Team:UGent/Project
From 2013.igem.org
| Line 44: | Line 44: | ||
[[File:UGent_2013_CIChE.jpg|thumb|400px|center|Tyo et al., 2009]] | [[File:UGent_2013_CIChE.jpg|thumb|400px|center|Tyo et al., 2009]] | ||
<html> | <html> | ||
| - | <p>This process is called chromosomal evolution. When the desired gene copy number is reached | + | <p>This process is called chromosomal evolution. When the desired gene copy number is reached <i>recA</i> is deleted, thereby fixing the copy number.</p> |
| + | <p>The use of CIChE-strains poses several advantages. They require no selection markers and can be cultured without the addition of antibiotics to the medium. This is in contrast to strains containing plasmids, which still need antibiotic selection. Also, in CIChE-strains yields can be tuned by varying the antibiotic concentration during chromosomal evolution.</p> | ||
</html> | </html> | ||
{{:Team:UGent/Templates/ToggleBoxEnd}} | {{:Team:UGent/Templates/ToggleBoxEnd}} | ||
| Line 56: | Line 57: | ||
<p>Antibiotic resistance is the resistance of a bacterium to an antibacterial medicine a.k.a. antibiotic to which it was originally sensitive. When a pathogenic bacterium has acquired resistance to a certain antibiotic, infections of this bacterium can no longer be treated with the antibiotic in question since the bacterium has become insensitive to the antibiotic. | <p>Antibiotic resistance is the resistance of a bacterium to an antibacterial medicine a.k.a. antibiotic to which it was originally sensitive. When a pathogenic bacterium has acquired resistance to a certain antibiotic, infections of this bacterium can no longer be treated with the antibiotic in question since the bacterium has become insensitive to the antibiotic. | ||
Overuse and misuse of antibiotics has accelerated the emergence and spread of resistant bacteria. Today antibiotic resistance has become a fairly well-known term as a result of the recurring appearance of multi-drug resistant bacteria such as MRSA and VRE in media, awareness campaigns etc. These bacteria have become resistant to the commonly used antibiotics. As a result infections are very difficult to treat. The emergence of these hard-to-kill pathogenic bacteria poses a serious risk to public health. | Overuse and misuse of antibiotics has accelerated the emergence and spread of resistant bacteria. Today antibiotic resistance has become a fairly well-known term as a result of the recurring appearance of multi-drug resistant bacteria such as MRSA and VRE in media, awareness campaigns etc. These bacteria have become resistant to the commonly used antibiotics. As a result infections are very difficult to treat. The emergence of these hard-to-kill pathogenic bacteria poses a serious risk to public health. | ||
| - | < | + | </p><p> |
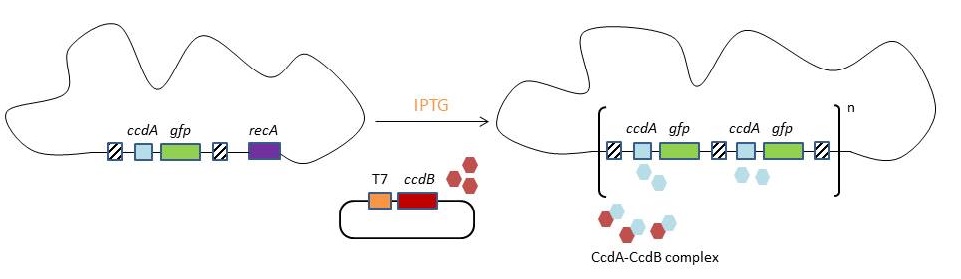
| - | In biotechnology antibiotic resistance genes (the genes that make a bacterium resistant to a certain antibiotic) are often used as selectable markers. The best known application is probably the selection of plasmids. In the original CIChE technique a chloramphenicol resistance gene is used as selectable marker. During chromosomal evolution the antibiotic resistance gene is duplicated since it is the driving pressure for the tandem duplication of the CIChE-construct. As a result the final bacterial strain carries multiple antibiotic resistance genes. The creation of such a strain raises several safety concerns as bacteria can pass resistance genes on to other, possibly pathogenic, bacteria in the environment through a process called horizontal gene transfer. Although | + | In biotechnology antibiotic resistance genes (the genes that make a bacterium resistant to a certain antibiotic) are often used as selectable markers. The best known application is probably the selection of plasmids. In the original CIChE technique a chloramphenicol resistance gene is used as selectable marker. During chromosomal evolution the antibiotic resistance gene is duplicated since it is the driving pressure for the tandem duplication of the CIChE-construct. As a result the final bacterial strain carries multiple antibiotic resistance genes. The creation of such a strain raises several safety concerns as bacteria can pass resistance genes on to other, possibly pathogenic, bacteria in the environment through a process called <i>horizontal gene transfer</i>. Although it is very unlikely to occur the possibility of horizontal gene transfer cannot be entirely ruled out. </p> |
| + | <p>The fact that the strain eventually contains multiple antibiotic resistance genes withholds this CIChE technique from being applicable in for example food industry, given several regulations as well as consumer’s mind-set. We will try to replace the use of antibiotic resistance genes with a toxin-antitoxin system. By eliminating the need for antibiotic resistance genes, CIChE could be applied in the industry without having to worry about the possibility of horizontal gene transfer and the spread of antibiotic resistance.</p> | ||
</html> | </html> | ||
Revision as of 12:33, 14 September 2013
|
Introduction: high gene expression in industrial biotechnologyThe main goal of industrial biotechnology is to increase the yield of biochemical products using microorganisms as production hosts. This includes engineering large synthetic pathways and improving their expression. Overexpression of genes has mainly been achieved by using high or medium copy plasmids. However, studies have demonstrated that plasmid-bearing cells lose their productivity fairly quickly as a result of genetic instability.
In industrial biotechnology, a common technique to express new synthetic products and pathways is the use of plasmids as vectors. Plasmids are easy to insert into cells and replicate independently from the genome, allowing strong gene expression. Overexpression is easily achieved by using plasmids with a medium or high copy number, different promoter systems, ribosome binding sites (RBS), etc. Thanks to plasmids, the industrial biotechnology has grown substantially over the past years. However, the use of plasmids entails some important disadvantages: plasmid maintenance imposes a metabolic burden on cells and plasmids suffer from genetic instability. Metabolic burdenWhen plasmids are present in cells and replicate, they create a metabolic burden. This is defined by Bentley et al. (1990) as “the amount of resources that are taken from the host cell metabolism for foreign DNA maintenance and replication”. As a result, metabolic load causes many alterations to the physiology and metabolism of the cell and reduces the cellular fitness. The most common change is a delayed growth. This can be caused by the fact that new pathways for energy generation are activated due to the competition between cell propagation and plasmid replication. This growth retardation leads to a lower yield of the desired product. Genetic instabilityPlasmids are genetically instable due to three processes: segregational instability, structural instability and allele segregation.
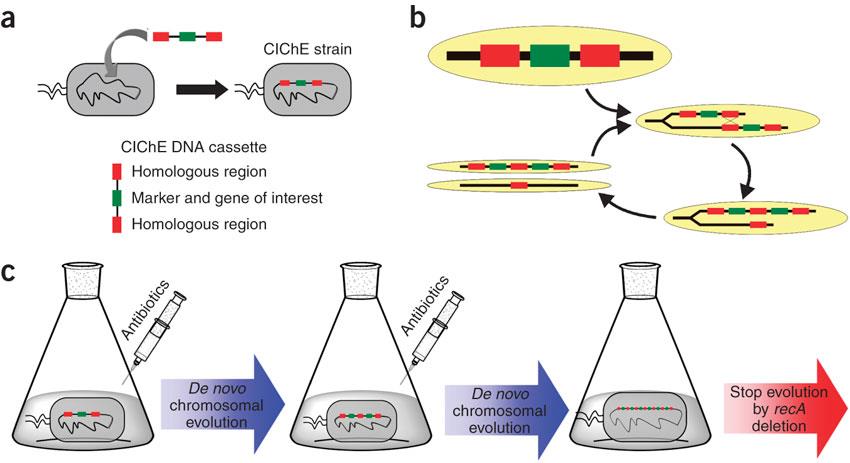
Therefore a new method was developed for the overexpression of a gene of interest in the bacterial chromosome: Chemically Inducible Chromosomal evolution (CIChE). In this technique the chromosome is evolved to contain a higher number of gene copies by adding a chemical inducer.
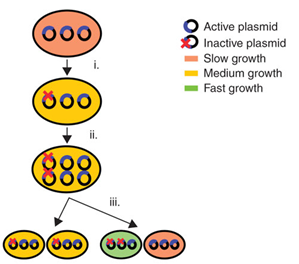
In 2009, Tyo et al. developed a technique for the stable, high copy expression of a gene of interest in E. coli without the use of high copy number plasmids, thus avoiding their previously stated negative characteristics. They called this plasmid-free, high gene copy expression system ‘chemically inducible chromosomal evolution’. In this method, the gene of interest is integrated in the microbial genome and then amplified to achieve multiple copies and reach the desired expression level. Genomic integration guarantees ordered inheritance, resolving the problem of allele segregation. CIChE works as follows: First a construct, containing the gene(s) of interest and the antibiotic marker chloramphenicol acetyl transferase (cat) flanked by homologous regions, is delivered to and subsequently integrated into the E. coli genome. The construct can be amplified in the genome through tandem gene duplication by recA homologous recombination. Then the strain is cultured in increasing concentrations of chloramphenicol, providing a growth advantage for cells with increased repeats of the construct and thereby selecting for bacteria with a higher gene copy number (Figure).
This process is called chromosomal evolution. When the desired gene copy number is reached recA is deleted, thereby fixing the copy number. The use of CIChE-strains poses several advantages. They require no selection markers and can be cultured without the addition of antibiotics to the medium. This is in contrast to strains containing plasmids, which still need antibiotic selection. Also, in CIChE-strains yields can be tuned by varying the antibiotic concentration during chromosomal evolution. CloseIt has been shown that approximately 40 and even up to 50 gene copies can be attained using chromosomal evolution. It has also been demonstrated that, while plasmid-bearing strains lose their productivity after 40 generations due to allele segregation, gene copy number and productivity of CIChE-strains remain stable even after 70 generations. This genetic stability is considered to be the most important asset of CIChE. The original model for CIChE, however, results in bacterial strains containing a large number of antibiotic resistance genes. To make this valuable technique more widely applicable in the industry, we developed a model for chromosomal evolution based on a toxin-antitoxin system instead of antibiotic resistance. | |||
 "
"