Team:Newcastle/Parts/HBsu-fp
From 2013.igem.org

Contents |
HBsu-xFP Fusion
Purpose and Justification
Bacillus subtilis contains HBsu, a non-specific DNA binding protein which is a homologue to eukaryotic histones. This protein is encoded by the gene hbs gene which is 263bp in length and involved not only in DNA binding but also SRP, DNA repair, HR, and pre-secretory protein translocation [http://www.ncbi.nlm.nih.gov/pubmed/10224127 (Kouji, et al., 1999)]. It binds to DNA by forming homodimer.
We conjugated the hbs gene with red fluorescent protein (RFP/GFP) in order to fluorescently tag the bacterial chromosome. We produced [http://parts.igem.org/Part:BBa_K1185001 BBa_K1185001] which codes for a HBsu-sfGFP conjugate and [http://parts.igem.org/Part:BBa_K1185002 BBa_K1185002] coding for a HBsu-RFP conjugate. Each bacterium was transformed with only one or the other. The expression of these BioBricks was regulated through an IPTG induce promoter (Pspac).
We fused one bacteria containing HBsu-RFP and another containing HBsu-sfGFP, and viewed the genomic shuffling via fluorescent microscopy. The formation of a colour mosaic allows us to confidently claim that recombination has occurred. Genomic shuffling has importance as it can be used to evolve and improve a cells phenotype.
Assembly
The BioBrick [http://parts.igem.org/Part:BBa_K1185001 BBa_K1185001] consists of superfolded green fluorescent protein (sfGFP) (BBa_E0040)conjugated to a HBsu protein.
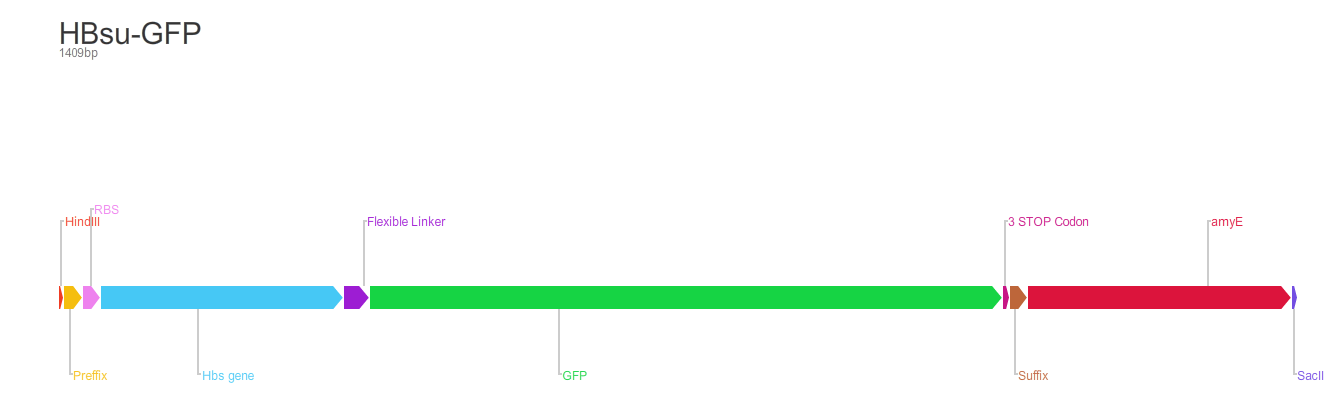
Whereas BioBrick [http://parts.igem.org/Part:BBa_K1185002 BBa_K1185002] contains red fluorescent protein(RFP) (BBa_E1010) conjugated to a HBsu.

HBsu is a non-specific DNA binding protein that binds to DNA as a homodimer. The HBsu is joined to the sfGFP/RFP through ten amino acid flexible linker sequence. This allows the observation of Bacillus subtilis DNA using fluorescence microscopy.
Integration
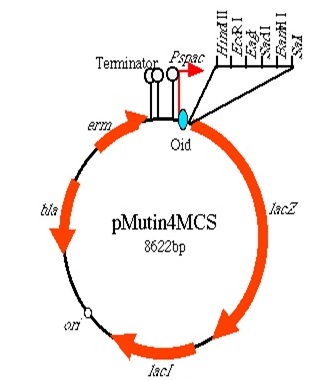
The integration strategy that we opted for was to clone in the BioBricks into the Multiple cloning site of pMutin4 backbones. First we attached HindIII restriction sites on 5'end and SacII restriction sites on 3'end of the BioBricks. We then cut the pMutin4 backbones and BioBricks using the previously mentioned restriction enzymes, this allowed us to ligate the plasmids and BioBricks together. We also attached a ~300bp amyE homology region onto the 3'end of the BioBricks after the SacII to allow the single cross over and integration of the plasmids into the genome. The pMutin4 plasmid contains a ery+ resistance marker for B.subtilis and amp+ for E.coli and also contains lacI, lacZ and Pspac promoter which is an IPTG induced promoter which regulated the transcription of the BioBricks. An alternative method to use this part would be to clone either BioBrick out and use any Assembly protocol to attach a desired promoter, RBS and antibiotic resistance genes.
Testing and Characterisation
References
[http://www.ncbi.nlm.nih.gov/pubmed/10224127 Kouji, N., Shou-ichi, Y., Takao, Y. & Kunio , Y., 1999. Bacillus subtilis Histone-like Protein, HBsu, Is an Integral Component of a SRP-like Particle That Can Bind the Alu Domain of Small Cytoplasmic RNA. The Journal of Biological Chemistry , Volume 274, pp. 13569-13576.]
 "
"