Team:Goettingen/Team/Reporter
From 2013.igem.org
FMCommmichau (Talk | contribs) |
FMCommmichau (Talk | contribs) |
||
| Line 55: | Line 55: | ||
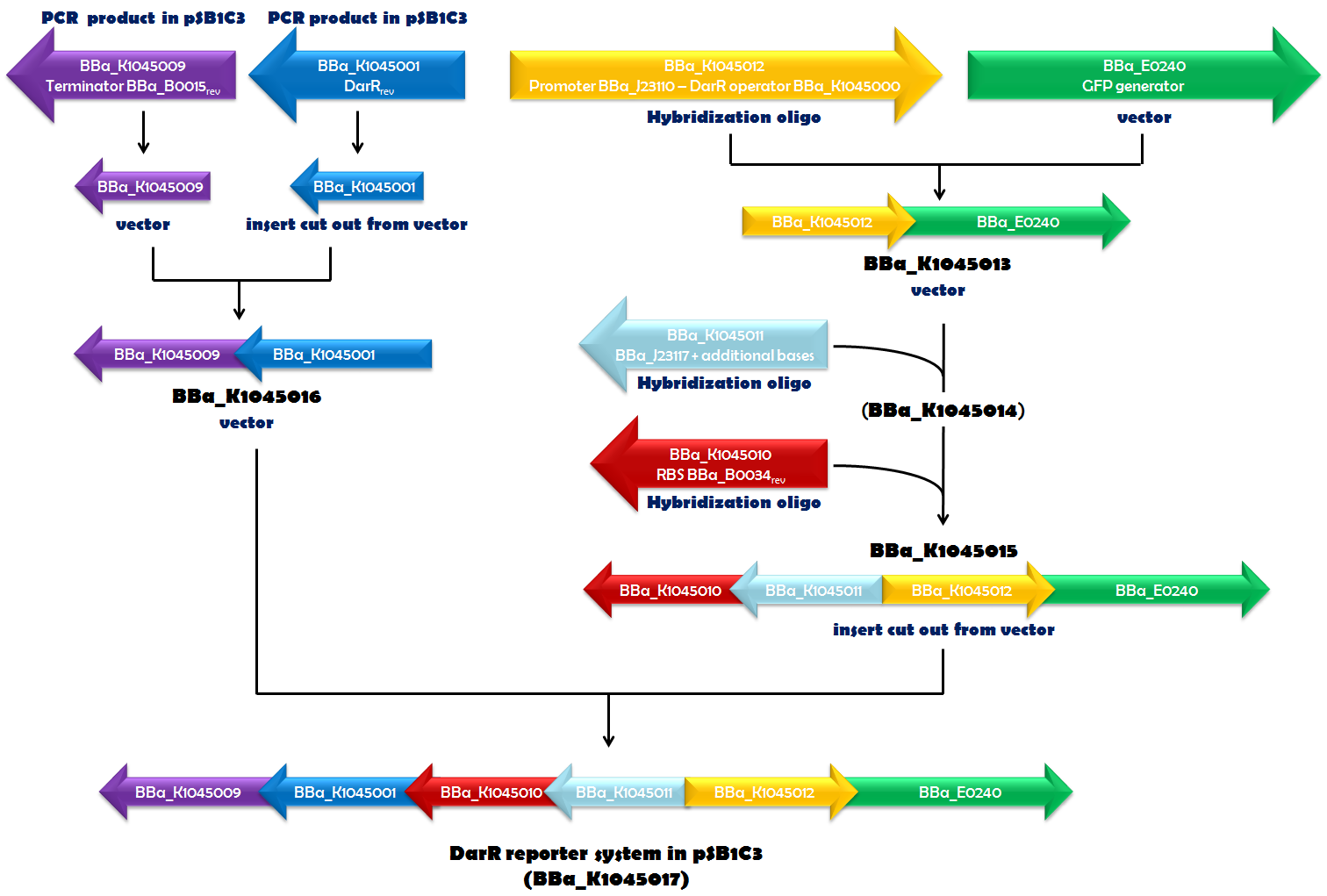
Recently, in ''Mycobacterium smegmatis'', a transcriptional repressor (DarR) was identified. It can bind to a specific DNA sequence, called the DarR operator. The binding efficiency of DarR is strongly enhanced by c-di-AMP. Since DarR is a c-di-AMP sensor, we intended to use it for our reporter system. We cloned DarR into pSB1C3 ([http://parts.igem.org/Part:BBa_K1045001 BBa_K1045001]) and also constructed the operator as a Biobrick ([http://parts.igem.org/Part:BBa_K1045000 BBa_K1045000]). We placed the DarR operator between a strong promoter ([http://parts.igem.org/Part:BBa_J23110 BBa_J23110]) and the GFP generator [http://parts.igem.org/Part:BBa_E0240 BBa_E0240]. In the very same plasmid but oriented in the opposite direction, we assembled a DarR expression unit. DarR expression was driven by a weak promoter based on [http://parts.igem.org/Part:BBa_J23117 BBa_J23117] and terminated by [http://parts.igem.org/Part:BBa_K1045009 BBa_K1045009]. This terminator is based on [http://parts.igem.org/Part:BBa_B0015 BBa_B0015], which is part of [http://parts.igem.org/Part:BBa_E0240 BBa_E0240], as well, and should ensure that the transcription of DarR and GFP did not influence each other. The RBS [http://parts.igem.org/Part:BBa_K1045010 BBa_K1045010] (derived from [http://parts.igem.org/Part:BBa_B0034 BBa_B0034]) was used as an RBS for DarR mRNA translation. Assembly of all those parts (Fig. 1.1) required a complex cloning process which finally resulted in part [http://parts.igem.org/Part:BBa_K1045017 BBa_K1045017], the DarR reporter system (Fig. 1.2). E. coli was then transformed with this construct to obtain the ''in vivo'' screening system for antibiotics directed against c-di-AMP. | Recently, in ''Mycobacterium smegmatis'', a transcriptional repressor (DarR) was identified. It can bind to a specific DNA sequence, called the DarR operator. The binding efficiency of DarR is strongly enhanced by c-di-AMP. Since DarR is a c-di-AMP sensor, we intended to use it for our reporter system. We cloned DarR into pSB1C3 ([http://parts.igem.org/Part:BBa_K1045001 BBa_K1045001]) and also constructed the operator as a Biobrick ([http://parts.igem.org/Part:BBa_K1045000 BBa_K1045000]). We placed the DarR operator between a strong promoter ([http://parts.igem.org/Part:BBa_J23110 BBa_J23110]) and the GFP generator [http://parts.igem.org/Part:BBa_E0240 BBa_E0240]. In the very same plasmid but oriented in the opposite direction, we assembled a DarR expression unit. DarR expression was driven by a weak promoter based on [http://parts.igem.org/Part:BBa_J23117 BBa_J23117] and terminated by [http://parts.igem.org/Part:BBa_K1045009 BBa_K1045009]. This terminator is based on [http://parts.igem.org/Part:BBa_B0015 BBa_B0015], which is part of [http://parts.igem.org/Part:BBa_E0240 BBa_E0240], as well, and should ensure that the transcription of DarR and GFP did not influence each other. The RBS [http://parts.igem.org/Part:BBa_K1045010 BBa_K1045010] (derived from [http://parts.igem.org/Part:BBa_B0034 BBa_B0034]) was used as an RBS for DarR mRNA translation. Assembly of all those parts (Fig. 1.1) required a complex cloning process which finally resulted in part [http://parts.igem.org/Part:BBa_K1045017 BBa_K1045017], the DarR reporter system (Fig. 1.2). E. coli was then transformed with this construct to obtain the ''in vivo'' screening system for antibiotics directed against c-di-AMP. | ||
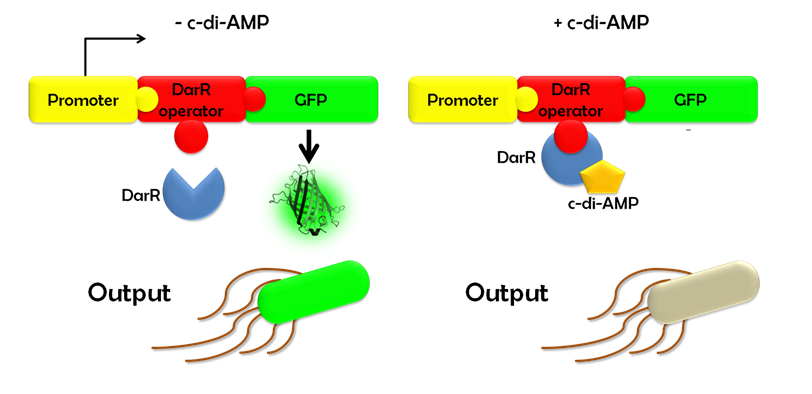
| - | Ideally, this c-di-AMP-sensing system works in the following way (Fig. 1.3): Without c-di-AMP, DarR is not bound to the DarR operator, so GFP is expressed resulting in green fluorescing cells. With c-di-AMP, DarR can bind to its binding sequence, repressing | + | Ideally, this c-di-AMP-sensing system works in the following way (Fig. 1.3): Without c-di-AMP, DarR is not bound to the DarR operator, so GFP is expressed resulting in green fluorescing cells. With c-di-AMP, DarR can bind to its binding sequence, repressing <i>gfp</i> transcription, leading to non-fluorescent cells. In the same way, the system might react to compounds similar to c-di-AMP. It can therefore be used to screen for compounds similar to c-di-AMP. It can be used in a high-throughput scale, as well. This way, the DarR reporter system will make it possible to find inhibitors and competitors which might be used as antibiotics. |
<html><a href="https://static.igem.org/mediawiki/2013/e/ea/Goe-rt-cloningSteps.png" target="_blank"><img src="https://static.igem.org/mediawiki/2013/e/ea/Goe-rt-cloningSteps.png" width="750"/></a></html> | <html><a href="https://static.igem.org/mediawiki/2013/e/ea/Goe-rt-cloningSteps.png" target="_blank"><img src="https://static.igem.org/mediawiki/2013/e/ea/Goe-rt-cloningSteps.png" width="750"/></a></html> | ||
| Line 68: | Line 68: | ||
| - | ''Figure 1.3: Without c-di-AMP, E. coli cells expressing the DarR reporter system | + | ''Figure 1.3: Without c-di-AMP, <i>E. coli</i> cells expressing the DarR reporter system will emit a green fluorescence signal. With c-di-AMP the will not fluoresce.'' |
<html> | <html> | ||
| Line 83: | Line 83: | ||
The <i>ydaO</i> riboswitch was, in addition to DarR, a perfect fit as a c-di-AMP sensor for our antibiotic detector. Hence, we constructed an alternative reporter system. | The <i>ydaO</i> riboswitch was, in addition to DarR, a perfect fit as a c-di-AMP sensor for our antibiotic detector. Hence, we constructed an alternative reporter system. | ||
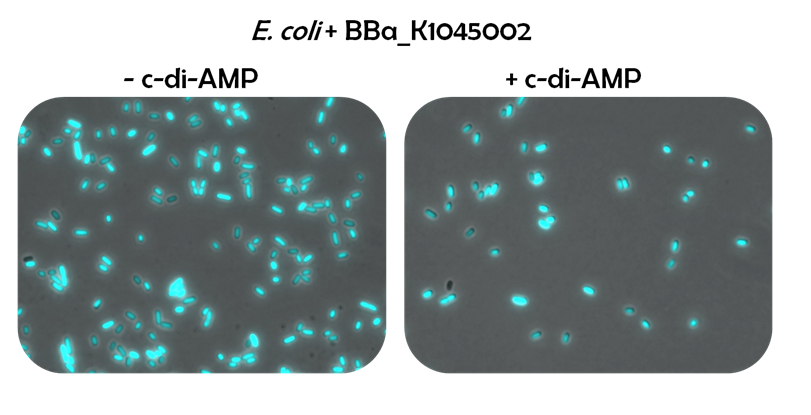
| - | Since it was unclear, where the exact beginning and ending of the ''ydaO'' promoter, the ''ydaO'' riboswitch and the ''ydaO'' RBS were, we created four different constructs (Fig. 2.; [http://parts.igem.org/Part:BBa_K1045004 BBa_K1045004], [http://parts.igem.org/Part:BBa_K1045005 BBa_K1045005], [http://parts.igem.org/Part:BBa_K1045006 BBa_K1045006], [http://parts.igem.org/Part:BBa_K1045007 BBa_K1045007]). The sequence of [http://parts.igem.org/Part:BBa_K1045004 BBa_K1045004] was cloned in front of the reporter gene CFP ([http://parts.igem.org/Part:BBa_E0020 BBa_E0020]). For the termination of transcription, we exploited the terminator on pSB1C3. In principle, the riboswitch reporter system might respond in exactly the same way to c-di-AMP, as the DarR reporter system does: Without c-di-AMP, E. coli cells harboring [http://parts.igem.org/Part:BBa_K1045002 BBa_K1045002] will fluoresce blue. In contrast, when c-di-AMP is present, the ''ydaO'' riboswitch will terminate transcription of ''cfp'' leading to non-fluorescent cells (Fig. 2.2). This way, we can scan substances which can act on the riboswitch as c-di-AMP does. This might lead to the identification of competitors or inhibitors for future antibiotics, as well. | + | Since it was unclear, where the exact beginning and ending of the ''ydaO'' promoter, the ''ydaO'' riboswitch and the ''ydaO'' RBS were, we created four different constructs (Fig. 2.; [http://parts.igem.org/Part:BBa_K1045004 BBa_K1045004], [http://parts.igem.org/Part:BBa_K1045005 BBa_K1045005], [http://parts.igem.org/Part:BBa_K1045006 BBa_K1045006], [http://parts.igem.org/Part:BBa_K1045007 BBa_K1045007]). The sequence of [http://parts.igem.org/Part:BBa_K1045004 BBa_K1045004] was cloned in front of the reporter gene CFP ([http://parts.igem.org/Part:BBa_E0020 BBa_E0020]). For the termination of transcription, we exploited the terminator on pSB1C3. In principle, the riboswitch reporter system might respond in exactly the same way to c-di-AMP, as the DarR reporter system does: Without c-di-AMP, <i>E. coli</i> cells harboring [http://parts.igem.org/Part:BBa_K1045002 BBa_K1045002] will fluoresce blue. In contrast, when c-di-AMP is present, the ''ydaO'' riboswitch will terminate transcription of ''cfp'' leading to non-fluorescent cells (Fig. 2.2). This way, we can scan substances which can act on the riboswitch as c-di-AMP does. This might lead to the identification of competitors or inhibitors for future antibiotics, as well. |
<html><a href="https://static.igem.org/mediawiki/2013/4/44/Goe-rt-riboswitchsystem.png" target="_blank"><img src="https://static.igem.org/mediawiki/2013/4/44/Goe-rt-riboswitchsystem.png" width="750"></a></html> | <html><a href="https://static.igem.org/mediawiki/2013/4/44/Goe-rt-riboswitchsystem.png" target="_blank"><img src="https://static.igem.org/mediawiki/2013/4/44/Goe-rt-riboswitchsystem.png" width="750"></a></html> | ||
Revision as of 13:25, 4 October 2013
 "
"