was induced by addition of isopropyl-ß-D-1-thiogalactopyranoside (IPTG); L: Lysate; FT: Flow-through; After wash 1 our protein was purified. | Fig. 3. '''SDS gel showing overexpression and purification of DacA.''' (A) Lane 1: Thermo Scientific PageRuler Plus Prestained Protein Ladder; NI: Non-induced, Clone contained the cyclase domain of which the activity was not induced; I: Induced, Clone contained the cyclase domain of which the activity was induced by addition of isopropyl-ß-D-1-thiogalactopyranoside (IPTG); L: Lysate; FT: Flow-through; After wash 1 our protein was purified. | ||
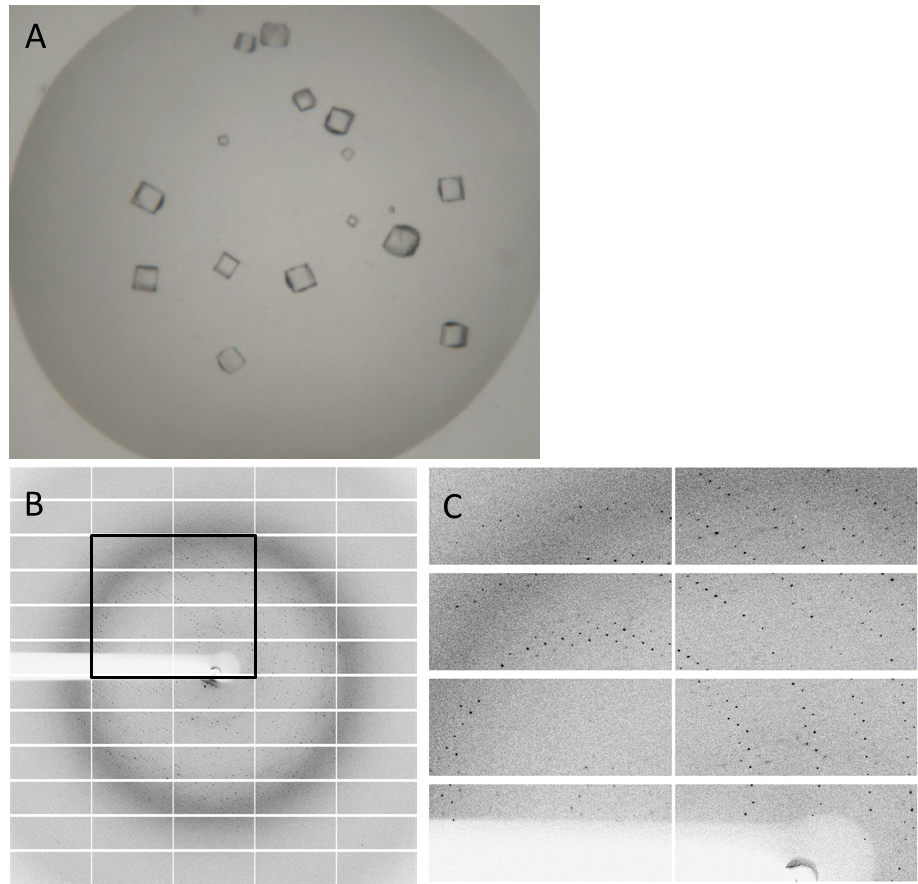
| - | Having dialyzed and concentrated the eluted protein, we obtained a protein concentration of about 10 mg/ml, which was sufficient to perform the crystallization reaction. Very nice crystals were obtained during trials when testing with different conditions (Fig. 4A). In order to find the perfect supplements for growing our crystals, the whole procedure was repeated. The crystals yielded an x-ray diffraction pattern, with a resolution of 2,8 Å (Fig. 4B,C). The dataset was measured at the EMBL Hamburg Beamline P13 at the PETRA III synchrotron on the DESY campus. | + | Having dialyzed and concentrated the eluted protein, we obtained a protein concentration of about 10 mg/ml, which was sufficient to perform the crystallization reaction. Very nice crystals were obtained during trials when testing with different conditions (Fig. 4A). In order to find the perfect supplements for growing our crystals, the whole procedure was repeated. The crystals yielded an x-ray diffraction pattern, with a resolution of 2,8 Å (Fig. 4B,C). The dataset was measured at the EMBL Hamburg Beamline P13 at the PETRA III synchrotron on the DESY campus, using a PILATUS2 6M X-ray detector (https://www.dectris.com/). |
Finally, we were also able to obtain the protein structure (Fig. 5)! The structure shows a globular protein with a distinct ATP-binding cleft. The ribbon model demonstrates the secondary structure composed of α-helices and β-sheets. | Finally, we were also able to obtain the protein structure (Fig. 5)! The structure shows a globular protein with a distinct ATP-binding cleft. The ribbon model demonstrates the secondary structure composed of α-helices and β-sheets. | ||
Revision as of 18:37, 27 October 2013
 "
"