Team:SydneyUni Australia/Project
From 2013.igem.org
(Difference between revisions)
m (→Overall project) |
m (→'Brief Project Description) |
||
| Line 33: | Line 33: | ||
| - | == '''Brief Project Description'' == | + | == '''Brief Project Description''' == |
Revision as of 12:17, 15 July 2013
This is a template page. READ THESE INSTRUCTIONS.
You are provided with this team page template with which to start the iGEM season. You may choose to personalize it to fit your team but keep the same "look." Or you may choose to take your team wiki to a different level and design your own wiki. You can find some examples HERE.
You MUST have all of the pages listed in the menu below with the names specified. PLEASE keep all of your pages within your teams namespace.
| Home | Team | Official Team Profile | Project | Parts Submitted to the Registry | Modeling | Notebook | Safety | Attributions |
|---|
Contents |
Brief Project Description
Background
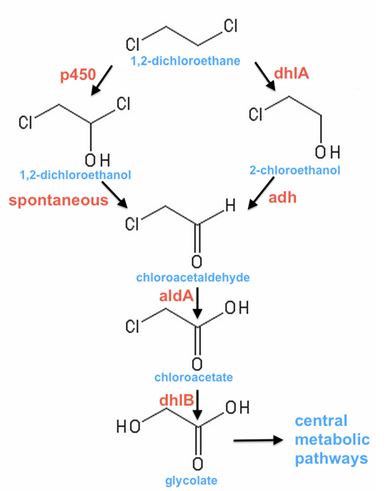
- 1,2-dichloroethane (DCA) is part of a family of chlorinated hydrocarbons that are primarily derived from industrial solvents such as tetrachloroethene (PCE) and trichloroethene (TCE). Some of these organochlorines are toxic, carcinogenic and generally nasty.
- DCA is a soluble and mobile contaminant of the groundwater in Botany Bay, Sydney, but also elsewhere around the world.
- Conventional treatment involves significant costs, such as the pumping and heat-stripping of groundwater at Botany Bay. A biological alternative may be cheaper and more effective.
- A suite of techniques (including sampling of contaminated sites, growth in bioreactors with selective conditions and protein engineering) have led to an understanding that there are two primary pathways of DCA-degradation (Fig. 1).
Goals
- Construct a BioBrick-compatible vector inspired by the broad host range vector, pBBR1MCS2.
- Construct and compare two of the proposed pathways of DCA biodegradation.
- Characterise the components of the DCA-degradation pathway for admission into the Registry of Standard Parts.
- (If we get time!) Demonstrate the integration of our the pathway in the chromosome of Pseudomonas stutzeri via natural transformation and site-specific recombination.
 "
"