Team:TU-Munich/Notebook/Labjournal
From 2013.igem.org
m (→Week 2) |
|||
| Line 34: | Line 34: | ||
= Labjournal = | = Labjournal = | ||
| - | |||
<div class="labbook"> | <div class="labbook"> | ||
Revision as of 12:08, 2 May 2013

Labjournal
Week 1
Monday, April 22nd
Transformation of E. coli XL1 blue with Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3)
Investigator: Jeff, Leonie, Rosario
Aim of the experiment: Transformation of Phytochrome B for protein fusion.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 2 µl of DNA was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of the cell suspension was plated on one chloramphenicol plate.
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on a new chlorampenicol plate.
Miniprep of pTUM100 with pGAL, pTEF1, pTEF2, pADH and RFC25 compatible RFP generator
Investigator: Jeff, Leonie, Rosario
Aim of the experiment: Miniprep of pTUM100 with pGAL, pTEF1, pTEF2, pADH and RFC25 compatible RFP generator
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
Sequencing of RFP-Generator (RFC25, pSB1C3)
Investigator: Jeff, Leonie, Rosario
Aim of the experiment: Sequencing of RFP-Generator (RFC25, pSB1C3)
Procedure:
Sequencing batch were prepared after manufacturer's protocol. (15 µl of plasmid DNA (50 - 100 ng) and 2 µl sequencing primer)
Tuesday, April 23rd
Picking of of E. coli XL1 blue with Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3)
Investigator: Jeff, Leonie, Rosario, Florian
Aim of the experiment: Picking of of E. coli XL1 blue with Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3)
Procedure:
- pSB1C3 plasmid with BBa_K801031 (PhyB 2 - 908 aa, RFC25): Colonies were picked from chloramphenicol plates.
- Picked pipette tips was transferred into cell-culture tubes with air-permeable, sterile cover. Each tube contain 4 mL of LB-medium + 4 µL chloramphenicol(1000x).
- 4 colonies were picked.
- These tubes were transferred in a cell culture shaker at 37 °C and were incubated overnight
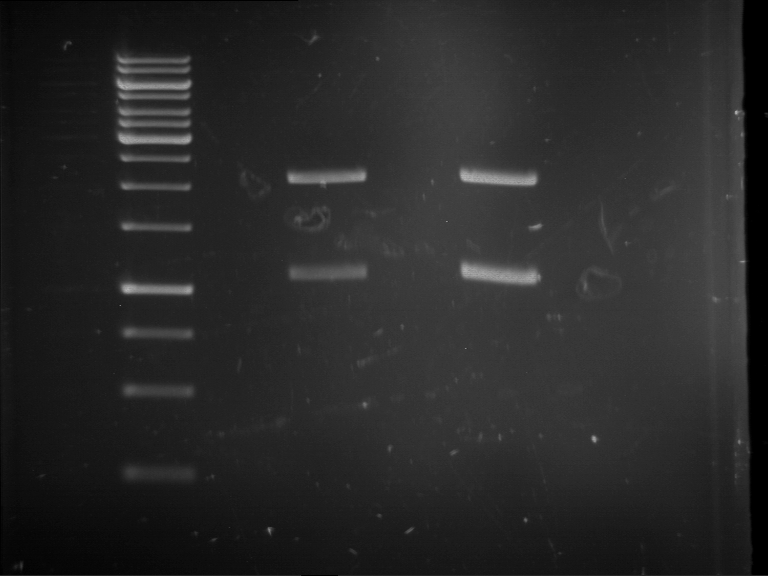
Analytical digestion and gelelectrophoresis of RFP-generator (RFC25, pSB1C3, P4 & P5)
Investigator: Jeff, Leonie, Rosario, Florian
Aim of the experiment: Analytical digestion and gelelectrophoresis of RFP-generator (RFC25, pSB1C3, P4 & P5).
Procedure:
- Batch for analytical digestion for P4 with NgoMIV+AgeI-HF
| volume | reagent |
| 2.5 µl | Plasmid DNA P4 |
| 2 µl | NEBuffer 4 (10x) |
| 0.25 µl | NgoMIV (10 U/µl) |
| 0.25 µl | AgeI-HF (20 U/µl) |
| 15 µl | ddH2O |
| =20 µl | TOTAL |
- Batch for analytical digestion for P5 with NgoMIV+AgeI-HF
| volume | reagent |
| 2.5 µl | Plasmid DNA P5 |
| 2 µl | NEBuffer 4 (10x) |
| 0.25 µl | NgoMIV (10 U/µl) |
| 0.25 µl | AgeI-HF (20 U/µl) |
| 15 µl | ddH2O |
| =20 µl | TOTAL |
- Incubation for 90 min at 37 °C.
- Analytical gelelectrophoresis was performed at 90 V for 60 min.
Results:
| 1 kbp ladder DNA ladder | P4 | P5 |
| Mutation successful | Mutation successful! |
- Parts are compliant and do not contain RFC25 forbidden restriction sites.
Sequencing of pTUM vectors with pGAL, pADH, pTEF1, pTEF2
Investigator: Jeff, Leonie, Rosario, Florian
Aim of the experiment: Sequencing of pTUM vectors with pGAL, pADH, pTEF1, pTEF2
Procedure:
Sequencing batch were prepared after manufacturer's protocol. (15 µl of plasmid DNA (50 - 100 ng) and 2 µl sequencing primer).
The different vectors we sequenced received the following barcodes:
- ADH in pTUM100: FR01002265
- TEF1 in pTUM100: FR01002266
- TEF2 in pTUM100: FR01002266
- GAL in pTUM100: FR01002268
Sequencing of TEF2 in pTUM100 was not interpretable. The other sequences were consistent with the sequences in the parts registry.
Wednesday, April 24th
Miniprep of Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3)
Investigator: Jeff, Leonie, Florian
Aim of the experiment: Miniprep of Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3).
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
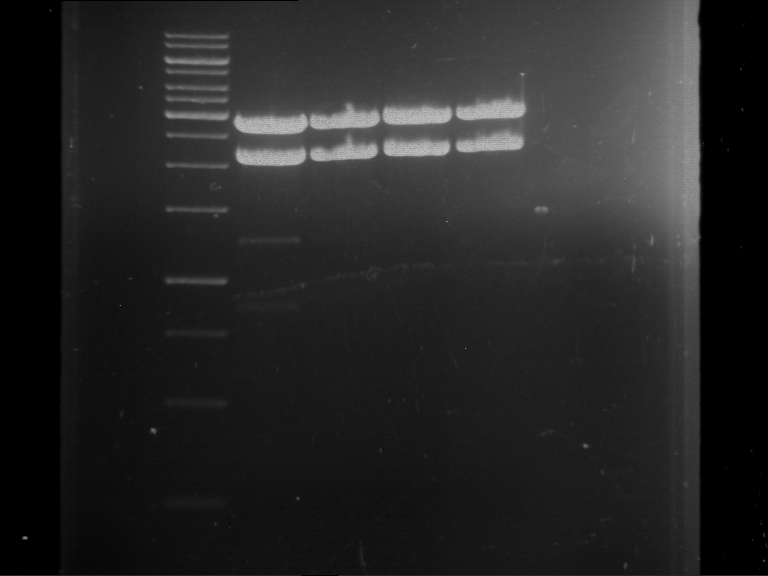
Analytical digestion and gelelectrophoresis of Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3), P7 - P10
Investigator: Jeff, Leonie, Florian
Aim of the experiment: Analytical digestion and gelelectrophoresis of Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3), P7 - P10.
Procedure:
- Batch for analytical digestion for P7 with NgoMIV+AgeI-HF
| volume | reagent |
| 2.5 µl | Plasmid DNA P7 |
| 2 µl | NEBuffer 4 (10x) |
| 0.25 µl | NgoMIV (10 U/µl) |
| 0.25 µl | AgeI-HF (20 U/µl) |
| 15 µl | ddH2O |
| =20 µl | TOTAL |
- Batch for analytical digestion for P8 with NgoMIV+AgeI-HF
| volume | reagent |
| 2.5 µl | Plasmid DNA P8 |
| 2 µl | NEBuffer 4 (10x) |
| 0.25 µl | NgoMIV (10 U/µl) |
| 0.25 µl | AgeI-HF (20 U/µl) |
| 15 µl | ddH2O |
| =20 µl | TOTAL |
- Batch for analytical digestion for P9 with NgoMIV+AgeI-HF
| volume | reagent |
| 2.5 µl | Plasmid DNA P9 |
| 2 µl | NEBuffer 4 (10x) |
| 0.25 µl | NgoMIV (10 U/µl) |
| 0.25 µl | AgeI-HF (20 U/µl) |
| 15 µl | ddH2O |
| =20 µl | TOTAL |
- Batch for analytical digestion for P10 with NgoMIV+AgeI-HF
| volume | reagent |
| 2.5 µl | Plasmid DNA P10 |
| 2 µl | NEBuffer 4 (10x) |
| 0.25 µl | NgoMIV (10 U/µl) |
| 0.25 µl | AgeI-HF (20 U/µl) |
| 15 µl | ddH2O |
| =20 µl | TOTAL |
- Incubation for 90 min at 37 °C.
- Analytical gelelectrophoresis was performed at 90 V for 60 min.
Results:
| 1 kbp ladder DNA ladder | P7 | P8 | P9 | P10 |
| Part is correct | Part is correct | Part is correct | Part is correct |
Transformation of E. coli XL1 blue with EreA and EreB (pDEST14)
Investigator: Jeff, Leonie, Florian
Aim of the experiment: Transformation of E. coli XL1 blue with EreA and EreB (pDEST14).
Procedure:
- Plasmid DNA was received dried in paper from McMaster University.
- DNA was resuspended in ddH2O
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 2 µl of DNA was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- The transformated cells were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated on chlorampenicol and ampicillin plates.
Week 2
Monday, April 29nd
Miniprep of pRK792, pRK793, pRIL, ereA, ereB
Investigator: Leonie
Aim of the experiment: Miniprep of pRK792, pRK793, pRIL, ereA, ereB
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
- Concentrations were determined by measuring the absorption:
| Plasmid | c [ng/µl] |
| pRK792 | 214,5 |
| pRK793 | 154,3 |
| pRIL | 6,1 |
| ereA | 183,4 |
| ereB | 98,3 |
- The procedure will have to be repeated for pRIL
Midiprep of RFC25 compatible RFP generator in (pSB1C3)
Investigator: Andreas, Florian
Aim of the experiment: Midiprep of RFC25 compatible RFP generator in (pSB1C3)
Procedure:
- Midiprep was performed after manufacturer's protocol (QIAprep Midiprep, QIAGEN)
Tuesday, April 30th
Preparative digestion and gelelectrophoresis of mINF1 and Leptin with HindIII and EheI
Investigator: Louise, Johanna
Aim of the experiment: Testing the enzymes, since the earlier digestion with probably older enzymes did not work (no bands on gel) .
Procedure:
- Batch for preparative digestion for mINF1 with HindIII and EheI
| volume | reagent |
| 10 µl | Plasmid DNA mINF11 |
| 5 µl | Tango Buffer (10x) |
| 2 µl | HindIII (10 U/µl) |
| 3 µl | EheI (10 U/µl) |
| 30 µl | ddH2O |
| =50 µl | TOTAL |
- Batch for preparative digestion for Leptin with HindIII and EheI
| volume | reagent |
| 20 µl | Plasmid DNA Leptin |
| 5 µl | Tango Buffer (10x) |
| 2 µl | HindIII (10 U/µl) |
| 3 µl | EheI (10 U/µl) |
| 20 µl | ddH2O |
| =50 µl | TOTAL |
- Incubation for 3 h at 37 °C.
Transformation of E. coli XL1 blue with PSH21 (P17)
Investigator: Johanna, Louise
Aim of the experiment: Transformation of E. coli XL1 blue with PSH21.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice for 10 minutes.
- 5 µl of DNA was added to 250 µl of competent cells and gently mixed.
- 60 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding the DNA to 2 ml LB-medium.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- The transformated cells were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated on chlorampenicol and ampicillin plates.
 "
"