Team:Newcastle/Project/L forms
From 2013.igem.org

Contents |
L-forms
Overview
We propose the use of L-form bacteria as a chassis for synthetic biology. L-form strains are derivatives of common cell-walled bacterial strains; however, L-forms are cell wall deficient. Many modern bacteria have the capacity to switch into L-form state, though specifically we are investigating L-forms in the model Gram-positive bacteria Bacillus subtilis. Unlike protoplasts, L-forms are still able to propogate and grow like their cell-walled counterparts. This growth and division does not occur in the same way as walled cells - L-forms have been described to undertake [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3603455/ membrane blebbing, tubulation and vesiculation].
As a result of their cell wall-deficiency L-forms are restricted by osmolarity much more then walled bacteria. L-forms require incubation for many generations in specific osmotically balanced conditions with enzymes and antibiotics that prevent invasion by walled contaminant cells. However, these specific osmotic demands of L-forms can also be seen as a kill-switch. Any bacteria that escape will no longer be in a maintained and osmotically balanced environment and will consequently not survive.
For conventional cell-walled bacterial strains to switch into wall-deficient L-form state the synthesis of the peptidoglycan cell wall must be disrupted. In B. subtilis this disruption can be achieved through controlling the expression of murE. The murE gene is responsible for the synthesis of a number of enzymes that are involved in the synthesis of a peptidoglycan precursor. When murE expression is down-regulated, this has a cascade effect leading to the down-regulation of peptidoglycan synthesis. It has been found that a mutation that spontaneously occurs in L-form B. subtilis cells is also necessary for the [http://www.ncbi.nlm.nih.gov/pubmed/19212404 survival of stable L-forms]. After selection for wall-deficient cells, survivors exhibit a mutation within the yqiD gene, which is similar to the Escherichia coli gene ispA. This mutation allows [http://www.ncbi.nlm.nih.gov/pubmed/23452849 for stabilisation of L-forms that are undergoing shape modulation due to excess of cell membrane].
The B. subtilis strain [http://www.ncbi.nlm.nih.gov/pubmed/23452849 LR2] - an L-form derivative of B. subtilis 168 created by the Errington research group at Newcastle University - contains a xylose-controlled promoter PxylR upstream of the murE gene on its chromosome. This allows for xylose-mediated control over murE expression. This strain also contains a chloramphenicol acetyl-transferase (cat) gene close to the locus of the PxylR promoter. This gene confers resistance to chloramphenicol and allows for selection for B. subtilis LR2 when in rod form. It is this region in the LR2 strain - containing the PxylR promoter and the cat gene - that has been utilised and altered to create the L-form switch BioBrick.

Characterisation
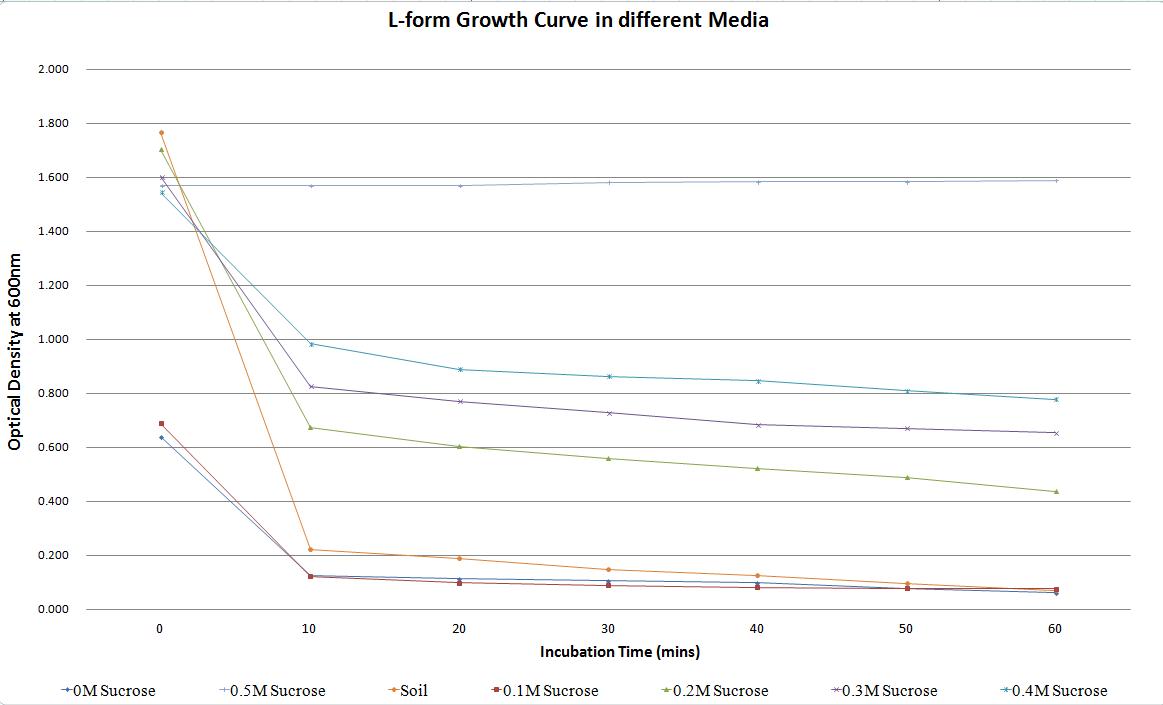
One way that L-forms differ from their cell walled counterparts is in their inability to survive in a wide range of osmotic pressures. This adds biosecurity that any L-forms that escape into the environment will not survive. This is one reason that genetically modified L-forms could be used in the agriculture industry as an alternative to other engineered bacteria. We conducted experiments to decipher what osmotic pressures are suitable for our B. subtilis L-forms. This included growing L-forms in 50/50 NB/MSM media with a variety of sucrose concentrations. A soil sample was also taken from outside the centre for bacterial cell biology, water was extracted. This was used to test the ability of L-forms to survive in soil water, in the event that they escape from a laboratory or a host plant.
| Sucrose Concentration (M) | Time (mins) | O.D (600) | Rate of Decrease (%) |
|---|---|---|---|
| 0 | 0.640 | ||
| 10 | 0.126 | ||
| 20 | 0.114 | ||
| 0 (positive control) | 30 | 0.106 | 90.31 |
| 40 | 0.100 | ||
| 50 | 0.080 | ||
| 60 | 0.062 | ||
| 0 | 0.691 | ||
| 10 | 0.123 | ||
| 20 | 0.100 | ||
| 0.1 | 30 | 0.091 | 88.71 |
| 40 | 0.083 | ||
| 50 | 0.080 | ||
| 60 | 0.078 | ||
| 0 | 1.705 | ||
| 10 | 0.676 | ||
| 20 | 0.605 | ||
| 0.2 | 30 | 0.559 | 74.25 |
| 40 | 0.523 | ||
| 50 | 0.491 | ||
| 60 | 0.439 | ||
| 0 | 1.604 | ||
| 10 | 0.828 | ||
| 20 | 0.772 | ||
| 0.3 | 30 | 0.729 | 59.16 |
| 40 | 0.685 | ||
| 50 | 0.672 | ||
| 60 | 0.655 | ||
| 0 | 1.546 | ||
| 10 | 0.984 | ||
| 20 | 0.891 | ||
| 0.4 | 30 | 0.865 | 49.55 |
| 40 | 0.847 | ||
| 50 | 0.810 | ||
| 60 | 0.780 | ||
| 0 | 1.768 | ||
| 10 | 0.222 | ||
| 20 | 0.190 | ||
| Soil | 30 | 0.150 | 95.93 |
| 40 | 0.128 | ||
| 50 | 0.096 | ||
| 60 | 0.072 | ||
| 0 | 1.569 | ||
| 10 | 1.569 | ||
| 20 | 1.569 | ||
| 0.5 (negative control) | 30 | 1.583 | -1.28 |
| 40 | 1.584 | ||
| 50 | 1.584 | ||
| 60 | 1.589 |
Table 1. Change in the optical density of L-forms in different media, over time. This highlights how a change in osmotic pressure can affect the propensity of L-forms to survive.
File:BareCillus Popping trim.mp4 Next page
Purpose
To create a BioBrick that enables the conversion of cell-walled B. subtilis cells into L-form cells that are cell wall deficient. L-forms should be able to be maintained in wall-less state once conversion from walled form is successful. The BioBrick should also facilitate the reversion back to walled cells when desired.
Aims
- Design a BioBrick which places murE under the control of a controllable promoter.
- Have this designed BioBrick synthesised.
- Integrate the designed BioBrick into B.subtilis.
- Determine the functionality of the BioBrick in removing the cell wall.
References
[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3603455/ Errington J. (2013) L-form bacteria, cell walls and the origins of life. Open Biology, 3, 120143.]
[http://www.ncbi.nlm.nih.gov/pubmed/19212404 Leaver M., Dominguez-CuevasP., Coxhead J.M., Daniel R.A. and Errington J. (2009) Life without a wall or division machine in Bacillus subtilis. Nature, 457, 849-853.]
[http://www.ncbi.nlm.nih.gov/pubmed/23452849 Mercier R., Kawai Y. and Errington J. (2013) Excess membrane synthesis drives a primitive mode of cell proliferation. Cell, 152, 997-1007.]
 "
"