Team:SydneyUni Australia/Project/SinceHK
From 2013.igem.org
| Line 27: | Line 27: | ||
*submit all of our genes as individually characterised parts that might be used by others in any other projects or context. | *submit all of our genes as individually characterised parts that might be used by others in any other projects or context. | ||
| - | == | + | ==Gibson Assembly Success== |
When assembled individually, we observed colonies from transformation of Gibson assembly produces for all our genes: | When assembled individually, we observed colonies from transformation of Gibson assembly produces for all our genes: | ||
* ([http://www.ncbi.nlm.nih.gov/protein/2660722 aldehyde dehydrogenase A] from "Xanthobacter autotrophicus". | * ([http://www.ncbi.nlm.nih.gov/protein/2660722 aldehyde dehydrogenase A] from "Xanthobacter autotrophicus". | ||
Revision as of 00:22, 29 October 2013


Design
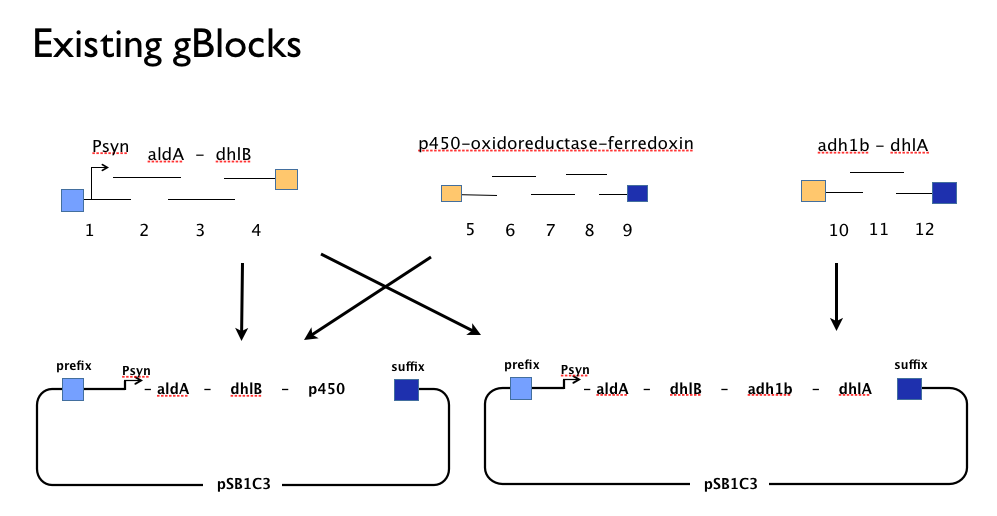
Throughout the year we’d been trying to assemble our DCA-degrading pathway in a single Gibson Assembly reaction. An approach with which we had found some troubles. We believe that the strong constitutive expression of our operon, especially some of our larger parts such as the cytochrome p450,-ferredoxin-reductase cluster may actively harm the cells or be metabollically taxing, effectively create a positive selection pressure for misassembled Gibson inserts. With this one-shot approach, we had left no flexibility in the face of failure: because we’d designed our gBlocks to assemble all at once, when this failed we couldn’t assemble the genes in our pathway from parts.
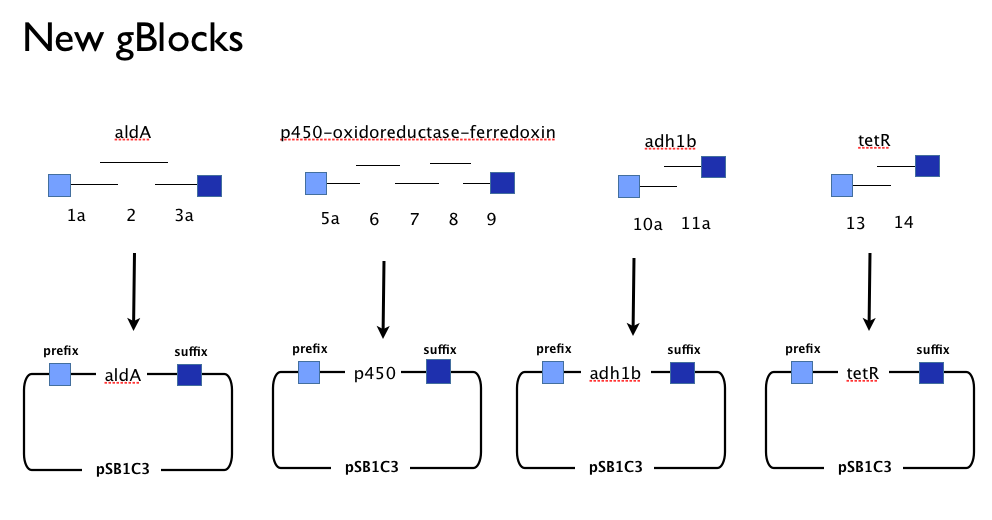
Having learned the virtues of modularity and flexibility the hard way, upon returning from the igem2013 Asia jamboree we designed replacement gBlocks for the start and end of each gene with the BioBrick prefix and suffix respectively. These new gBlocks, along with our old intermediate gBlocks could be gibson assembled with the Registry’s shipping vector, pSB1C3 seperately. Because we still suspected that our pathway, or parts-of, might be toxic in "E.coli" we designed an inducible promoter system consiting of the Tetracycline resistance operon promoter preceded by a constitutive tetracycline repressor protein generator.
This approach would allow us to:
- use the more time-consuming but flexible of [http://parts.igem.org/Assembly:Standard_assembly conventional BioBrick assembly].
- identify which, if any, genes present a toxic or metabolic burden to E. coli.
- characterise each gene individually to inform our model.
- optimise the order of genes in our pathway.
- submit all of our genes as individually characterised parts that might be used by others in any other projects or context.
Gibson Assembly Success
When assembled individually, we observed colonies from transformation of Gibson assembly produces for all our genes:
- ([http://www.ncbi.nlm.nih.gov/protein/2660722 aldehyde dehydrogenase A] from "Xanthobacter autotrophicus".
- [http://www.ncbi.nlm.nih.gov/protein/91700989 cytochrome p450]-[http://www.ncbi.nlm.nih.gov/protein/91700987 ferredoxin]-[http://www.ncbi.nlm.nih.gov/protein/91700988 reductase] from "Polararomonas JS666"
- Our Tet-based inducible promoter system.
- Not adh1b2 because we forgot to order it! Oops.
The cytochrome p450,-ferredoxin-reductase cluster transformants yielded comparatively fewer colonies than the other two. This may be in-part the result of a more complex assembly (five blocks compared to two or three) and in-part a result of the suspected high metabolic load the cluster places on cells. To circumvent the low yield of clones we performed successive round of transformations from the gibson reaction product.
We genotypically screened around 40 colonies for each construct initially by junction PCR, followed by diagnostic digests of plasmid preparations of promising colonies.
Cloning
We attempted to take a short cut in constructing our pathway by inserting a new restriction enzyme recognition site at the end of our Promoter system (just upstream of the suffix) and at the start of our transcriptional unit parts (just downstream of the prefix to avoid incorporation of forbidden sites in later constructs). However in our haste to order new gBlocks post-Hong Kong, we had selected NheI which, whilst unique in our coding sequence and pSB1C3, creates a CTAG overhang that is compatible with both XbaI and SpeI site digests (Oops)!
Results
For the Friar Tuck,
Talk about the magic we made -
- GA of parts
- cloning with inducible system
- two p450 clones look promising on cl- assay (the GC experiment had way higher conc. tet and way less time with tet)
- ...yet to be characterised...
Rob
 "
"