09/08/13
From 2013.igem.org
(Difference between revisions)
(→Running an agarose gel) |
(→Running an agarose gel) |
||
| Line 51: | Line 51: | ||
[[File:lim_chlorodig_sacI_090813.jpg]] | [[File:lim_chlorodig_sacI_090813.jpg]] | ||
| + | |||
| + | *The lanes are as expected. | ||
| + | *Samples 5.1, 10.1 and 10.3 were expected to show 3 fragments, sized 2070, 1890 and 1529bp. This is what is shown on the gel. | ||
| + | *Sample 10.3 only has one SacI restriction site and when cut, a band of 2070bp is expected. That is also shown on the gel. | ||
==Glycerol stocks== | ==Glycerol stocks== | ||
Revision as of 09:29, 12 August 2013
| Home | Team | Official Team Profile | Project | Parts Submitted to the Registry | Modeling | Notebook | Safety | Attributions |
|---|
Contents |
Isolating plasmid
- From overnight culture, took 3ml of culture and centrifuged into pellet of samples 5.1, 10.1, 10.2, 10.3
- Isolated the plasmid using omega bio-tek Plasmid mini kit I
- Concentrations from nano drop are shown in the table below
| Sample | Volume(ul) | Conc.(ng/ul) | 260/280 | 260/230 |
| 5.1 | 92 | 65.8 | 1.87 | 1.75 |
| 10.1 | 88 | 36.9 | 1.80 | 1.77 |
| 10.2 | 89 | 48.3 | 1.79 | 1.98 |
| 10.3 | 94 | 17.4 | 1.84 | 1.59 |
Digesting the plasmids for restriction mapping
- Digesting 200ng of DNA with SacI
- 1ul of SacI
- 2ul of NEB buffer 1.1
- DNA volumes added for samples 5.1 to 10.3:
- 3ul; 5ul; 4ul; 11.5ul
- Water added for samples 5.1 to 10.3:
- 14ul; 12ul; 13ul; 5.5ul
- Digestion in 37C water bath for 30mins
- Heat kill at 80C for 20mins
Running an agarose gel
- Add 5ul of 6x orange G to each sample
- 1kb marker is used
- Wells are loaded in the following order:
- 5ul of marker, 25ul of samples 5.1; 10.1; 10.2; 10.3, 5ul of marker
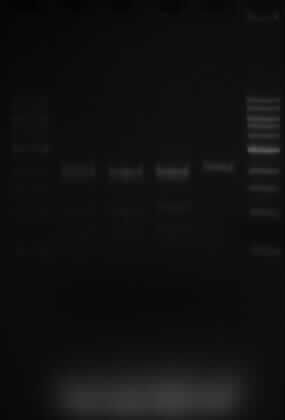
- The lanes are as expected.
- Samples 5.1, 10.1 and 10.3 were expected to show 3 fragments, sized 2070, 1890 and 1529bp. This is what is shown on the gel.
- Sample 10.3 only has one SacI restriction site and when cut, a band of 2070bp is expected. That is also shown on the gel.
Glycerol stocks
- Took 750ul from overnight culture and centrifuged
- Supernatant was removed
- Pellet resuspended in 375ul of HMFM
- samples were then frozen at -80
Sonicating Herring sperm DNA
- The herring Sperm was to viscous, so to reduce viscosity the sonicator was used
- 1ml of core DNA used in 2 tubes, 1 control and 1 to go in the sonicator
- The control tube was left on the bench at room temperature
- The 2nd tube was put in the sonicator for the folowing times: 2mins, 2mins, 5mins, 5mins, 20mins, 20mins, 30mins
- Each time both tubes were filmed to compare viscosity
 "
"