DAC Team
Introduction
In order to find a novel target that can be inhibited by new antibacterial compounds, we focused on the recently discovered signaling molecule bis-(3’,5’)-cyclic dimeric adenosine monophosphate (c-di-AMP). c-di-AMP was shown to be an essential second messenger in many important pathogenic Gram-positive bacteria (Witte et al., 2008). Moreover, c-di-AMP has a crucial function in cell wall synthesis and spore formation in Bacillus subtilis (Oppenheimer-Shaanan et al., 2011; Mehne et al., 2013). Interestingly, both lack and excess of c-di-AMP have detrimental effects on cell growth and morphology (Luo and Helmann, 2012; Mehne et al., 2013).
Based on these observations, it makes sense that we take a closer look at the enzyme, the diadenylate cyclase (DAC), which produces c-di-AMP. The DAC CdaA is conserved among several Gram-positive bacteria like B. subtilis, and in the important pathogenic bacteria Streptococcus pneumoniae, Staphylococcus aureus and Listeria monocytogenes (Corrigan and Gründling, 2013). So far, the DAC DisA from B. subtilis has been purified and crystallized (Witte et al., 2008). However, this protein is not present in all Gram-positive bacteria as well as in many pathogenic bacteria. Therefore we decided to concentrate on the DAC CdaA from L. monocytogenes, which is - as mentioned above - well-conserved in all bacteria that need c-di-AMP for growth!
Results and Discussion

 Unfortunately, cloning of the full-length gene, encoding the membrane-bound DAC, DacA (Lmo2120) using Escherichia coli failed. Therefore, we decided to clone a truncated dacA gene, that encodes a DAC enzyme, lacking the trans-membrane domain. In this protein the first 100 amino acids are missing. Nevertheless, the resulting truncated part still included the essential DAC domain, and therefore represents one of our favorite BioBricks: [http://parts.igem.org/Part:BBa_K1045003 BBa_K1045003]!
Unfortunately, cloning of the full-length gene, encoding the membrane-bound DAC, DacA (Lmo2120) using Escherichia coli failed. Therefore, we decided to clone a truncated dacA gene, that encodes a DAC enzyme, lacking the trans-membrane domain. In this protein the first 100 amino acids are missing. Nevertheless, the resulting truncated part still included the essential DAC domain, and therefore represents one of our favorite BioBricks: [http://parts.igem.org/Part:BBa_K1045003 BBa_K1045003]!
By conducting several experiments, we proved that the truncated DacA protein (BBa_K1045003) was not only active in vivo, but also did it's job in vitro. Moreover, we were able to purify the DAC domain in large scale for determining its 3D structure! Using the structure data, one can now search for chemical compounds that interfere with the activity of the cyclase either by testing with available chemical libraries or by computational modeling. In the following text, the experiments will be explained in more detail. However, if you wish to get even more details, please visit the Parts Registry or our LabBook.
The truncated DacA protein ([http://parts.igem.org/Part:BBa_K1045003 BBa_K1045003]) was extended with an N-terminal Step-tag allowing the rapid purification using the Strep-tag:Streptactin purification system. Synthesis of our protein is driven by a T7 promoter. This promoter is recognized by the T7 polymerase, which is encoded in the genome of the E. coli strain BL21. Synthesis of the T7 polymerase can be controlled by Isopropyl-β-D-thiogalactopyranoside (IPTG). The generated plasmid was then used to transform the E. coli strain BL21, which is a powerful strain for synthesis of recombinant proteins. In contrast to Gram-positive bacteria, the Gram-negative bacterium E. coli does not produce c-di-AMP and growth is not affected by the signaling molecule.
In order to analyze the DAC activity in vivo, DAC production by the E. coli clones was induced by adding IPTG. The cells were then lysed to extract c-di-AMP from the cells.
By performing the SDS PAGE, we could nicely show that the desired protein was well-produced (Fig. 1). Furthermore, the presence of c-di-AMP in the supernatant of the lysed bacteria was confirmed using LC-MS/MS. Thus, we can conclude that the truncated DacA protein codes for an DAC domain that is active in vivo.

Fig. 1. Confirmation of DAC production. The SDS PAGE revealed high-level DAC production by three biological replicates. As expected, the DAC domain has a molecular wight of about 20 kDa; Lane 1: Thermo Scientific Page Ruler Plus Prestained Protein Ladder.
DACs catalyze the condensation reaction of two molecules of ATP to a single molecule c-di-AMP, while releasing two pyrophosphate (PP) molecules, consisting of the β-γ-phosphates (P) of each ATP (Fig. 2). In order to analyze the DAC activity in vitro, we performed an assay in which the generation of PP molecules served as an indicator for c-di-AMP production. Among other things such as malachite-green and molybdate, the assay included a PP phosphatase that cleaves the PP molecule into free P molecules. Malachite-green forms a complex with free P molecules and molybdate stabilizes the complex. The product absorption can be measured at a wavelength of 630 nm. With this in vitro assay, the concentration of free P molecules and thus the conversion rate of ATP to c-di-AMP was analyzed.
Our results confirmed that the DAC domain of the DacA enzyme (Lmo2120) is active in the presence of a divalent cation, ATP and a buffer system at pH 8 (in vitro), however, the catalysis rate in vivo appears to be much more efficient.
Fig. 2. c-di-AMP production and degradation. c-di-AMP is produced from two molecules of ATP by DACs and degraded to pApA by phosphodiasterases. (Edited from Corrigan and Gründling, 2013)
Finally, we are coming to the core of our project, the protein structure of the DacA DAC domain! It is very helpful to have the molecular structure of a DAC in hands because it allows to perform in silico experiments. These in silico experiments can lead to the discovery of new antibacterial substance classes, that specifically inhibit DAC activity. The final goal is to use potential antibiotics to treat patients that suffer from infection with pathogenic Gram-positive bacteria.
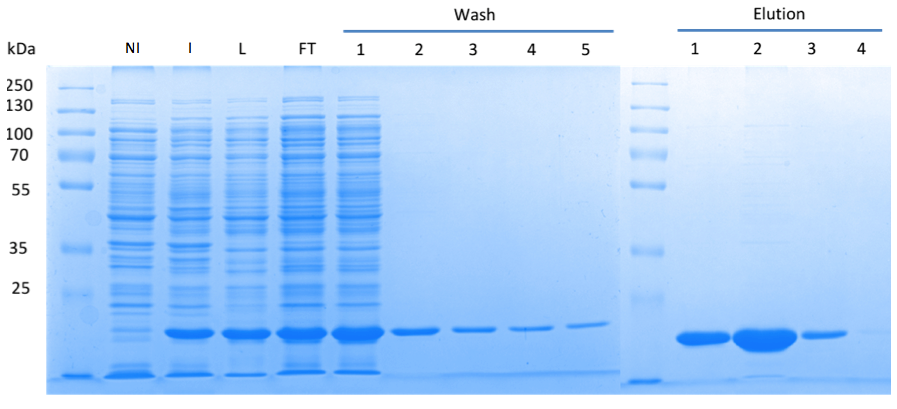
In order to purify a large amount of this protein, we used our BioBrick [http://partsregistry.org/Part:BBa_K1045003 BBa_K1045003] to produce the N-terminally Strep-tagged DAC domain in E. coli. Expression was confirmed by SDS PAGE and the protein showed a molecular weight of about 20 kDa (Fig. 3). The E. coli strain was grown in a large scale (10 liters), the cells were harvested by centrifugation, lysed and the protein was purified by affinity purification (Fig. 3).
Fig. 3. SDS PAGE showing high-level production and purification of the DAC domain. (A) Lane 1: Thermo Scientific PageRuler Plus Prestained Protein Ladder; NI: Non-induced clone, the cells did not produce the DAC domain and c-di-AMP; I: Induction of dac expression with IPTG, the clone synthesized the DAC domain and c-di-AMP was detected; L: Lysate; FT: Flow-through; the protein eluted in the first elution fraction.
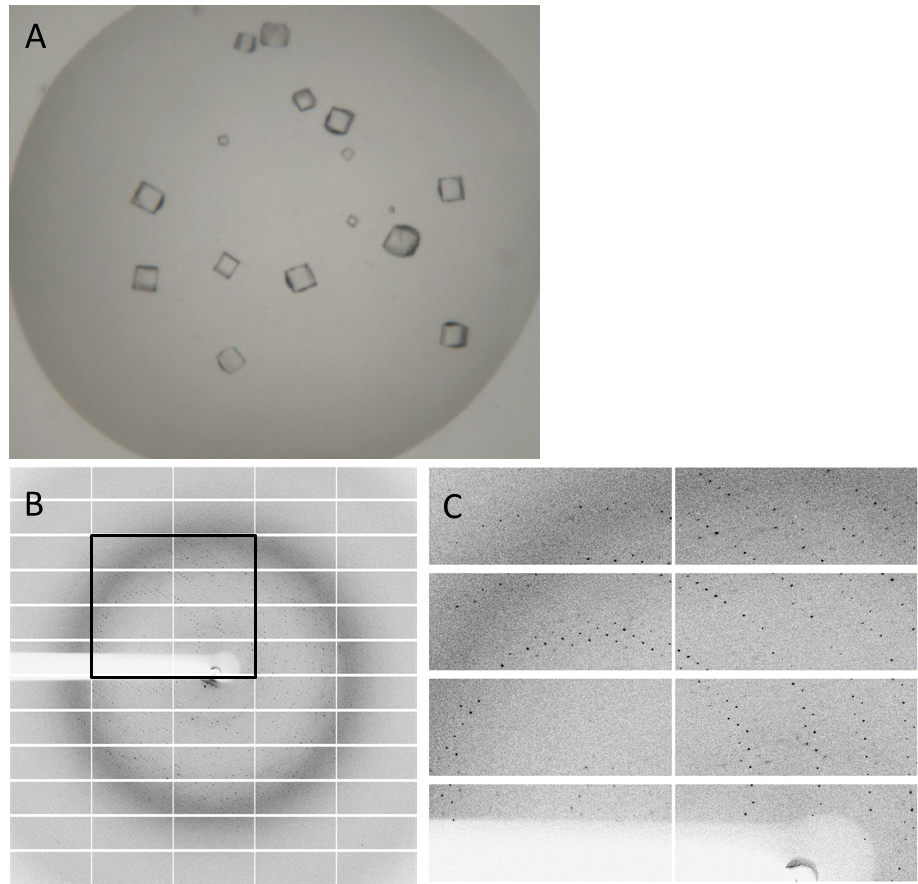
Having dialyzed and concentrated the purified protein, we obtained a protein solution with a concentration of about 10 mg/ml, which was sufficient to perform a crystallization screening. Luckily, we have obtained very nice crystals in the initial crystallization screen (Fig. 4A). In order to find the perfect supplements for growing our crystals, the whole procedure was repeated and the conditions were refined. The crystals yielded an x-ray diffraction pattern, with a resolution of 2,8 Å (Fig. 4B,C). The dataset was measured at the EMBL Hamburg Beamline P13 at the PETRA III synchrotron on the DESY campus, using a PILATUS2 6M X-ray detector (https://www.dectris.com/).
Finally, we were also able to obtain the protein structure (Fig. 5)! The structure shows a globular protein with a distinct ATP-binding cleft. The ribbon model demonstrates the secondary structure composed of α-helices and β-sheets.
Fig. 4. Crystals and diffraction pattern. Nice crystals were obtained with a medium concentration of alcohol and other supplements (confidential :-])(A). X-ray diffraction image of the DacA DAC domain crystals(B); the highlighted box is shown enlarged (C). The dataset was measured at the EMBL Hamburg Beamline P13 at the PETRA III synchrotron on the DESY campus.

Fig. 6. Protein structure of the DacA DAC domain. (A, B) Ribbon model of the DAC domain in its ATP-bound state. (C, D) Surface structure of the DAC domain and the ATP-binding pocket. (E) Magnified view into the ATP-binding pocket
References
Corrigan RM and Gründling A. (2013) Cyclic di-AMP: another second messenger enters the fray. Nat Rev Microbiol. 11(8):513-524
Luo Y and Helmann JD. (2012) Analysis of the role of Bacillus subtilis σ(M) in β-lactam resistance reveals an essential role for c-di-AMP in peptidoglycan homeostasis. Mol Microbiol. 83(3):623-639
Mehne FM, Gunka K, Eilers H, Herzberg C, Kaever V, Stülke J. (2013) Cyclic di-AMP homeostasis in Bacillus subtilis: both lack and high level accumulation of the nucleotide are detrimental for cell growth. J Biol Chem. 288(3):2004-2017
Oppenheimer-Shaanan Y, Wexselblatt E, Katzenhendler J, Yavin E, Ben-Yehuda S. (2011) c-di-AMP reports DNA integrity during sporulation in Bacillus subtilis. EMBO Rep. 12(6):594-601
Witte G, Hartung S, Büttner K, Hopfner KP. (2008) Structural biochemistry of a bacterial checkpoint protein reveals diadenylate cyclase activity regulated by DNA recombination intermediates. Mol Cell. 30(2):167-178
 "
"