Team:Newcastle/Notebook/calendar
From 2013.igem.org
| Line 197: | Line 197: | ||
==== Details ==== | ==== Details ==== | ||
<div> | <div> | ||
| - | |||
| - | We | + | We started preparing for our project presentation at UCL, and we have also started to prepare a presentation for our workshop about BioNetGen modelling. |
</div> | </div> | ||
Revision as of 12:58, 26 June 2013

June
| Sun | Mon | Tue | Wed | Thu | Fri | Sat |
|---|---|---|---|---|---|---|
|
|
|
|
|
|
|
1
|
|
2
|
3
|
4
|
5
|
6
|
7
|
8
|
|
9
|
10
|
11
|
12
|
13
|
14
|
15
|
|
16
|
17
|
18
|
19
|
20
|
21
|
22
|
|
23
|
24
|
25
|
26
|
27
|
28
|
29
|
|
30
|
|
|
|
|
|
|
July
| Sun | Mon | Tue | Wed | Thu | Fri | Sat |
|---|---|---|---|---|---|---|
|
|
1
|
2
|
3
|
4
|
5
|
6
|
|
7
|
8
|
9
|
10
|
11
|
12
|
13
|
|
14
|
15
|
16
|
17
|
18
|
19
|
20
|
|
21
|
22
|
23
|
24
|
25
|
26
|
27
|
|
28
|
29
|
30
|
31
|
|
|
|
August
| Sun | Mon | Tue | Wed | Thu | Fri | Sat |
|---|---|---|---|---|---|---|
|
|
|
|
|
1
|
2
|
3
|
|
4
|
5
|
6
|
7
|
8
|
9
|
10
|
|
11
|
12
|
13
|
14
|
15
|
16
|
17
|
|
18
|
19
|
20
|
21
|
22
|
23
|
24
|
|
25
|
26
|
27
|
28
|
29
|
30
|
31
|
September
| Sun | Mon | Tue | Wed | Thu | Fri | Sat |
|---|---|---|---|---|---|---|
|
1
|
2
|
3
|
4
|
5
|
6
|
7
|
|
8
|
9
|
10
|
11
|
12
|
13
|
14
|
|
15
|
16
|
17
|
18
|
19
|
20
|
21
|
|
22
|
23
|
24
|
25
|
26
|
27
|
28
|
|
29
|
30
|
|
|
|
|
|
Contents |
6
10
Titles
- First Day of Training
Details
Morning
We started a discussion of our sub-project, doing some research into project presentation: wiki layout, t-shirt design and possible team logos. We looked at precedents for inspirational ideas.,Afternoon
Learn about Computational Modelling and BioBrick concepts. Decided to start with biophysical modelling of the lipid membrane of Bacillus subtilis and modelling of lipid synthetic pathway. Later in the day, we had a team meeting to discuss aims and progress of our work.
6
11
Modelling Training
- Modelling Training 1
Details
Morning
Modelling Exercise. Identifying components and species of subtilin biosynthesis pathway. We also sketched down some potential logo ideas and created a shortlist of favorites.
Afternoon
In the afternoon we learned about the differences and similarities between stochastic and deterministic modeling. We also used rule based modeling such as RuleBender and BioNetGen. We changed the parameters and rates of reaction and observed the effects.
6
12
Modelling Training 2
- Modelling Training 2
Details
we continued learning about stochastic and deterministic modeling and we looked at converting the rate parameters in RuleBender. We continued working on the logo.
6
13
Basic Lab Skills 1
- Basic Lab Skills 1
Details
Morning
On our first day of lab, we were briefed on various lab health and safety policies to ensure we maintained a safe working environment. We were then led by our supervisor on a tour around the lab to inform us about the layout. Once we have familiarised with everything, we officially started our first lab session.
We started by callibrating our pipettes and learning how to operate them, then we were shown how to innoculate bacteria to a liquid culture. Finally, before we took a break, we prepared bacteria streak plates.
Afternoon
In the afternoon, we extracted GFP and RFP bricks from the iGEM 2013 biobrick plate and transformed it into competent Escherichia coli cells. These procedures were performed using the following protocols (). Then plate those cells onto LB with chloramphenicol selection marker plates.
6
14
Basic Lab Skills 2
- Basic Lab Skills 2
Details
Morning
When we first arrived in the lab, we were all excited to check on our plates to see if anything grew. The RFP plates showed a lot of collonies, while the GFP plates did not work as expected. The next thing we did was too view the plates under the fluorescent inverted microscope.
Afternoon
We had an introductory lecture on basic molecular biology and biobrick design. It was particullarly helpful for the non-biologist of the team.
6
17
Basic Lab Skills 3
- Basic Lab Skills 3
Details
Morning
The first thing we did was to transfer collonies of bacteria into LB. Then we extracted the plasmids via (plasmid prep) protocol, and check the concentration of plasmids through the use of nanodrop spectrophotometry. Next,we prepared 1% Agarose and whilst waiting for it to set we performed a single digest with (Xbal or Pstl) and double digest with both restriction digest. Finally, we load our samples to the gel in addition with DNA ladder and control.

6
18
Basic Lab Skills 3
- Wiki work
Details
Morning
We came up with a script, story board and a list of materials needed for the introductory video on our wiki. We had fun learning to crochet in order to create characters for the video. Furthermore, we updated the team profile page on the wiki by adding individual pictures of all team members.
6
19
Cloning strategies 1
- Cloning strategies 1
Details
Morning
The group started thinking about their cloning strategies for each theme of the project. We also discussed idea for human practices and came up with the idea of an educational game based on synthetic biology. At the end of the day we had a group meeting to discuss our progress and our ideas for the modelling workshop for the UCL meet up.
6
20
Cloning strategies 2
- Cloning strategies 2
Details
Morning
We continued work on the cloning strategies as well as starting modelling the subtilin two component system for the UCL workshop and the gus reporter system using BioNetGen. We also researched how L-Forms are made through research papers.
6
21
Cloning strategies 3
- Cloning strategies 3
Details
Morning
We further developed our cloning strategies and improved the logo so it's ready for T-shirt printing.
6
24
Progress
- Progress
Details
Morning
We performed a mini prep of the Groningen 2012 integration plasmid DNA grown in E.coli overnight at 37 degrees. The purity was tested using a NanoDrop.
Afternoon
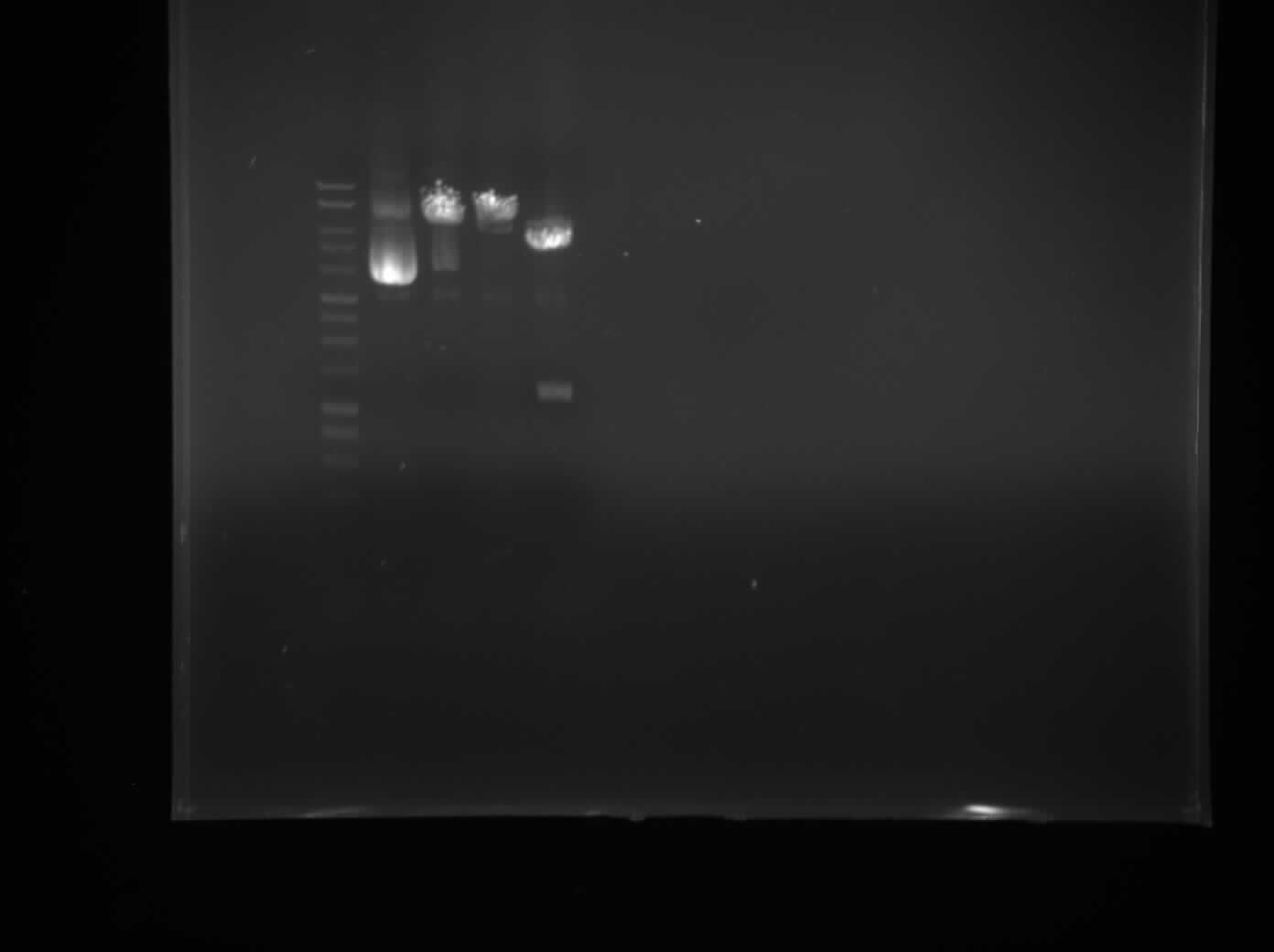
Introduction to microfluidics. Start work on designing Master Molds. Check the extracted pDNA to confirm the outcome of the purification of plasmid using the electrophoresis.
The plasmid has been cut with fast digest XbaI and BamHI to confirm the presence of the plasmid in the purified pDNA. Plasmid which has been cut by XbaI or BamHI only gives a linear piece of DNA. The gel shows that well with the DNA cut with both enzymes contains two pieces of DNA. One - plasmid backbone and the other - RFP reporter gene previously inserted into the plasmid. This confirms its identity.
6
25
Modelling
- Modelling
Details
We started preparing for our project presentation at UCL, and we have also started to prepare a presentation for our workshop about BioNetGen modelling.
-->
 "
"