Team:Paris Bettencourt/Project/Target
From 2013.igem.org
| Line 24: | Line 24: | ||
<h2>BioBricks</h2> | <h2>BioBricks</h2> | ||
<ol> | <ol> | ||
| - | <li><a href=" | + | <li><a href="http://parts.igem.org/Part:BBa_K1137000">BBa_K1137000 (SirA)</a></li> |
<li><a href="" target="_blank">BBa_K1137001 (FprA)</a></li> | <li><a href="" target="_blank">BBa_K1137001 (FprA)</a></li> | ||
<li><a href="" target="_blank">BBa_K1137002 (FdxA)</a></li> | <li><a href="" target="_blank">BBa_K1137002 (FdxA)</a></li> | ||
Revision as of 21:56, 4 October 2013

Background
SirA is an essential gene in latent tuberculosis infections
Results
- Produced an E. coli strain which relies upon mycobacterial sirA, fprA and fdxA genes to survive in M9 minimal media
- Demonstrated that E. coli can survive with mycobacterial sulfite reduction pathway with Flux Balance Analysis
- Located drug target sites on sirA as well as identified high structural similarity between cysI and sirA through structural anaylsis
Aims
To perform an drug screen targeted at the sirA gene from mycobacteria
Skip to Introduction
Skip to Modeling
Skip to Design
Skip to Results
Introduction
We have designed a drug screen assay to specifically target the mycobacterial sulfite reductase protein SirA, using M.Smegmatis’ synthetic sulfite reduction pathway cloned into an E. coli chassi. SirA is essential for M. tuberculosis persistence phenotype as sulfur containing amino acids are particularly sensitive to oxidative stress within the macrophage and must regularly be replaced. Currently, there are no drug candidates that are known to specifically inhibit SirA and conventional drug screens involve do not provide information regarding the mechanism of drug action nor do compounds that inhibit exponential growth necessarily have an effect on persistent TB. Our model overcomes the problem of the long doubling time of M. tuberculosis and informs us of specific inhibition of the sulfite reduction pathway. We achieve this by comparing a drug screen of our E. coli construct against its parent E. coli prior to the deletion of the native sulfite reduction pathway. Any drug candidates that have activity against both the wild-type E. coli and our construct are non-specific inhibitors of E. coli growth. However, any drug candidates that inhibit only the growth of our E. coli construct will have activity specific for our cloned pathway.

Flux Balance Analysis of Sulfite Reduction Pathway
We used an E. coli model iJR904 obtained from BiGG database as a starting model and obtained a growth rate represented by the f value of 0.9129. We then deleted the reaction ‘SULR’ which encodes for the sulphite reduction pathway involving cysI and obtained a f value of -8.63596783409936e-13 indicating that the sulphite reduction pathway is required for growth. Finally we introduced two new reactions for sirA and fprA and a new species fdxA. We found that growth was restored with the mycobacteria pathway resulting in a f value of 0.9105.
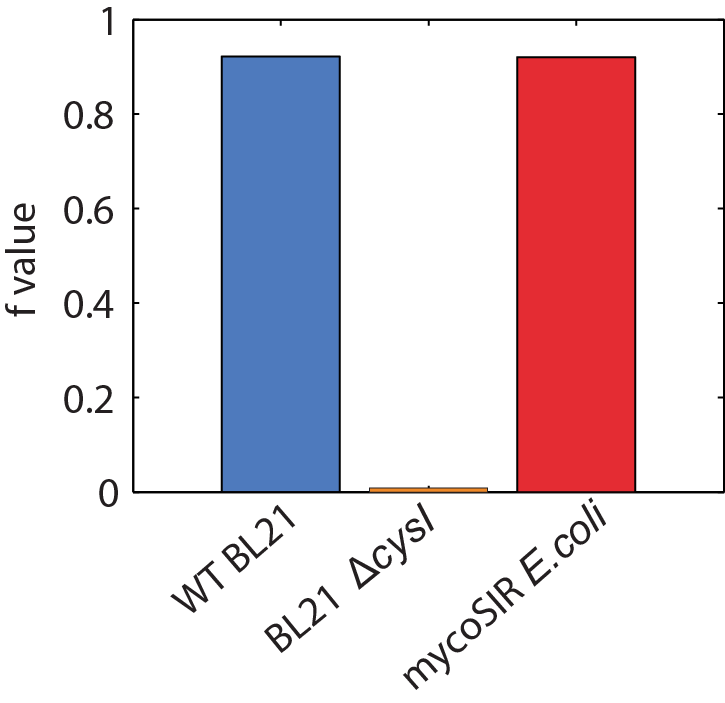
Biomass Flux through E. coli and mycoSIR E. coli
Structural Analysis of SirA
Our analysis has shown that the structures of M.tuberculosis SirA and E.coli CysI and particularly the active sites are very similar. Both proteins have the same symmetry (psuedo 2 fold) indicative of a common evolutionary origin. Superimposition analysis highlighted important conserved residues, involved in substrate binding to be Arg97, Arg130, Arg166, Lys207. These positively charged residues are conserved in the sulphite/nitrite reductase family. In addition, 4 Cys residues are conserved for iron-sulphur binding.
The most profound structural differences between SirA and E.coli are found in the ferredoxin binding site and SirA's most C terminal residues and several surface loop regions due to deletions or insertions. A stark difference is a covalent bond formed between Cys161 (thiolate) and Tyr69 (C carbon atom) found adjacent to the redox center (Cu ions) in SirA. The covalently bound residues act as a secondary cofactor in tyrosyl radical stabilization.

The superimposed 3D protein structures of SirA and CysI.
Synthetic Mycobacteria Pathway
We designed a synthetic M.smegmatis derived sulfite reduction pathway containing SirA; the sulfite reductase, and two supporting genes that are required for its function in E.coli; FdxA and FprA. FdxA is a mycobacterial Ferredoxin cofactor which is oxidised by SirA during the sulfite reduction reaction and FprA is a Ferredoxin-NADPH reductase use replenish the reduced Fdx pool. The sequence used as a template was taken from literature in which these genes were previously expressed in E. coli for purification and in vitro characterization of the enzyme kinetics with restriction sites removed and then codon optimized for expression in E. coli. These genes were then cloned into two Duet expression vectors, one containing sirA and one containing the supporting genes before being transformed into our knock out mutant strains of E. coli.

Growth curves:
Creation of Knock out Mutants
We prepared two strains of E. coli which have the sulfite reduction pathway deleted: BL21 (DE3) ΔCysI Δfpr ΔydbK and BL21 (AI) ΔCysI. CysI is responsible for sulfite reduction in E. coli, while fpr and ydbK are two non-essential genes that consume ferredoxin. These two genes are deleted, as sulfite reduction in mycobacteria is ferredoxin dependent in comparison to E. coli in which it is NADPH dependant. To ensure that these two genes do not interfere with our system, we deleted these genes as well.
Synthetic Corn Pathway
Additionally we prototyped the system with a reconstruction of a sulphite reduction pathway previously designed and published by the silver group (2011 Barstow et al). In place of CysI, a corn (Zea mays) derived sulfite reductase (zmSIR) was used. Two additional genes were included: Spinach ferredoxin (soFD), and corn derived ferredoxin NADP+ reductase (zmFNR). These genes, respectively, are required for production of the ferredoxin cofactor and the NADP+ ferredoxin reductase and are required for sulfite reductase (zmSIR) to function within E. coli.

Growth curves:
Results
Upon successful cloning of the three genes into our E. coli deletion strain, we continued to confirm that all three genes are required for growth on minimal media. Our two synthetic pathways were found to rescue growth on a sulfurless amino acid supplemented minimal media. We hope that this technique of using synthetic biology to overcome problems faced in naturally occurring systems will be both a large boon to the pursuit of finding novel drug candidates in M. tuberculosis and more broadly as this technique can be used for high-throughput screening of any pathway that can be constructed to be essential for growth in E. coli.

Growth of zmSIR E.coli on minimal media.
 "
"







 +33 1 44 41 25 22/25
+33 1 44 41 25 22/25


