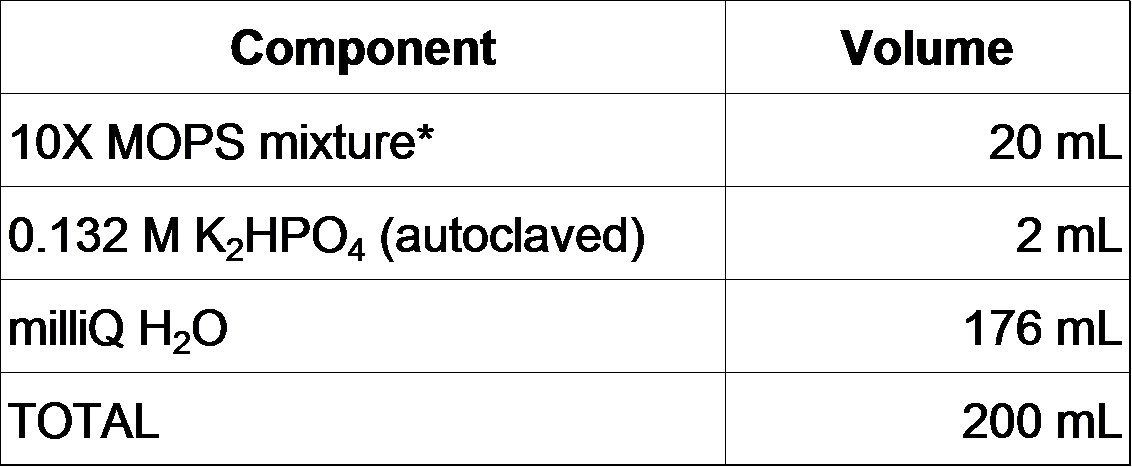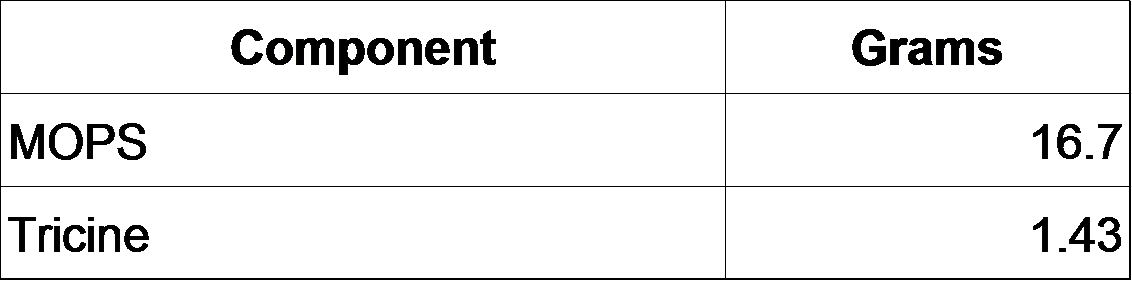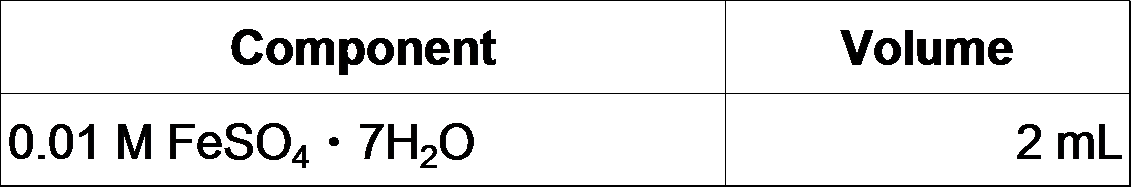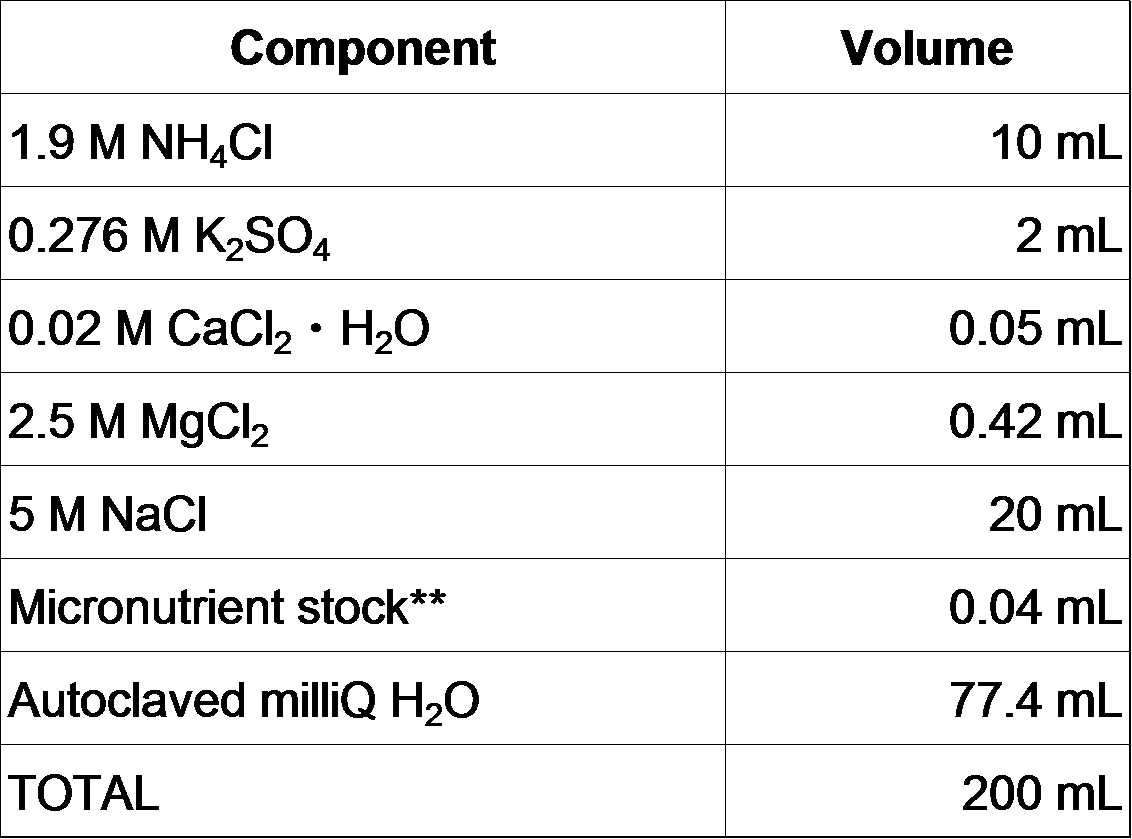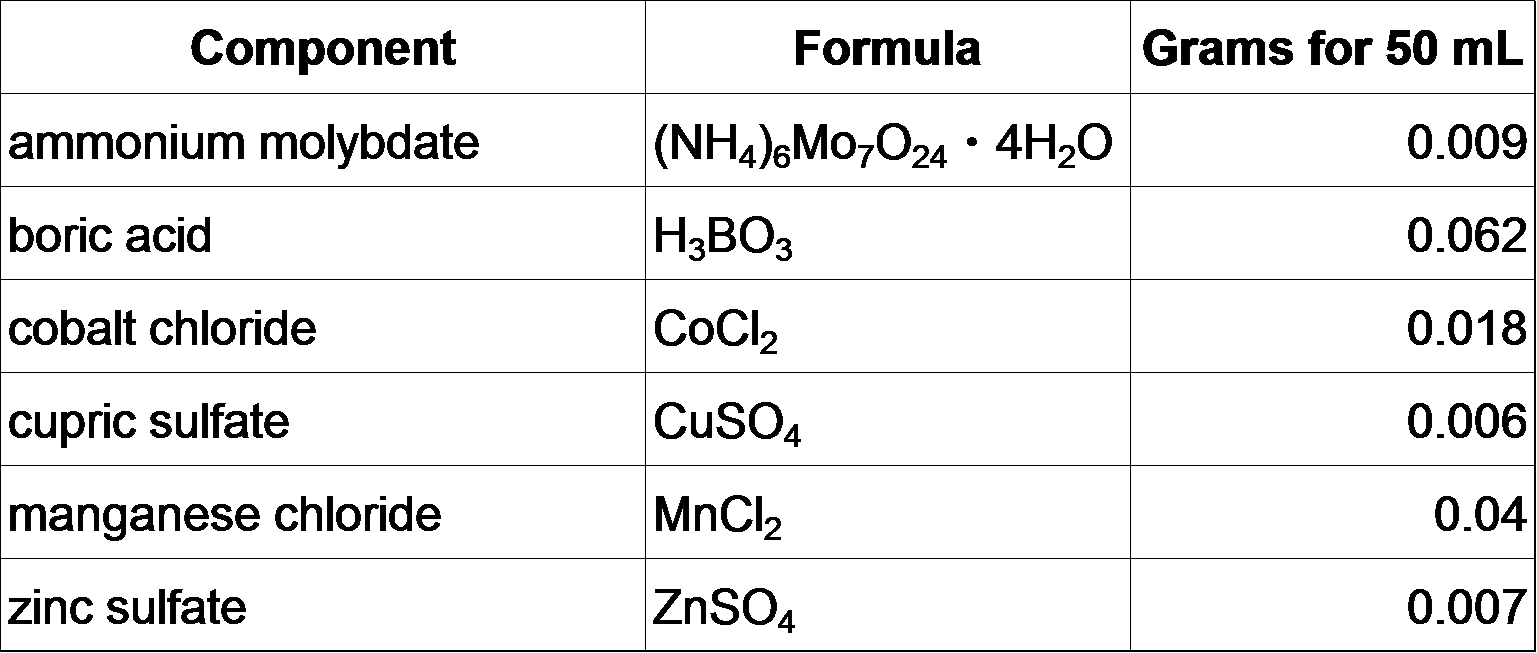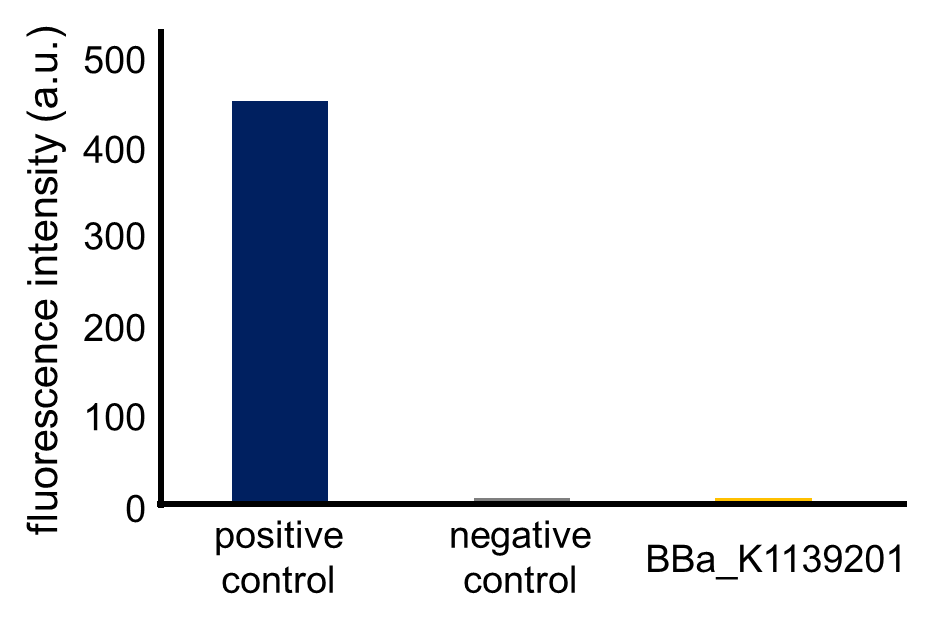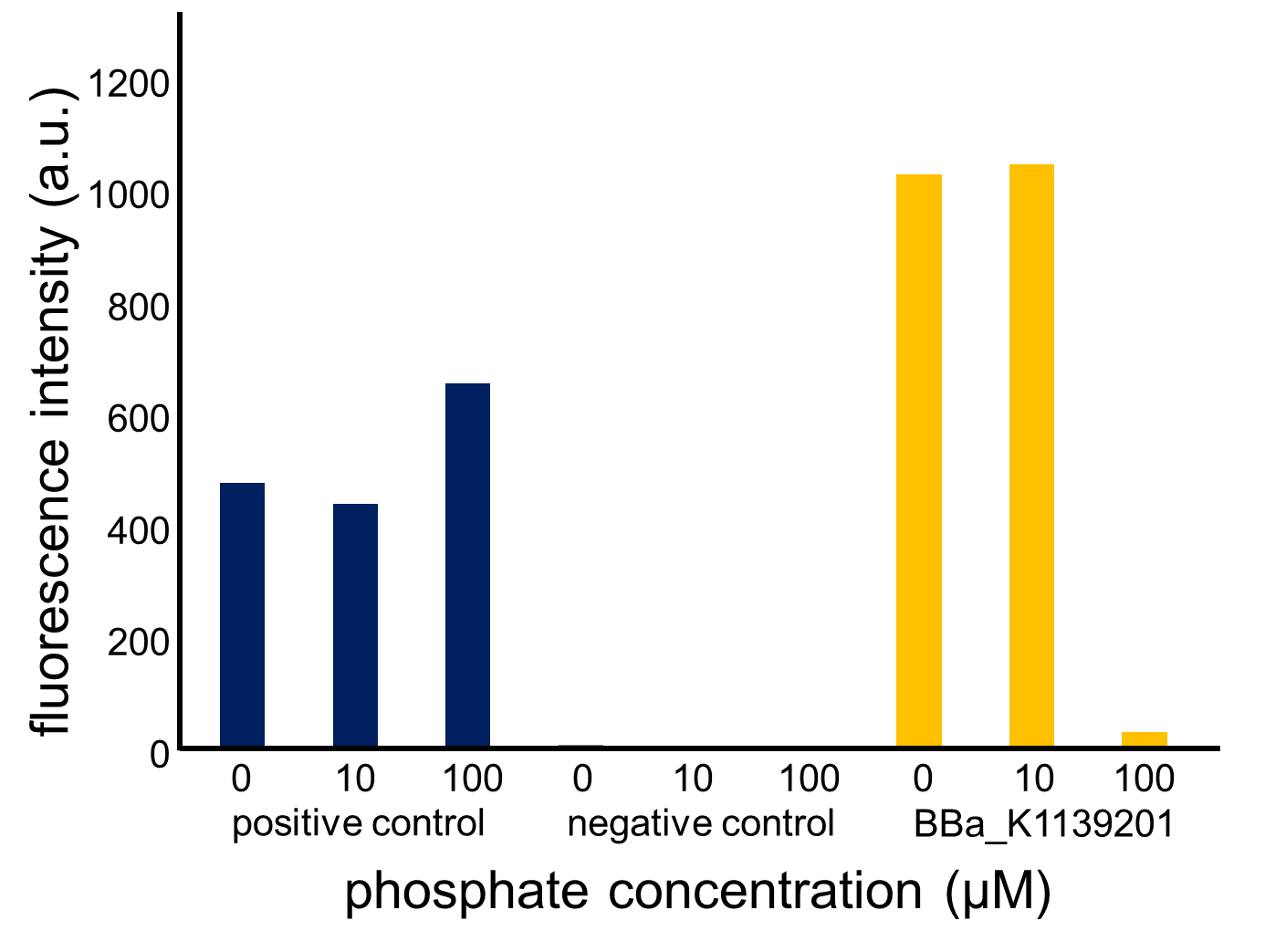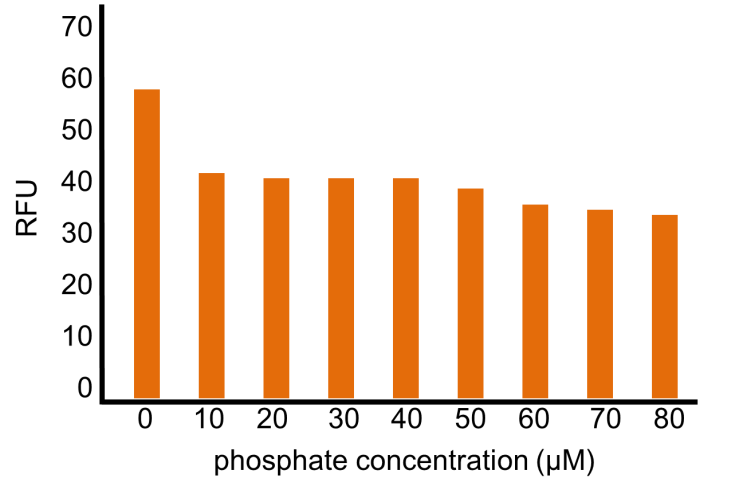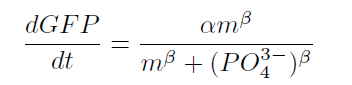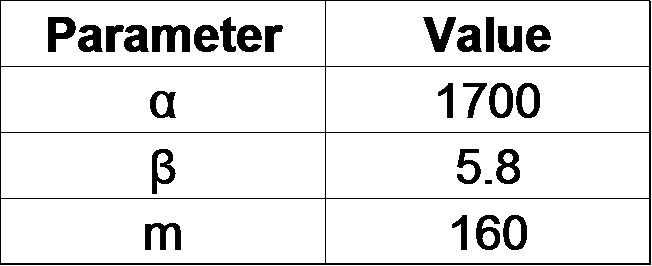Team:Tokyo Tech/Experiment/phoA Promoter Assay
From 2013.igem.org
| Line 1: | Line 1: | ||
{{tokyotechmenudark}} | {{tokyotechmenudark}} | ||
<div id="text-area"><br> | <div id="text-area"><br> | ||
| + | <div class="box" id="title"> | ||
| + | <p style="line-height:0em;"><i>phoA</i> Promoter Assay</p> | ||
| + | </div> | ||
<div class="box"> | <div class="box"> | ||
| - | + | <h1>1. Introduction </h1> | |
| - | <h1> | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | 1. Introduction | + | |
| - | </ | + | |
<h2> | <h2> | ||
<p> | <p> | ||
| Line 18: | Line 15: | ||
</h2> | </h2> | ||
| - | < | + | <h1>2. Materials and Methods</h1> |
| - | 2. Materials and Methods | + | |
| - | </ | + | |
<h2> | <h2> | ||
| - | < | + | <h3>2-1. Construction </h3> |
| - | 2-1. Construction | + | |
| - | </ | + | |
<div style="margin-left:3em; text-indent:0em;"> | <div style="margin-left:3em; text-indent:0em;"> | ||
<br> | <br> | ||
| Line 43: | Line 36: | ||
[[Image:Titech2013_phoa_Fig_3-5-6.png|450px|thumb|center|Fig. 3-5-6. Steps for PCR of <i>phoA</i> promoter]] | [[Image:Titech2013_phoa_Fig_3-5-6.png|450px|thumb|center|Fig. 3-5-6. Steps for PCR of <i>phoA</i> promoter]] | ||
| - | < | + | <h3>2-2. Assay Protocol</h3> |
| - | 2-2. Assay Protocol | + | |
| - | </ | + | |
<div style="margin-left:50px;"> | <div style="margin-left:50px;"> | ||
2-2-0. Prepare MOPS minimal medium as follows (F. Neidhardt et al., 1974).<br> | 2-2-0. Prepare MOPS minimal medium as follows (F. Neidhardt et al., 1974).<br> | ||
| Line 109: | Line 100: | ||
</div> | </div> | ||
</h2> | </h2> | ||
| - | < | + | <h1>3. Results</h1> |
<h2> | <h2> | ||
<p>3-1. Before inducing by phosphate concentration | <p>3-1. Before inducing by phosphate concentration | ||
| Line 123: | Line 114: | ||
[[Image:Titech2013_phoa_Fig_3-5-14.png|450px|thumb|center|Fig. 3-5-14. Picture of fluorescence of the induced cells]] | [[Image:Titech2013_phoa_Fig_3-5-14.png|450px|thumb|center|Fig. 3-5-14. Picture of fluorescence of the induced cells]] | ||
</h2> | </h2> | ||
| - | < | + | <h1>4. Discussion</h1> |
| - | 4. Discussion | + | |
| - | </ | + | |
<h2> | <h2> | ||
<p>We confirmed that the increase in phosphate concentration repressed the <i>phoA</i> promoter. Compared to OUC-China’s phosphate sensor part including <i>phoB</i> promoter (Fig. 3-5-16), our phosphate sensor part shows clearer result (Fig. 3-5-15). | <p>We confirmed that the increase in phosphate concentration repressed the <i>phoA</i> promoter. Compared to OUC-China’s phosphate sensor part including <i>phoB</i> promoter (Fig. 3-5-16), our phosphate sensor part shows clearer result (Fig. 3-5-15). | ||
| Line 133: | Line 122: | ||
<br><br><br><br><br><br> | <br><br><br><br><br><br> | ||
</h2> | </h2> | ||
| - | < | + | <h1>5. Modeling</h1> |
| - | 5. Modeling | + | |
| - | </ | + | |
<h2> | <h2> | ||
<p>From our results explained above, we determined parameters for the induction mechanism. By fitting the results to the following Hill equation (Fig. 3-5-17), we identified m and the hill coefficient. Those parameters (Fig. 3-5-18) will be used in our future modeling. Plants are reported to be in phosphate starvation under the concentration of 1 mM (D. Hoagland et al., 1950). Our part can also sense the concentration below 1 mM (Fig. 3-5-19). Therefore, our improved part is useful for our farming circuit. We also identified maximum GFP production rate in this construct. | <p>From our results explained above, we determined parameters for the induction mechanism. By fitting the results to the following Hill equation (Fig. 3-5-17), we identified m and the hill coefficient. Those parameters (Fig. 3-5-18) will be used in our future modeling. Plants are reported to be in phosphate starvation under the concentration of 1 mM (D. Hoagland et al., 1950). Our part can also sense the concentration below 1 mM (Fig. 3-5-19). Therefore, our improved part is useful for our farming circuit. We also identified maximum GFP production rate in this construct. | ||
| Line 143: | Line 130: | ||
[[Image:Titech2013_phoa_Fig_3-5-19.png|450px|thumb|center|Fig. 3-5-19. A model with fitting the results of our assay]] | [[Image:Titech2013_phoa_Fig_3-5-19.png|450px|thumb|center|Fig. 3-5-19. A model with fitting the results of our assay]] | ||
</h2> | </h2> | ||
| - | < | + | <h1>6. Reference</h1> |
| - | </ | + | |
<h2> | <h2> | ||
<p>1. M. Dollard et al. “Whole-cell bacterial sensors for the monitoring of phosphate availability” Journal of Microbiological Methods (2003) | <p>1. M. Dollard et al. “Whole-cell bacterial sensors for the monitoring of phosphate availability” Journal of Microbiological Methods (2003) | ||
Revision as of 03:59, 27 September 2013
phoA Promoter Assay
Contents |
1. Introduction
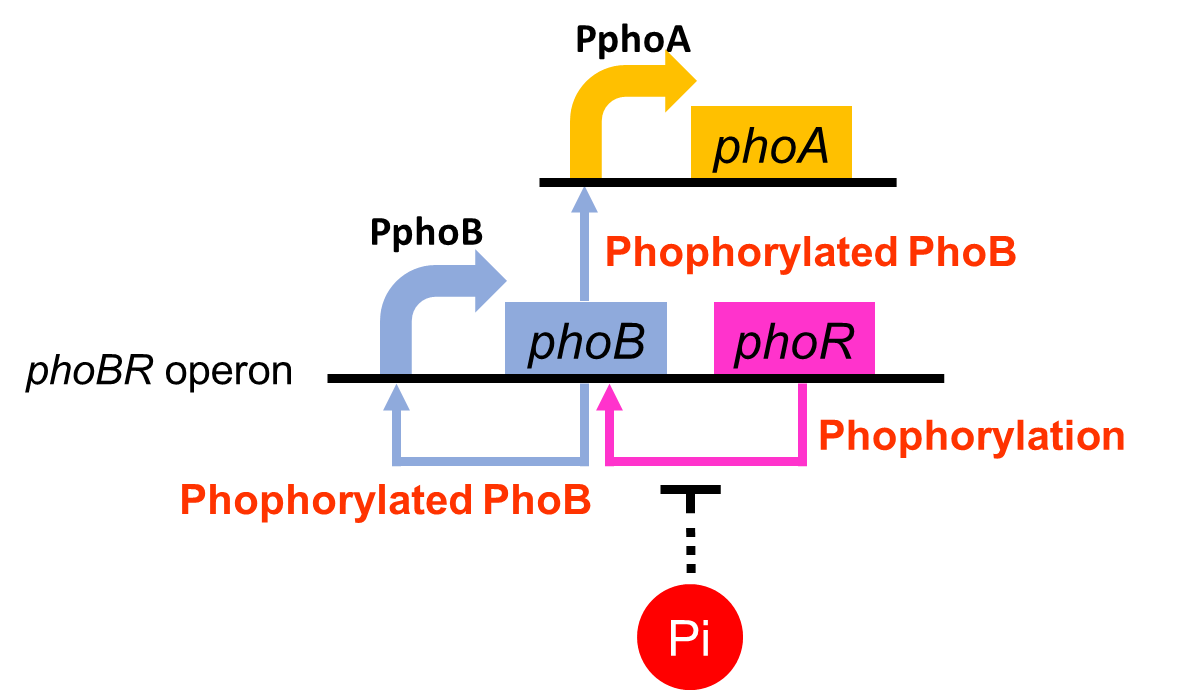
To realize our farming project, we improved a phosphate sensor part ([http://parts.igem.org/Part:BBa_K1139201 BBa_ K1139201], Fig. 3-5-1) since the existing phosphate sensor part (OUC-China 2012, [http://parts.igem.org/Part:BBa_K116401 BBa_K116401]) does not have sufficient data. Our part includes the inducible promoter of the alkaline phosphatase gene (phoA) derived from E.coli (M. Dollard et al., 2003). The phoA promoter is repressed by high concentration phosphate. A brief system of the phoA promoter is shown in Fig. 3-5-2. The phoA gene is regulated by phoB and phoR, which belong to pho regulon (H. Shinagawa et al., 1983). Phosphorylated PhoB activates the expression of phoA. Under conditions of phosphate limitation, PhoR phosphorylates PhoB. On the other hand, under conditions of high phosphate concentrations, PhoR dephosphorylate phospho-PhoB (Y. Hsieh et al., 2010). We amplified the phoA promoter from E. coli (MG1655). We ligated this promoter upstream of GFP part and introduced this part into E.coli (MG1655). By following induction assay, we confirmed that the phoA promoter was actually repressed by the increase in phosphate concentration
2. Materials and Methods
2-1. Construction
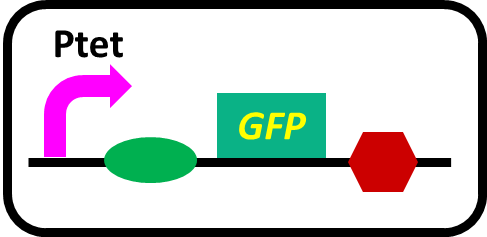
-pSB6A1-PphoA-GFP (MG1655)…BBa_K1139201
-pSB6A1-Ptet-GFP (MG1655)… positive control
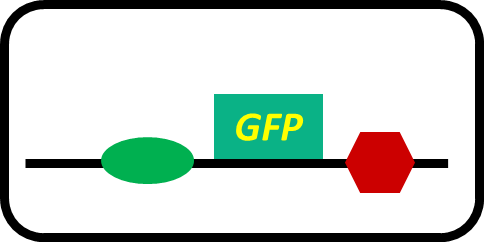
-pSB6A1-promoterless-GFP (MG1655)… negative control
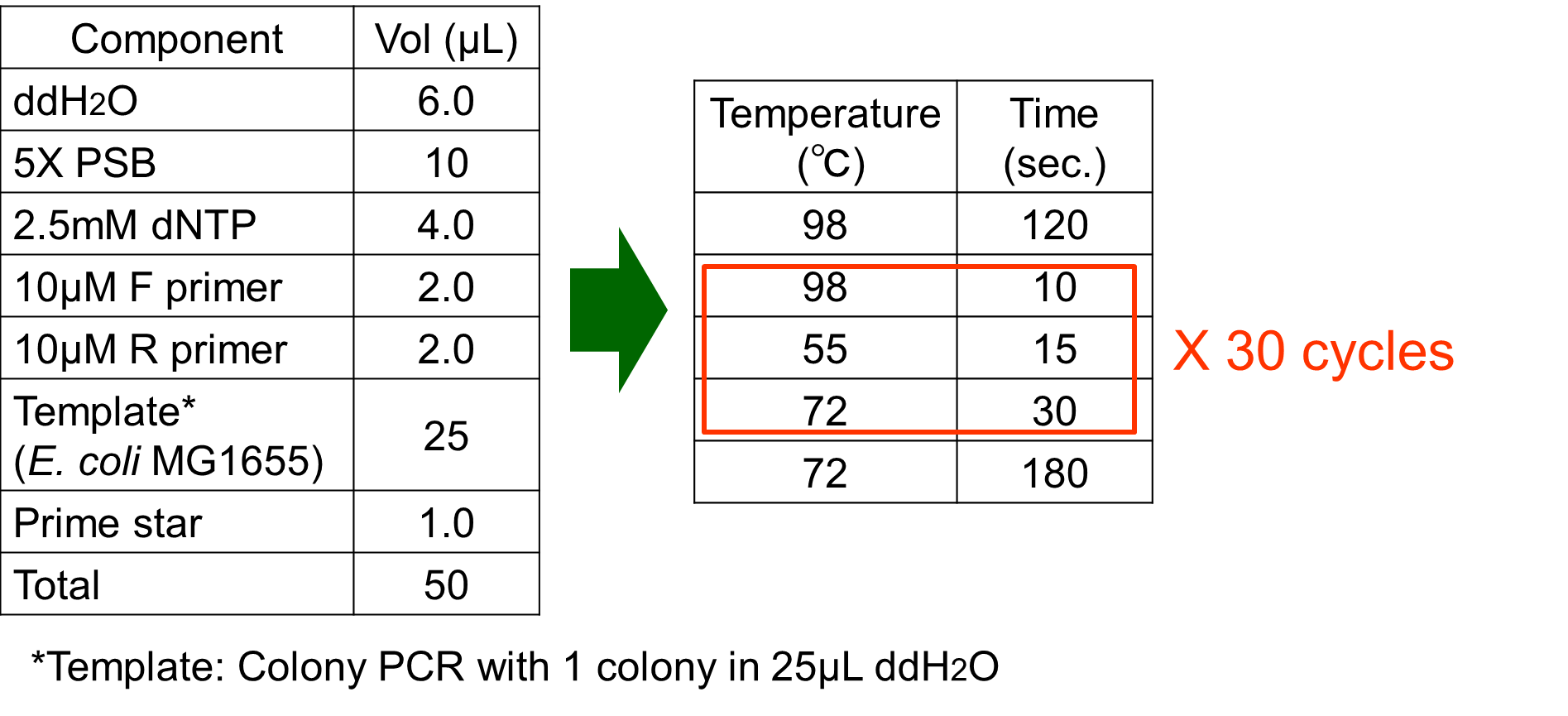
The phoA promoter region of E. coli was amplified from MG1655 genomic DNA by PCR using upstream primer (5’-acgtgaattcgcggccgcttctagagaaagttaatcttttcaacagctgtcataaag-3’) and downstream primer (5’ccgctactagtaaatacattaaaaaataaaaacaaagcgactataagtctc-3’). Amplification was carried out with the steps shown in Fig. 3-5-6.
2-2. Assay Protocol
2-2-0. Prepare MOPS minimal medium as follows (F. Neidhardt et al., 1974).
Also, prepare a series of phosphate concentration gradient 1 X MOPS by changing the volume of K2HPO4 (We prepared the series as 0, 10, 30, 100, 300, 1000 microM).
[Prepare MOPS minimal medium]
200 mL of 1 X MOPS is prepared as follows.
1. Mix ingredients above and adjust the pH to 7.2 with 5 M NaOH.
2. Filter sterilize. It can be stored at 4°C for up to 1 month.
3. Before use, add carbon source (we used a final concentration of 0.1% glucose).
*10 X MOPS mixture (200 mL)
1. Add the following to ~60 mL milliQ H2O:
2. Add 5 M KOH to a final pH of 7.4.
3. Bring total volume to 88 mL.
4. Make fresh FeSO4 solution and add it to the MOPS/Tricine solution:
5. Add the following solutions to the MOPS/Tricine/FeSO4 solution.
(Mix in the order shown)
6. Filter sterilize with 0.2 micron filter.
7. Aliquot into sterile plastic bottle and freeze at -20°C.
**Micronutrient stock (50 mL)
Mix everything together in 40 mL autoclaved milliQ H2O, bring up total volume to 50 mL.
Store at room temperature.
2-2-1. Prepare overnight culture of BBa_K1139201, positive control and negative control, each in MOPS medium (including 1.32 mM K2HPO4) containing ampicillin (50 microg/mL) at 37°C.
2-2-2. Dilute the overnight cultures to an OD600 of 0.1 in fresh MOPS medium (3 mL) containing ampicillin (50 microg/mL). (→fresh cultures)
2-2-3. Incubate the fresh cultures until the observed OD600 reaches 0.4-0.6.
2-2-4. Centrifuge the cells at 6000g, 25°C, 10 minutes, wash twice with MOPS minimal medium without phosphate containing ampicillin (50 microg/mL), and then suspend in the same medium to obtain a final OD600 of 10.
2-2-5. Add 300 microL of prepared cell suspension to 2.7 mL of test solution, the series of phosphate concentration gradient 1 X MOPS, containing ampicillin (50 microg/mL).
2-2-6. Incubate the cells for 140 minutes at 26°C.
2-2-7. 1 mL of each culture was harvested by centrifugation and suspended by adding 1 mL of PBS (phosphate-buffered saline). Dilute the suspension to obtain a final OD600 of around 0.2 by PBS.
2-2-8. Dispense 600 microL of each suspension into a disposable tube through a cell strainer. Fluorescence intensity was measured with a flow cytometer of Becton, Dickinson and Company.
-pSB6A1-PphoA-GFP (MG1655)…BBa_K1139201
-pSB6A1-Ptet-GFP (MG1655)… positive control
-pSB6A1-promoterless-GFP (MG1655)… negative control
The phoA promoter region of E. coli was amplified from MG1655 genomic DNA by PCR using upstream primer (5’-acgtgaattcgcggccgcttctagagaaagttaatcttttcaacagctgtcataaag-3’) and downstream primer (5’ccgctactagtaaatacattaaaaaataaaaacaaagcgactataagtctc-3’). Amplification was carried out with the steps shown in Fig. 3-5-6.
2-2-0. Prepare MOPS minimal medium as follows (F. Neidhardt et al., 1974).
Also, prepare a series of phosphate concentration gradient 1 X MOPS by changing the volume of K2HPO4 (We prepared the series as 0, 10, 30, 100, 300, 1000 microM).
[Prepare MOPS minimal medium]
200 mL of 1 X MOPS is prepared as follows.
1. Mix ingredients above and adjust the pH to 7.2 with 5 M NaOH.
2. Filter sterilize. It can be stored at 4°C for up to 1 month.
3. Before use, add carbon source (we used a final concentration of 0.1% glucose).
*10 X MOPS mixture (200 mL)
1. Add the following to ~60 mL milliQ H2O:
2. Add 5 M KOH to a final pH of 7.4.
3. Bring total volume to 88 mL.
4. Make fresh FeSO4 solution and add it to the MOPS/Tricine solution:
5. Add the following solutions to the MOPS/Tricine/FeSO4 solution. (Mix in the order shown)
6. Filter sterilize with 0.2 micron filter.
7. Aliquot into sterile plastic bottle and freeze at -20°C.
**Micronutrient stock (50 mL)
Mix everything together in 40 mL autoclaved milliQ H2O, bring up total volume to 50 mL. Store at room temperature.
2-2-1. Prepare overnight culture of BBa_K1139201, positive control and negative control, each in MOPS medium (including 1.32 mM K2HPO4) containing ampicillin (50 microg/mL) at 37°C.
2-2-2. Dilute the overnight cultures to an OD600 of 0.1 in fresh MOPS medium (3 mL) containing ampicillin (50 microg/mL). (→fresh cultures)
2-2-3. Incubate the fresh cultures until the observed OD600 reaches 0.4-0.6.
2-2-4. Centrifuge the cells at 6000g, 25°C, 10 minutes, wash twice with MOPS minimal medium without phosphate containing ampicillin (50 microg/mL), and then suspend in the same medium to obtain a final OD600 of 10.
2-2-5. Add 300 microL of prepared cell suspension to 2.7 mL of test solution, the series of phosphate concentration gradient 1 X MOPS, containing ampicillin (50 microg/mL).
2-2-6. Incubate the cells for 140 minutes at 26°C.
2-2-7. 1 mL of each culture was harvested by centrifugation and suspended by adding 1 mL of PBS (phosphate-buffered saline). Dilute the suspension to obtain a final OD600 of around 0.2 by PBS.
2-2-8. Dispense 600 microL of each suspension into a disposable tube through a cell strainer. Fluorescence intensity was measured with a flow cytometer of Becton, Dickinson and Company.
3. Results
3-1. Before inducing by phosphate concentration
Fig. 3-5-12 shows the fluorescence intensity of the fresh cultures (the MOPS medium which contains 1.32 mM K2HPO4) incubated until the observed OD600 reached 0.4-0.6 (Assay protocol 2-2-3). The phoA promoter was repressed because the MOPS medium contained 1.32 mM phosphate, which was a high concentration for phoA promoter.
3-2. After inducing by phosphate concentration
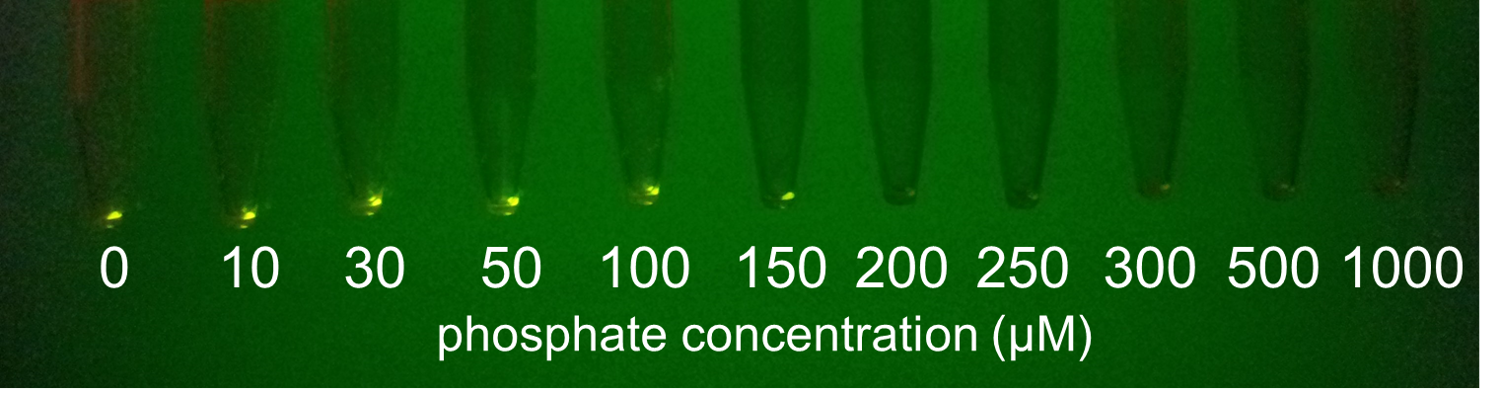
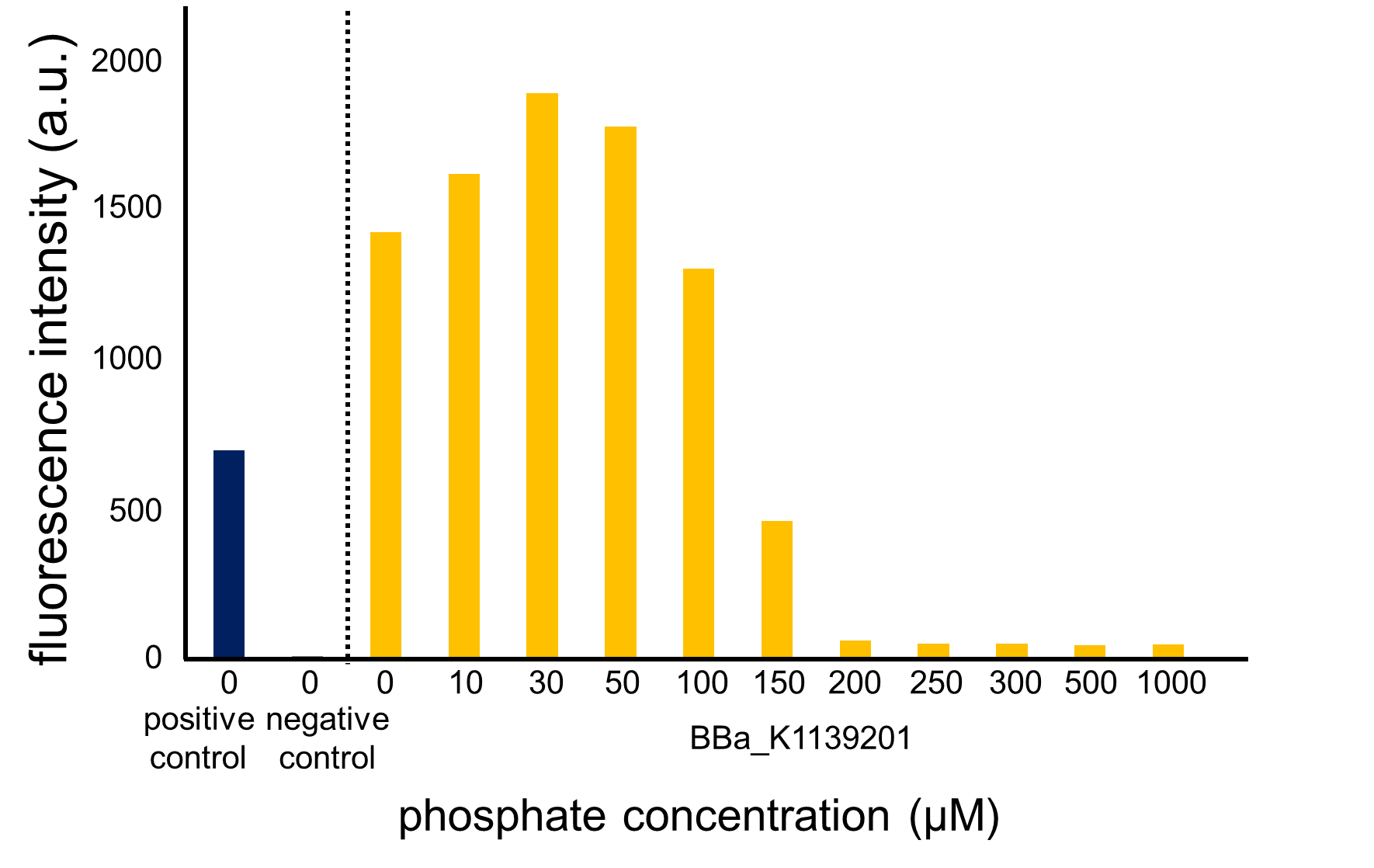
Fig. 3-5-13 shows the fluorescence intensity of the induced cells by phosphate concentration. Fig. 3-5-14 shows the picture of fluorescence of the induced cells. The increase in phosphate concentration repressed the phoA promoter. Especially, we saw that the phoA promoter was drastically repressed at phosphate concentrations of 100 to 300 microM.
4. Discussion
We confirmed that the increase in phosphate concentration repressed the phoA promoter. Compared to OUC-China’s phosphate sensor part including phoB promoter (Fig. 3-5-16), our phosphate sensor part shows clearer result (Fig. 3-5-15).
5. Modeling
From our results explained above, we determined parameters for the induction mechanism. By fitting the results to the following Hill equation (Fig. 3-5-17), we identified m and the hill coefficient. Those parameters (Fig. 3-5-18) will be used in our future modeling. Plants are reported to be in phosphate starvation under the concentration of 1 mM (D. Hoagland et al., 1950). Our part can also sense the concentration below 1 mM (Fig. 3-5-19). Therefore, our improved part is useful for our farming circuit. We also identified maximum GFP production rate in this construct.
6. Reference
1. M. Dollard et al. “Whole-cell bacterial sensors for the monitoring of phosphate availability” Journal of Microbiological Methods (2003)
2. H. Shinagawa et al. “Regulation of the pho Regulon in Escherichia coli K-12” J. Mol. Biol. (1983)
3. Y. Hsieh et al. “Global regulation by the seven-component Pi signaling system” Current Opinion in Microbiology (2010)
4. F. Neidhardt et al. “Culture Medium for Enterobacteria” Journal of Bacteriology (1974)
5. CCG/UNAM. Regulon DB. http://regulondb.ccg.unam.mx/index.jsp (accessed 2013-09-25)
6. D. R. Hoagland and D. I. Arnon (1950), The water-culture method for growing plants without soil. California agrigultual experiment station circular, 347, 1-32
 "
"