Team:Newcastle/Parts/e-coli swich
From 2013.igem.org
YDemyanenko (Talk | contribs) m |
|||
| Line 4: | Line 4: | ||
</html> | </html> | ||
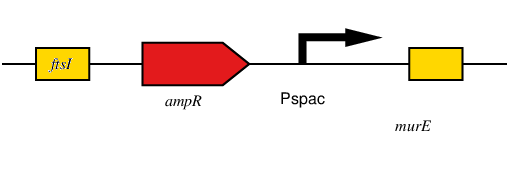
| - | This is an engineered BioBrick derived from the BBa_K1185000 (L-form switch for ''Bacillus subtilis'')which can be used as a switch to enable the model Gram-negative bacteria ''Escherichia coli'' to switch between cell-walled rod form and wall-less L-form (BBa_K1185005). The BioBrick contains homologous regions to the ftsI (homolog to pbpb in Bacillus subtilis)and murE genes to allow integration into the chromosome of E. coli via homologous recombination. Within the BioBrick there is a IPTG inducible promoter Pspac. This promoter is upstream of the murE coding sequence when integrated into the chromosome of E. coli, placing murE operon expression under the control of IPTG. The murE operon is responsible for the synthesis of enzymes involved in production of peptidoglycan synthesis. Disruption of this pathway can be utilised to down-regulate production of the cell wall. | + | This is an engineered BioBrick derived from the BBa_K1185000 (L-form switch for ''Bacillus subtilis'')which can be used as a switch to enable the model Gram-negative bacteria ''Escherichia coli'' to switch between cell-walled rod form and wall-less L-form (BBa_K1185005). The BioBrick contains homologous regions to the ftsI (homolog to pbpb in Bacillus subtilis)and ''murE'' genes to allow integration into the chromosome of ''E. coli'' via homologous recombination. Within the BioBrick there is a IPTG inducible promoter ''Pspac''. This promoter is upstream of the ''murE'' coding sequence when integrated into the chromosome of ''E. coli'', placing murE operon expression under the control of IPTG. The ''murE'' operon is responsible for the synthesis of enzymes involved in production of peptidoglycan synthesis. Disruption of this pathway can be utilised to down-regulate production of the cell wall. |
After integration of the BioBrick, the expression of murE operon is controlled by the presence, or absence, of IPTG(via the Pspacpromoter). When IPTG is present, murE operon genes are expressed and functional cell wall is produced. When IPTG is not present the cell wall is no longer produced and cells can transition to the cell wall-less L-form phenotype. | After integration of the BioBrick, the expression of murE operon is controlled by the presence, or absence, of IPTG(via the Pspacpromoter). When IPTG is present, murE operon genes are expressed and functional cell wall is produced. When IPTG is not present the cell wall is no longer produced and cells can transition to the cell wall-less L-form phenotype. | ||
| Line 10: | Line 10: | ||
The BioBrick also contains a cat gene, conferring Ampicilin resistance. This can be used to select for transformants in rod form. | The BioBrick also contains a cat gene, conferring Ampicilin resistance. This can be used to select for transformants in rod form. | ||
| - | This BioBrick only functions in enabling switching between walled cells and wall-deficient L-forms in E. coli. We are giving opportunities for future iGEM team to work with cell wall-less E. coli. | + | This BioBrick only functions in enabling switching between walled cells and wall-deficient L-forms in ''E. coli''. We are giving opportunities for future iGEM team to work with cell wall-less ''E. coli''. |
This BioBrick should work in a similar manner as the BBa_K1185000 (B. subtilis Switch BioBrick). The characterization of this BioBrick is something that future iGEM can do. | This BioBrick should work in a similar manner as the BBa_K1185000 (B. subtilis Switch BioBrick). The characterization of this BioBrick is something that future iGEM can do. | ||
Revision as of 03:24, 29 October 2013

Switch BioBrick for Escherichia coli
This is an engineered BioBrick derived from the BBa_K1185000 (L-form switch for Bacillus subtilis)which can be used as a switch to enable the model Gram-negative bacteria Escherichia coli to switch between cell-walled rod form and wall-less L-form (BBa_K1185005). The BioBrick contains homologous regions to the ftsI (homolog to pbpb in Bacillus subtilis)and murE genes to allow integration into the chromosome of E. coli via homologous recombination. Within the BioBrick there is a IPTG inducible promoter Pspac. This promoter is upstream of the murE coding sequence when integrated into the chromosome of E. coli, placing murE operon expression under the control of IPTG. The murE operon is responsible for the synthesis of enzymes involved in production of peptidoglycan synthesis. Disruption of this pathway can be utilised to down-regulate production of the cell wall.
After integration of the BioBrick, the expression of murE operon is controlled by the presence, or absence, of IPTG(via the Pspacpromoter). When IPTG is present, murE operon genes are expressed and functional cell wall is produced. When IPTG is not present the cell wall is no longer produced and cells can transition to the cell wall-less L-form phenotype.
The BioBrick also contains a cat gene, conferring Ampicilin resistance. This can be used to select for transformants in rod form.
This BioBrick only functions in enabling switching between walled cells and wall-deficient L-forms in E. coli. We are giving opportunities for future iGEM team to work with cell wall-less E. coli.
This BioBrick should work in a similar manner as the BBa_K1185000 (B. subtilis Switch BioBrick). The characterization of this BioBrick is something that future iGEM can do.
Figure 1, shows the SBOL Visual of this BioBrick design.
 "
"