Team:Imperial College/Modelling P3HBsyn
From 2013.igem.org
Contents |
Modelling: P(3HB) synthesis module
Specifications
- Input = monomer (3HB)
- Output = polymer (P(3HB))
Deterministic model
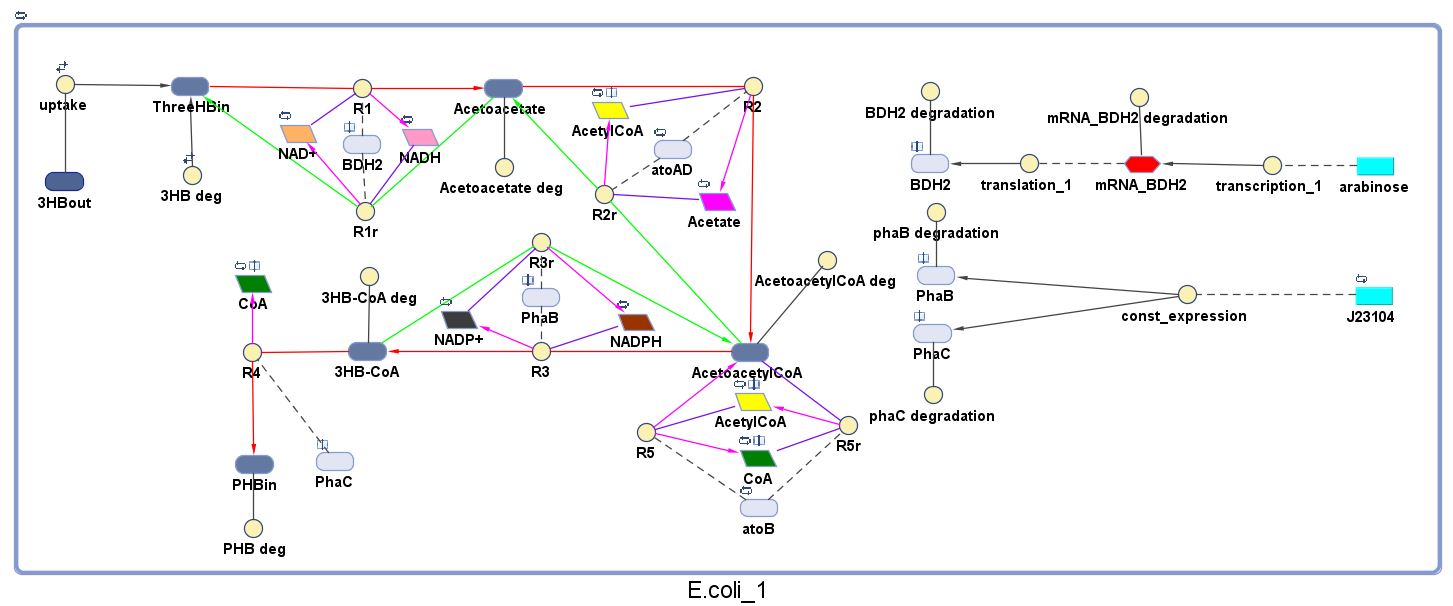
The model was built and simulated in Simbiology, a Matlab package designed for modelling biological systems. In our model we have a compartment (labelled "E. coli_1") that represents our engineered cell. The compartment contains the reactions and species that interact with the polymer synthesis pathway. The pathways on the right half of the cell are genetic expressions of the 3 enzymes involved in the plastic synthesis pathway. The pathways on the left represent the actual P(3HB) production pathway.
ODEs in Matlab Simbiology model
Genetic regulations and expressions
BDH2 – 3-hydroxybutyrate dehydrogenase
phaB – Acetoacetyl-CoA reductase
phaC – P(3HB) synthase
Enzyme kinetics
BDH2 – 3-hydroxybutyrate dehydrogenase
atoAD – Acetyl-CoA:acetoacetyl-CoA transferase (α and β subunits)
phaB – Acetoacetyl-CoA reductase
phaC – P(3HB) synthase
Simulation results
P(3HB) production
Model-guided design and optimisation
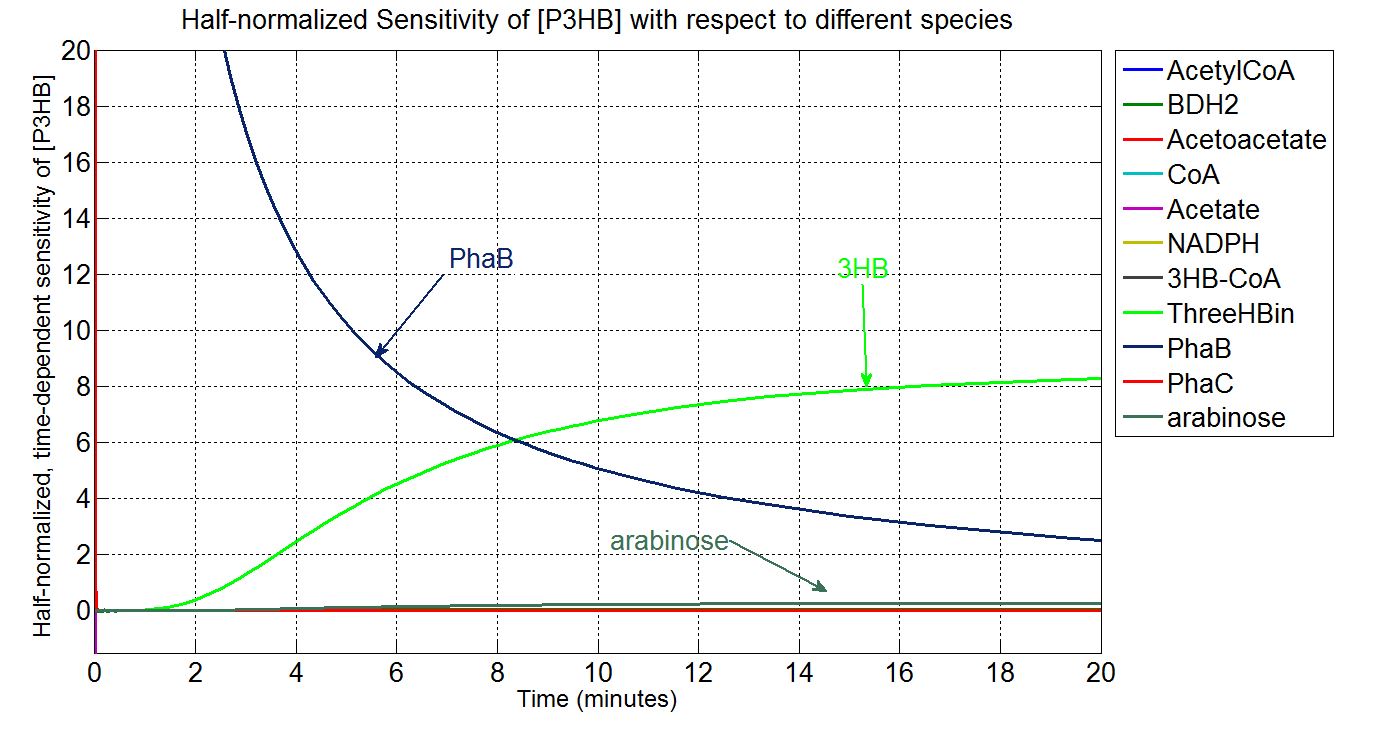
Sensitivity analysis: species concentrations
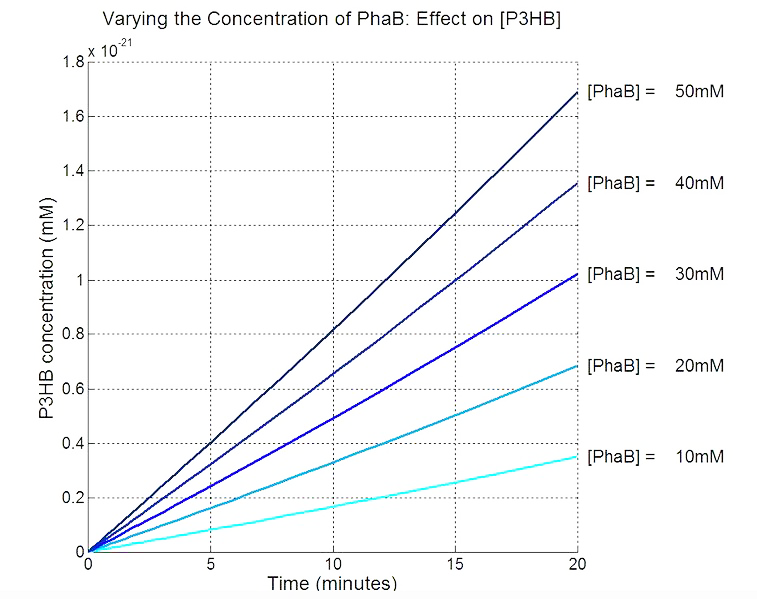
Scan with different levels of PhaB
Difference between promoter expressions after optimisation
Metabolic considerations
The metabolic model predicts the interaction of our pathway with endogenous pathways. In synthetic biology, an ideal system is fully orthogonal. However, it can be the case, such as here that coenzymes are needed for the reaction steps in a metabolic pathway. When we realised that our pathway uses a lot of Co-A, we constructed a model to see how this can be recycled and predict whether it would negatively effect the chassis. Tell that the Co-A is regenerated at the same speed as it is used and therefore there is not a huge amount of Co-A "stuck" in the pathway at any one time. (Bobby, how much Co-A is that exactly?) This informed us about that the cells should be able to bear the burden of the pathway.
Metabolic model to optimise PHB production. Tell us how you predicted that phaB is the key enzyme and justify why the increase in enzyme level will increase the amount of PHB. This informed our design and we are changing the promoter in front of the phaCAB operon. Tell that if we can predict that it would be useful to downregulate any genes in the native metabolism of E.coli.
Effect on growth and interference with metabolism
Initial concentrations of metabolites
 "
"