Team:TU-Munich/Notebook/Labjournal
From 2013.igem.org
}$(document).ready(function(){
// put the footer in the right place
$("#footer-box").prepend($("#social-footer"));
// implement image preloading
var images = new Array()
function preload() {
for (i = 0; i < preload.arguments.length; i++) {
images[i] = new Image()
images[i].src = preload.arguments[i]
}
}
// preload menu backgrounds
preload( " ",
",
"", "
", "
", "
", "
" );
// preload team pictures
if ( $("div#teamfield").length > 0 ) {
preload( " ",
",
"", "
", "
", "
", "
", "
", "
", "
", "
", // Katrin "
", "
", "
", "
", "
", "
", "
", "
", "
", // Rosario "
", "
", "
", "
", "
", "
", "
", "
", "
", // Fabian "
", "
", "
", "
", "
", "
", "
", "
", "
", // Andreas "
", "
", "
", "
", "
", "
", "
", "
", "
", // Louise "
", "
", "
", "
", "
", "
", "
", "
", "
", // Johanna "
", "
", "
", "
", "
", "
", "
", "
", "
", // Meike "
", "
", "
", "
", "
", "
", "
", "
", "
", // Volker "
", "
", "
", "
", "
", "
", "
", "
", "
", // Polte "
", "
", "
", "
", "
", "
", "
", "
", "
", // Leonie "
", "
", "
", "
", "
", "
", "
", "
", "
", // Philipp "
", "
", "
", "
", "
", "
", "
", "
", "
", // Jeff "
", "
", "
", "
", "
", "
", "
", "
", "
", // Chris "
", "
", "
", "
", "
", "
", "
", "
", "
", // Flo "
", "
", "
", "
", "
", "
", "
", "
" );
}
// Slideshows
$('.bxslider').bxSlider({
responsive: false, auto: true, autoHover: true
});
$('.bxgallery').bxSlider({
slideMargin: 10, minSlides: 3, maxSlides: 3, moveSlides: 1, slideWidth: 5000
});
$("ul.bxgallery img").slimbox({ loop: true }, function(el) { url = el.src; url = url.substring(0, url.lastIndexOf('/')); url = url.replace('/thumb/', '/'); // description = $(el).parents("div.thumbinner").children("div.thumbcaption").text(); return [url, ]; }, function(el) { return (this == el) || (this.parentNode.parentNode && (this.parentNode.parentNode == el.parentNode.parentNode)); });
// Counter and Countdown
function render_counter(c) { i = 0; iid = window.setInterval(function(){ if ( (c-i) > (c/200) ) { $('span#counter').html(i); i += Math.round(c/200); } else { $('span#counter').html(c); window.clearInterval(iid); } }, 10); }
if ($('span#counter').length > 0) { $.ajax({ url: "https://2013.igem.org/Special:PopularPages", success: function( html ) { dom = $.parseHTML(html); visitors = $(dom).find('a[title="Team:TU-Munich"]').parent().text(); visitors = visitors.substring(visitors.indexOf('(')+1); visitors = visitors.substring(0, visitors.indexOf(' ')); visitors = visitors.replace(',', ); render_counter(visitors); }, error: function( xhr, status ) { render_counter(4700); } }); }
if ($('span#countdown) { clock = window.setInterval(function(){ jetzt = new Date(); time_left = Date.UTC(2013, 9, 5, 4, 0, 0) - Date.UTC(jetzt.getUTCFullYear(), jetzt.getUTCMonth(), jetzt.getUTCDate(), jetzt.getUTCHours(), jetzt.getUTCMinutes(), jetzt.getUTCSeconds()); left_sec = (time_left/1000)%60; left_sec = (left_sec < 10) ? "0" + left_sec : left_sec; left_min = Math.floor(time_left/60000)%60; left_min = (left_min < 10) ? "0" + left_min : left_min; left_h = Math.floor(time_left/3600000)%24; left_h = (left_h < 10) ? "0" + left_h : left_h; left_d = Math.floor(time_left/86400000); left_d = (left_d == 1) ? left_d + " day" : left_d + " days"; $('span#countdown').html(left_d + " " + left_h + ":" + left_min + ":" + left_sec); }, 1000); }
// Animate teamfield
if ( $("div#teamfield").length > 0 ) {
var $members = $("div#teamfield a");
$("body").mousemove(function(event){ for (i=0; i<$members.length; i++) {
if ( $members.eq(i).offset().left > event.pageX ) {
if ( $members.eq(i).offset().top > event.pageY ) {
$members.eq(i).removeClass();
$members.eq(i).addClass("top-left");
} else if ( $members.eq(i).offset().top <= event.pageY && ( $members.eq(i).offset().top + $members.eq(i).height() ) >= event.pageY ) {
$members.eq(i).removeClass();
$members.eq(i).addClass("left");
} else if ( ( $members.eq(i).offset().top + $members.eq(i).height() ) < event.pageY ) {
$members.eq(i).removeClass();
$members.eq(i).addClass("bottom-left");
}
} else if ( $members.eq(i).offset().left <= event.pageX && ( $members.eq(i).offset().left + $members.eq(i).width() ) >= event.pageX ) {
if ( $members.eq(i).offset().top > event.pageY ) {
$members.eq(i).removeClass();
$members.eq(i).addClass("top");
} else if ( $members.eq(i).offset().top <= event.pageY && ( $members.eq(i).offset().top + $members.eq(i).height() ) >= event.pageY ) {
$members.eq(i).removeClass();
$members.eq(i).addClass("front");
} else if ( ( $members.eq(i).offset().top + $members.eq(i).height() ) < event.pageY ) {
$members.eq(i).removeClass();
$members.eq(i).addClass("bottom");
}
} else if ( ( $members.eq(i).offset().left + $members.eq(i).width() ) < event.pageX ) {
if ( $members.eq(i).offset().top > event.pageY ) {
$members.eq(i).removeClass();
$members.eq(i).addClass("top-right");
} else if ( $members.eq(i).offset().top <= event.pageY && ( $members.eq(i).offset().top + $members.eq(i).height() ) >= event.pageY ) {
$members.eq(i).removeClass();
$members.eq(i).addClass("right");
} else if ( ( $members.eq(i).offset().top + $members.eq(i).height() ) < event.pageY ) {
$members.eq(i).removeClass();
$members.eq(i).addClass("bottom-right");
}
}
} });
}
});
Labjournal
Week 1
Monday, April 22nd
Transformation of E. coli XL1 blue with Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3)
Investigator: Jeff, Rosario
Aim of the experiment: Transformation of Phytochrome B for protein fusion.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 2 µl of DNA was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of the cell suspension was plated on one chloramphenicol plate.
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on a new chlorampenicol plate.
Miniprep of pTUM100 with pGAL, pTEF1, pTEF2, pADH and RFC25 compatible RFP generator
Investigator: Jeff, Rosario
Aim of the experiment: Miniprep of pTUM100 with pGAL, pTEF1, pTEF2, pADH and RFC25 compatible RFP generator
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
Sequencing of RFP-Generator (RFC25, pSB1C3)
Investigator: Jeff, Rosario
Aim of the experiment: Sequencing of RFP-Generator (RFC25, pSB1C3)
Procedure:
Sequencing batch were prepared after manufacturer's protocol. (15 µl of plasmid DNA (50 - 100 ng) and 2 µl sequencing primer)
Tuesday, April 23rd
Picking of of E. coli XL1 blue with Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3)
Investigator: Jeff, Rosario, Florian
Aim of the experiment: Picking of of E. coli XL1 blue with Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3)
Procedure:
- pSB1C3 plasmid with BBa_K801031 (PhyB 2 - 908 aa, RFC25): Colonies were picked from chloramphenicol plates.
- Picked pipette tips was transferred into cell-culture tubes with air-permeable, sterile cover. Each tube contain 4 mL of LB-medium + 4 µL chloramphenicol(1000x).
- 4 colonies were picked.
- These tubes were transferred in a cell culture shaker at 37 °C and were incubated overnight
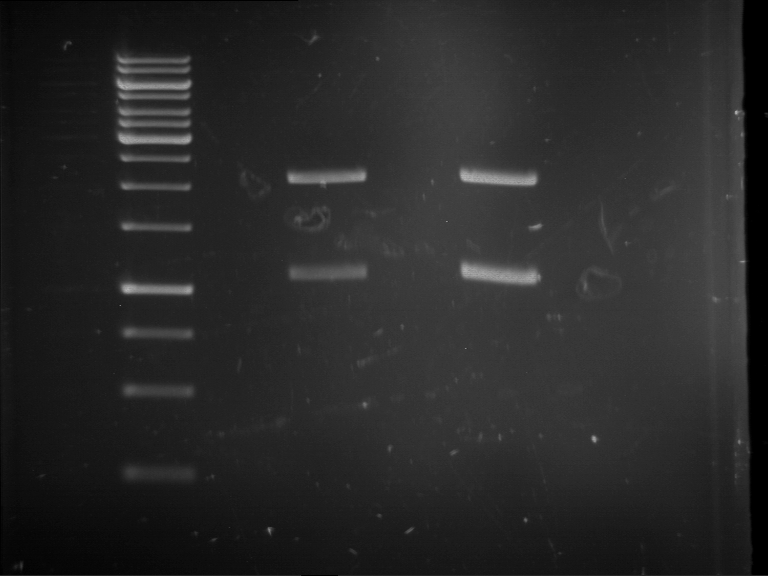
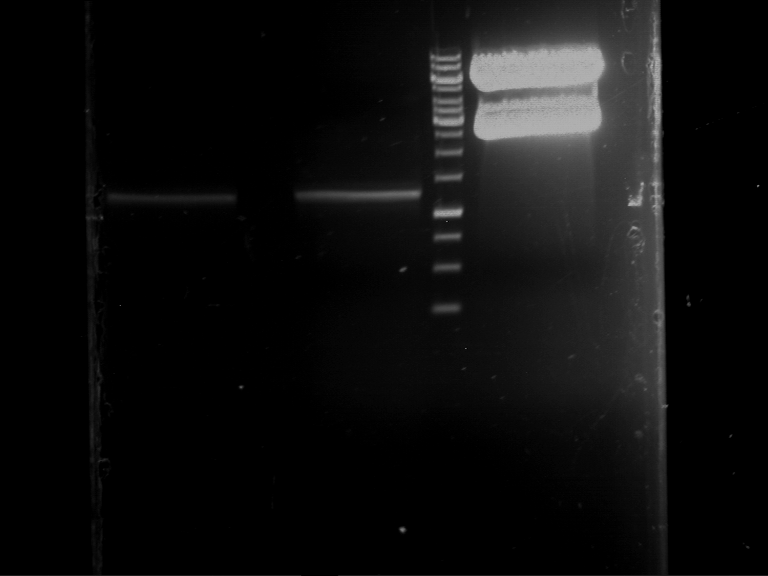
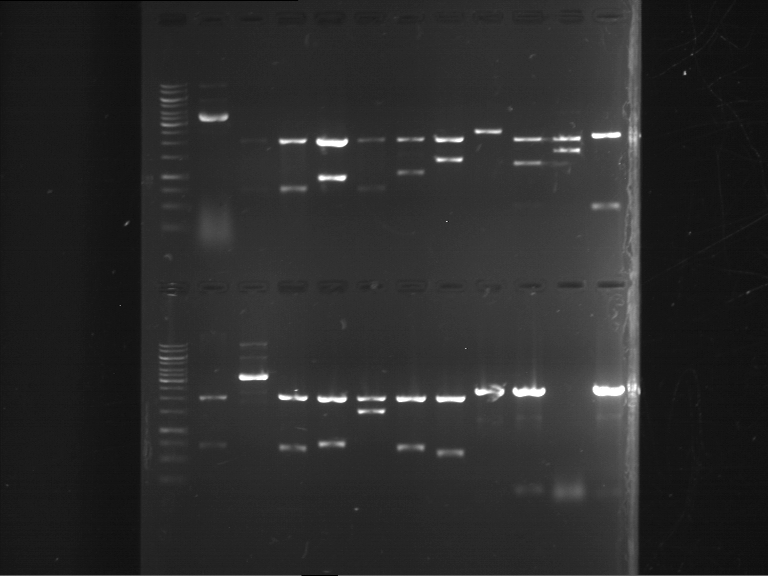
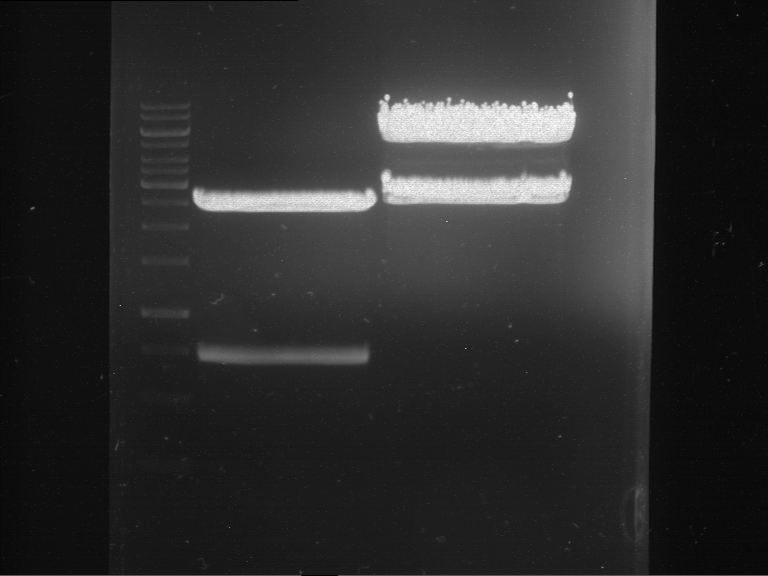
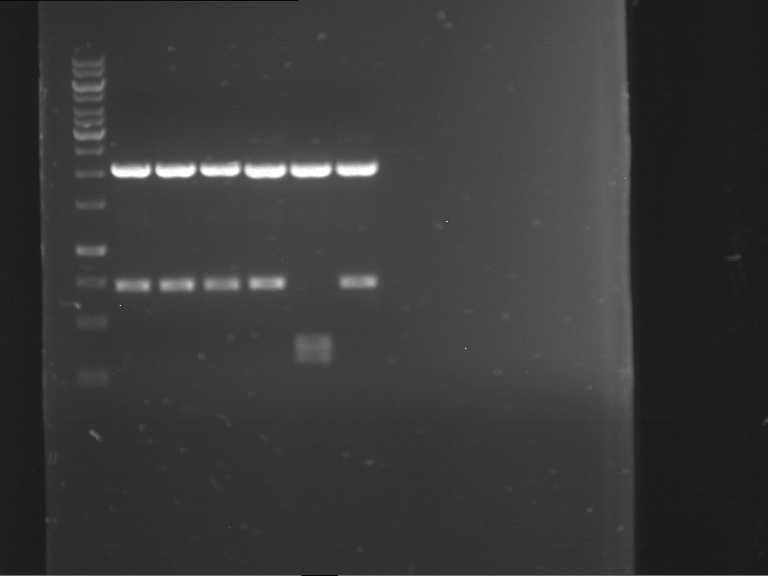
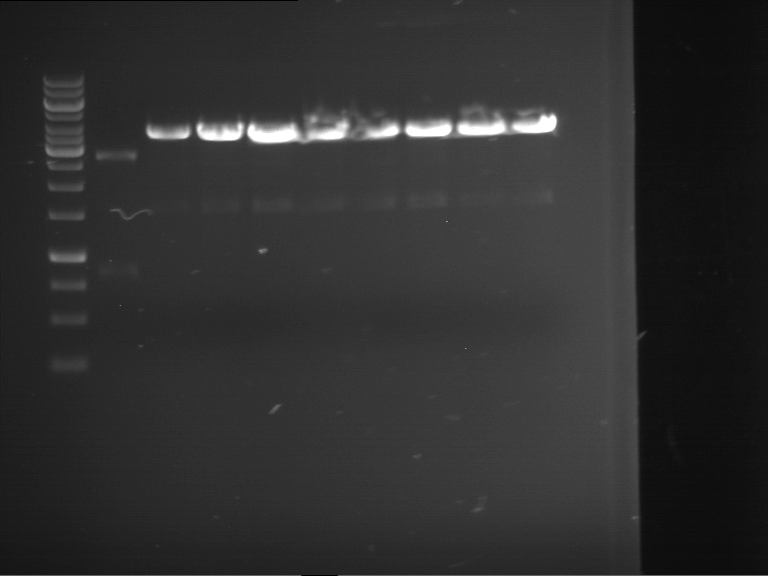
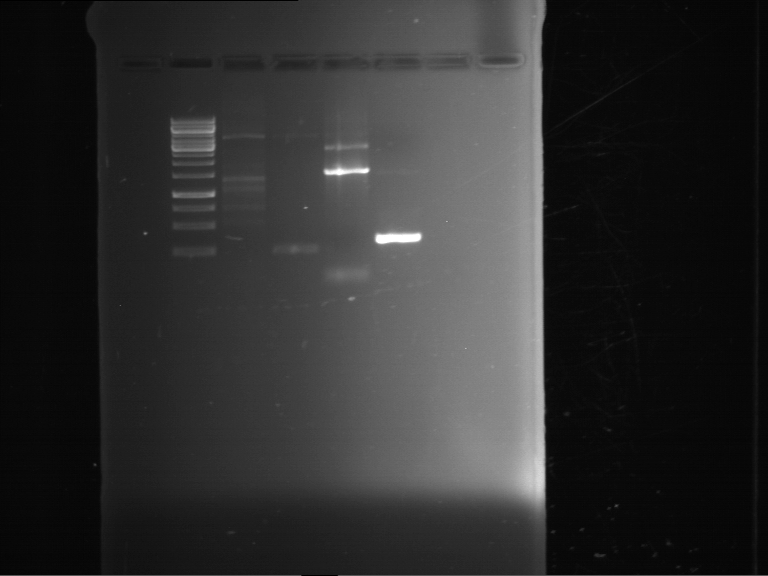
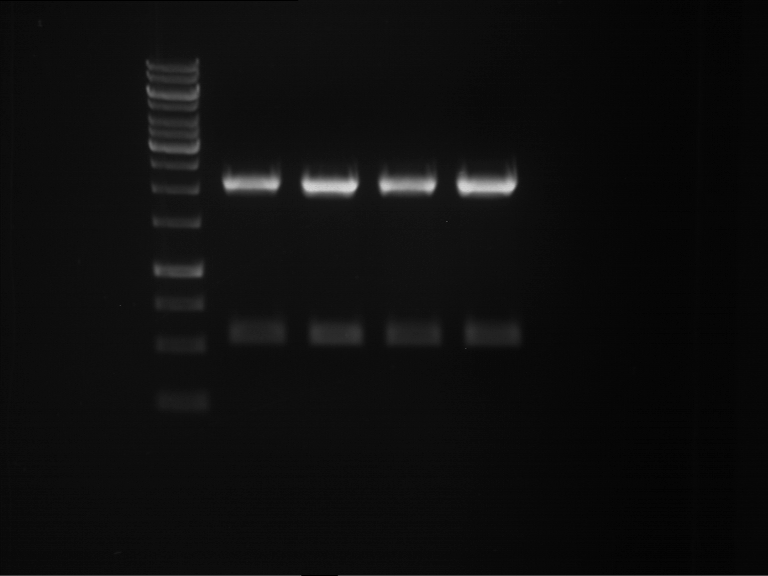
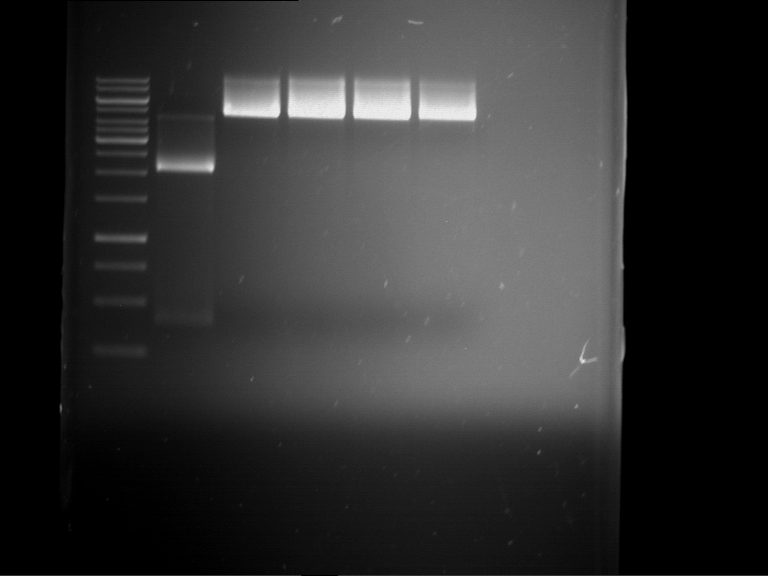
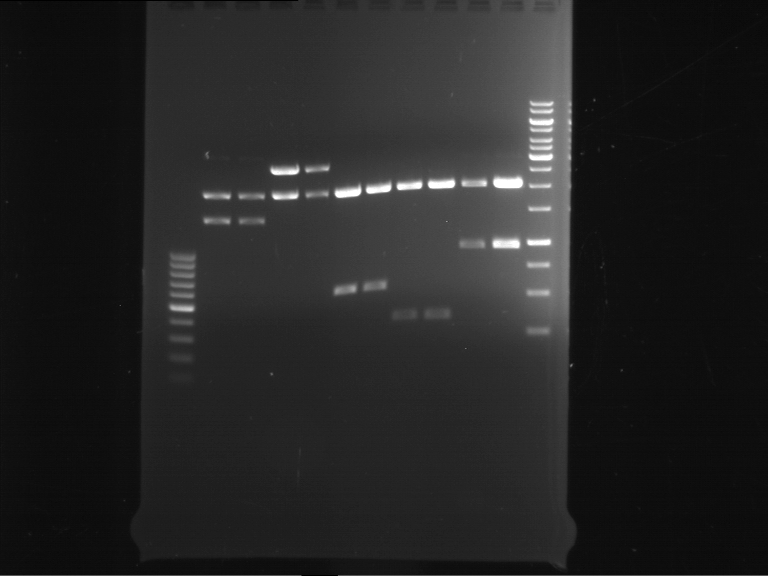
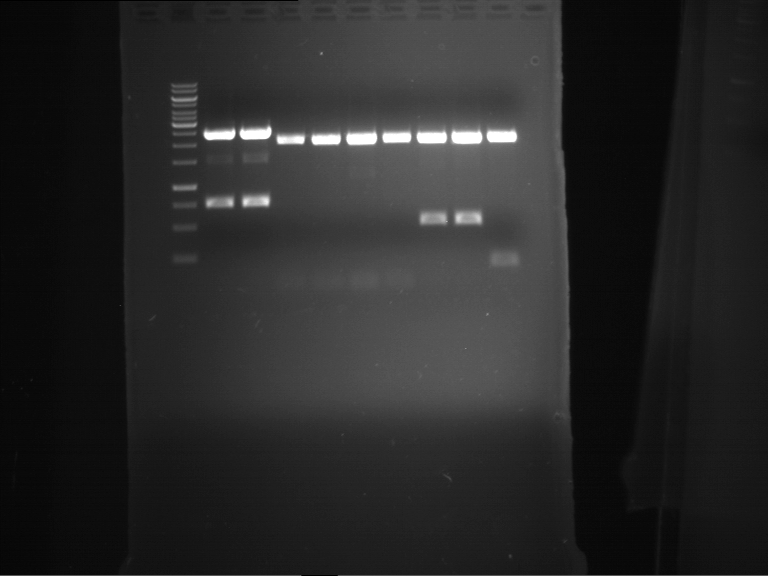
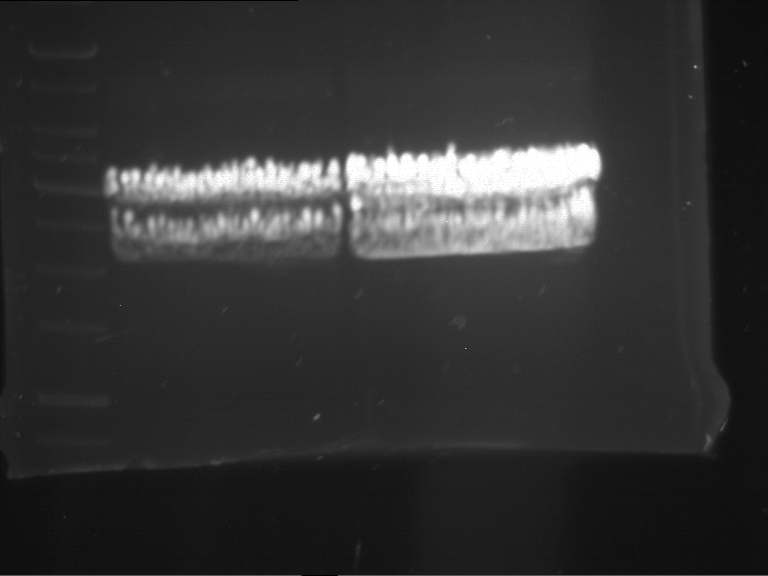
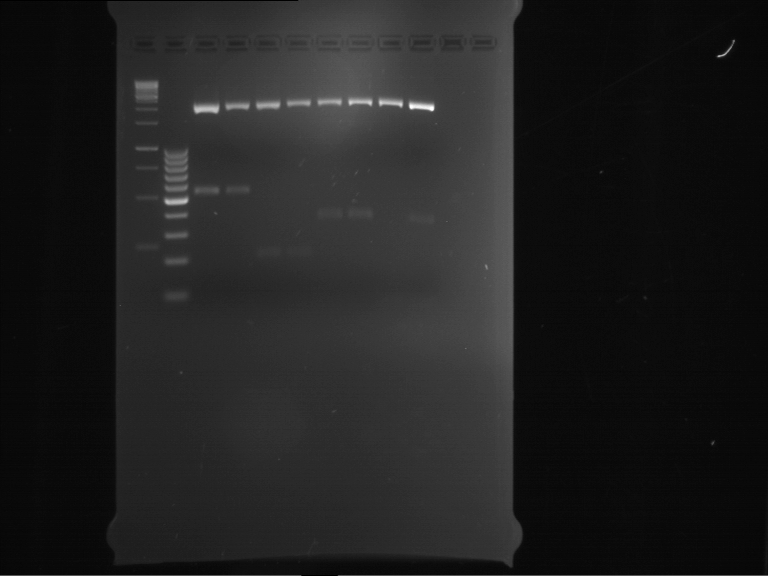
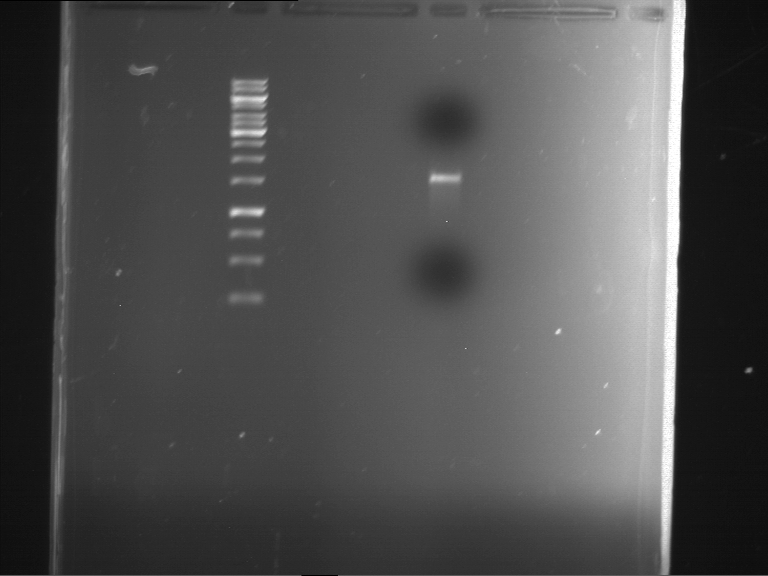
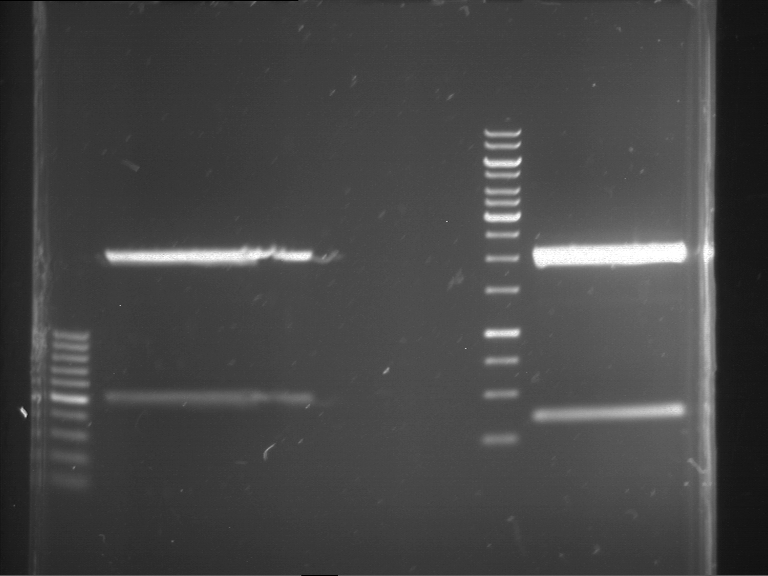
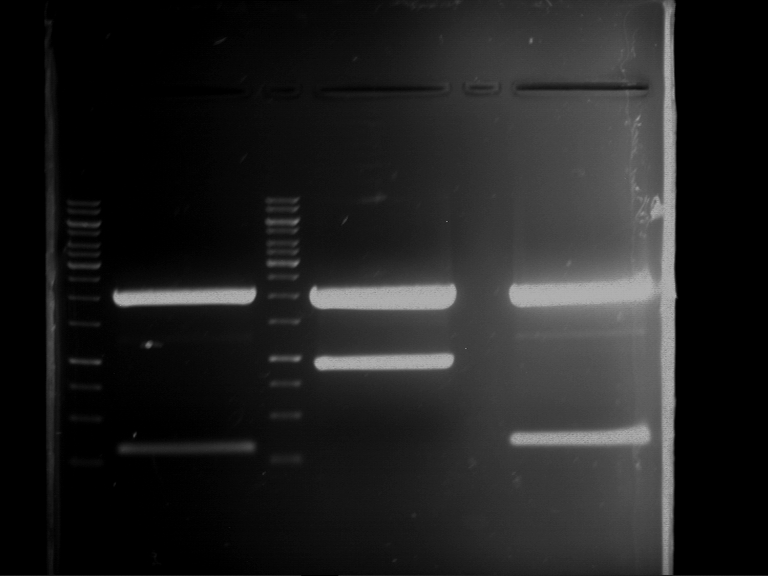
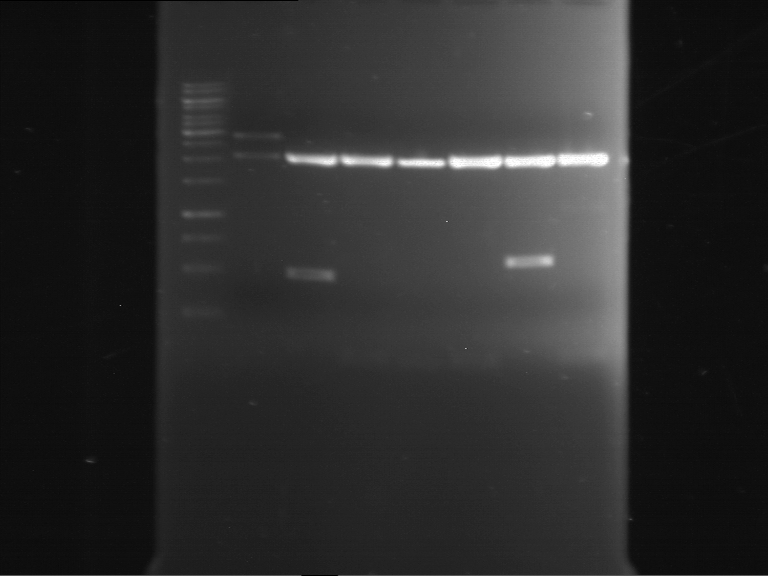
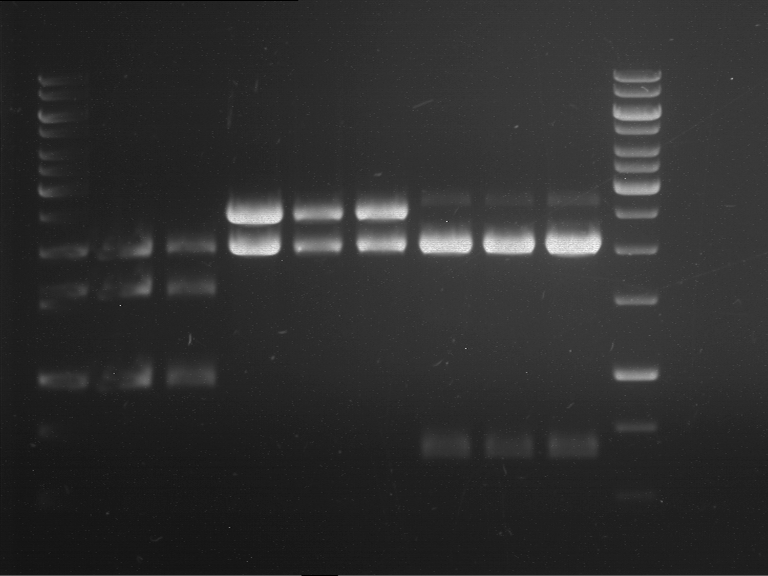
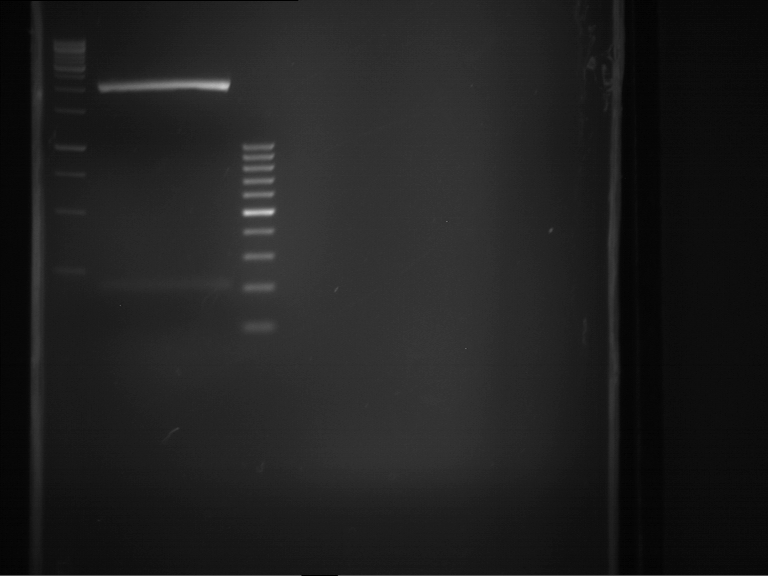
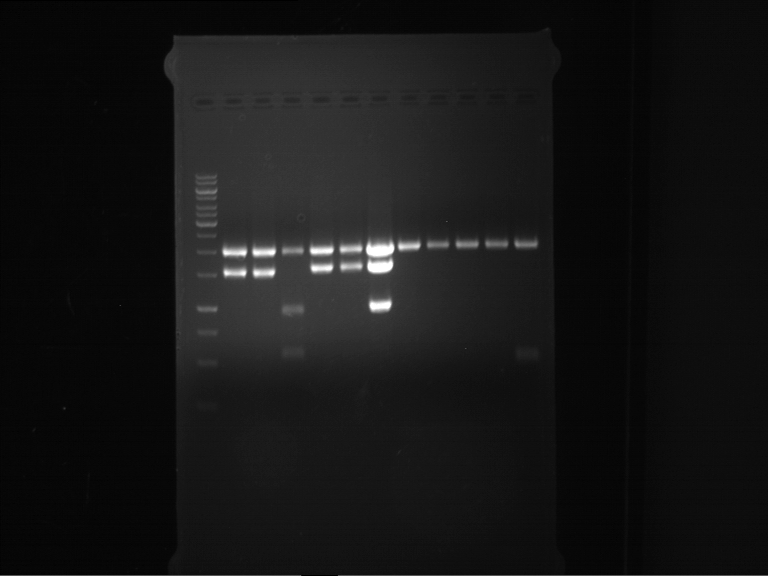
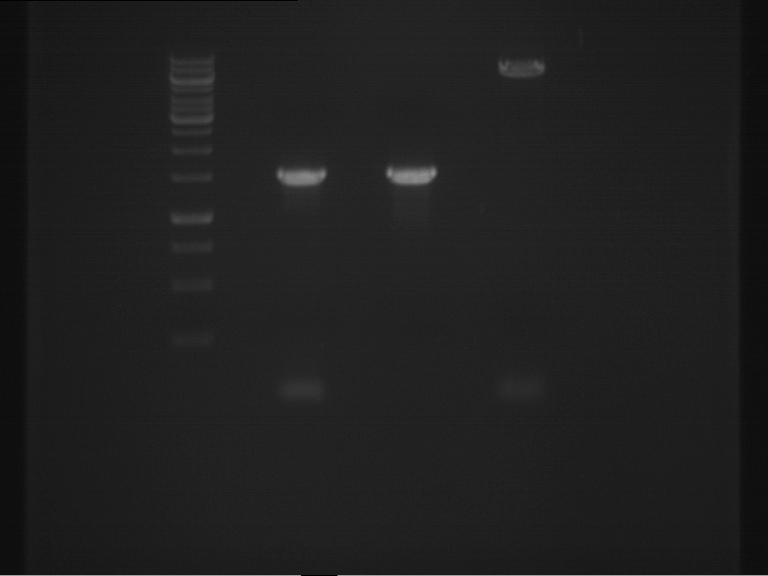
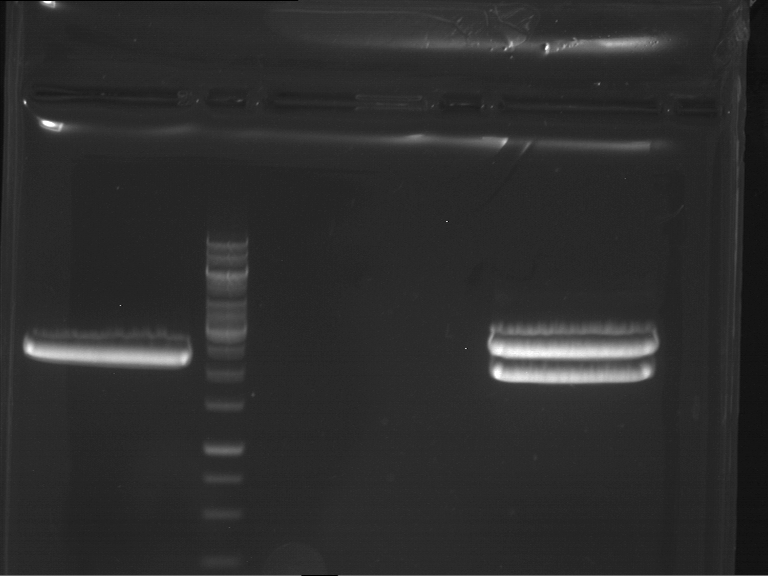
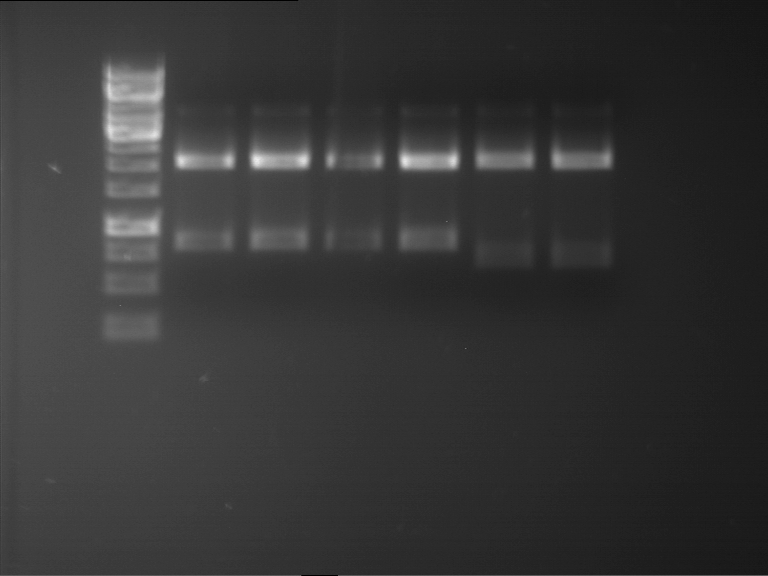
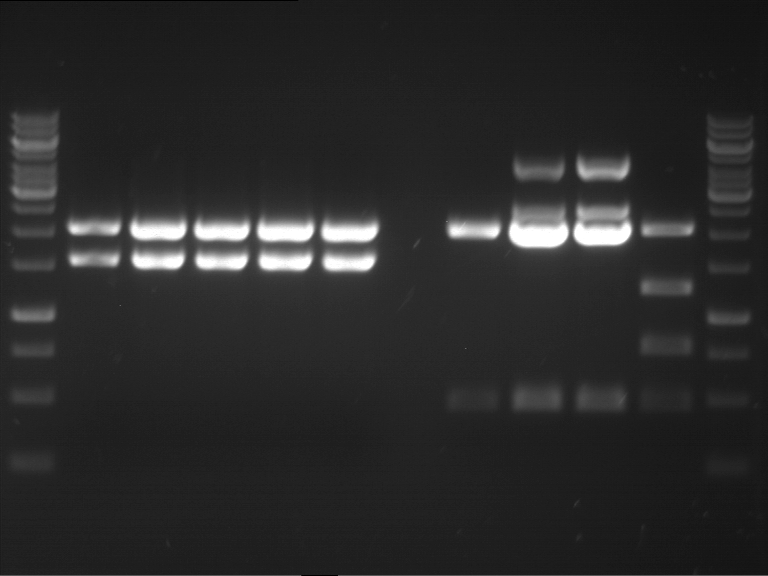
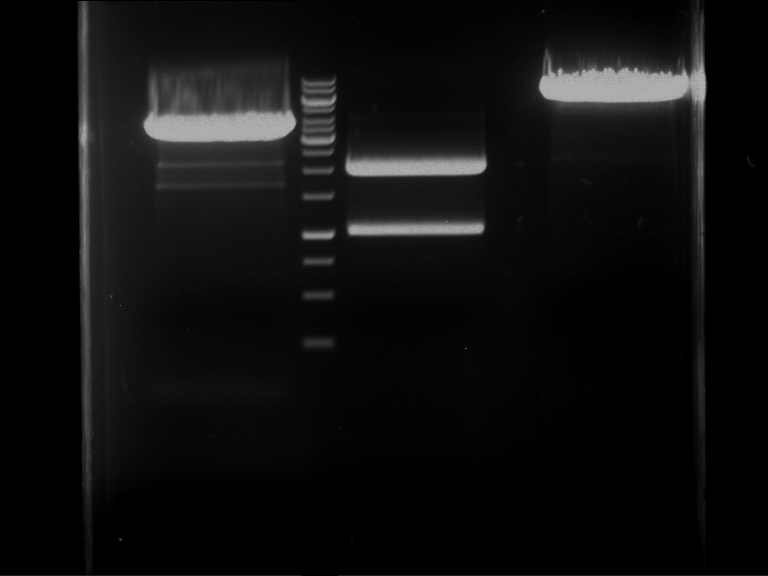
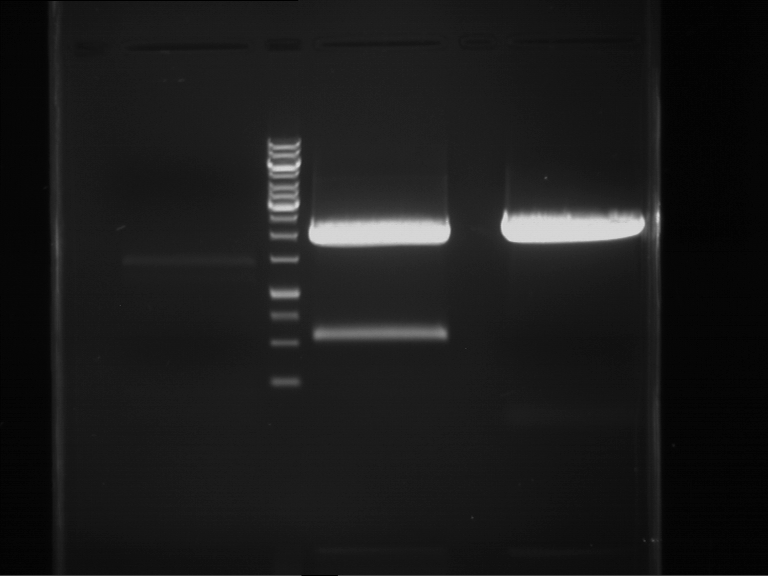
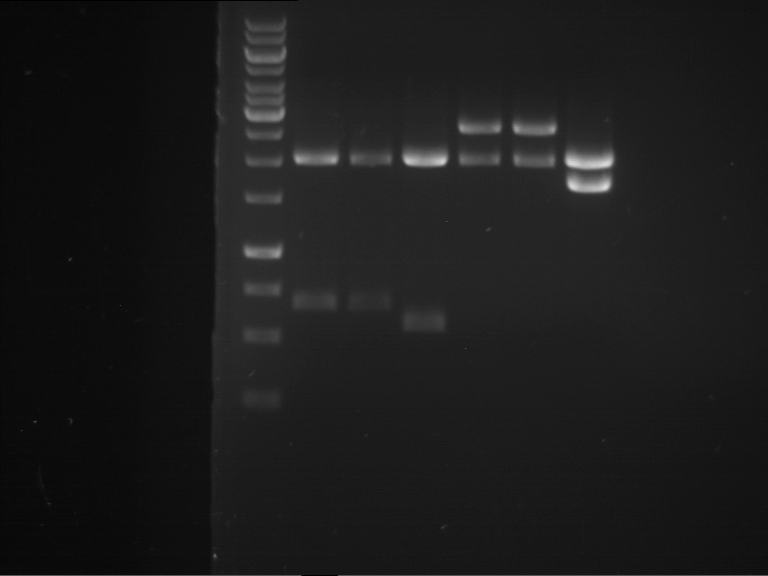
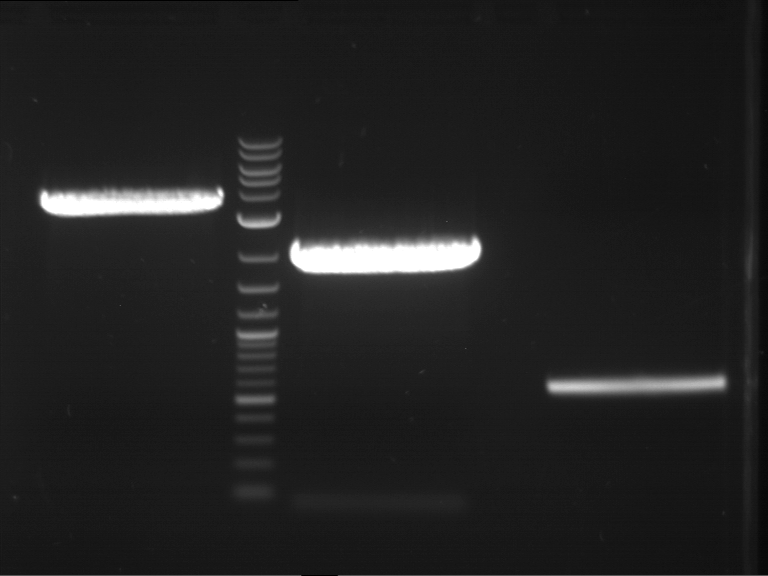
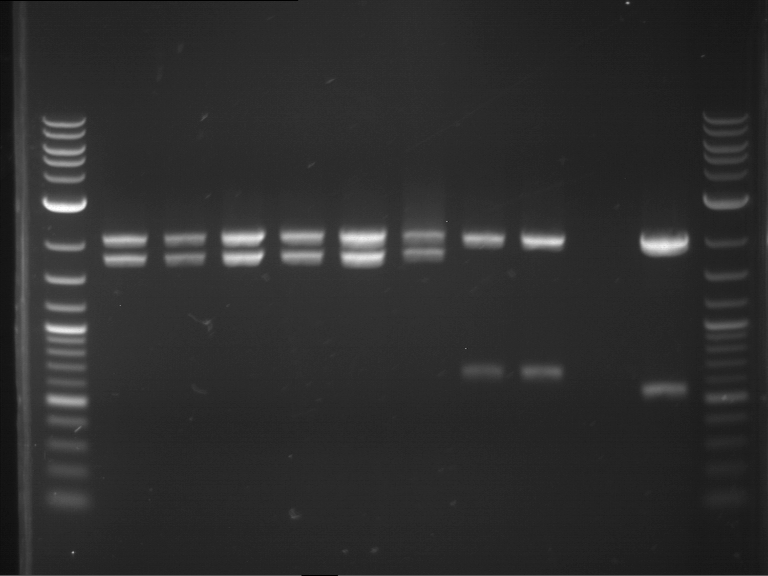
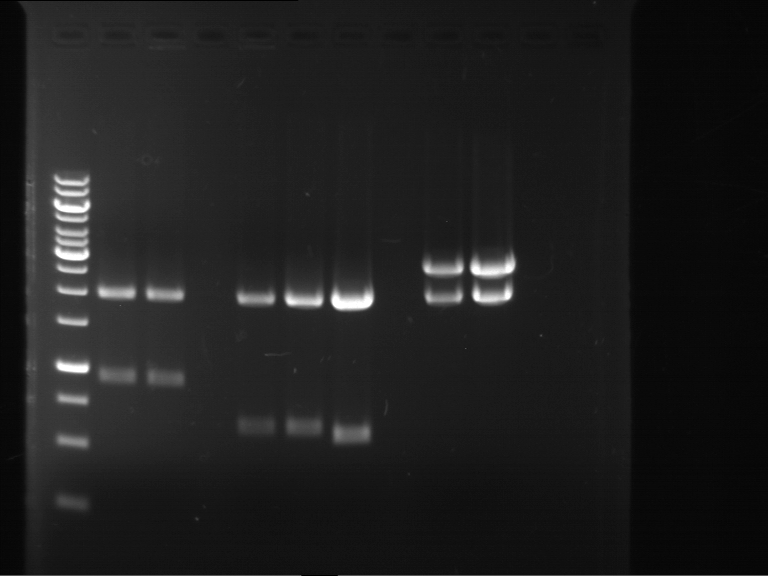
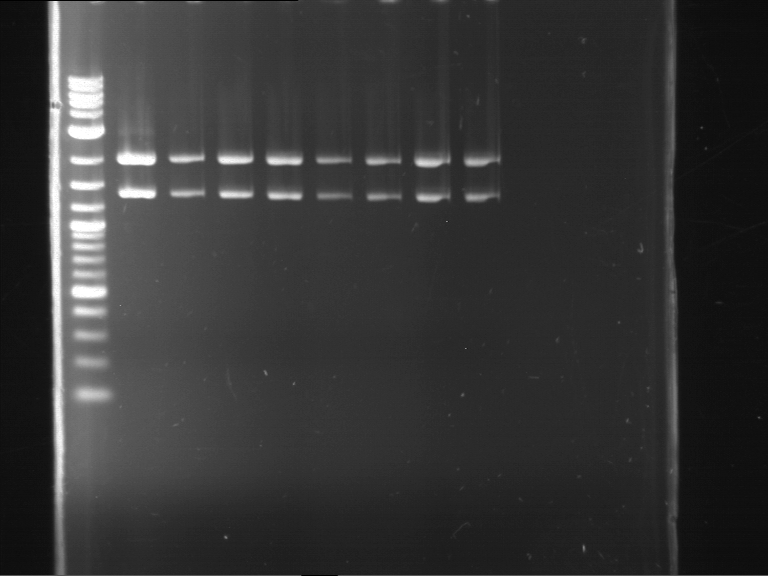
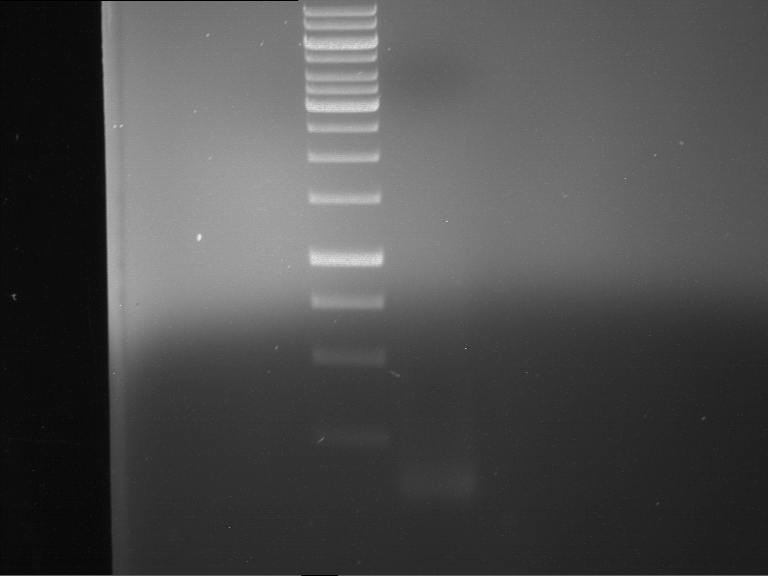
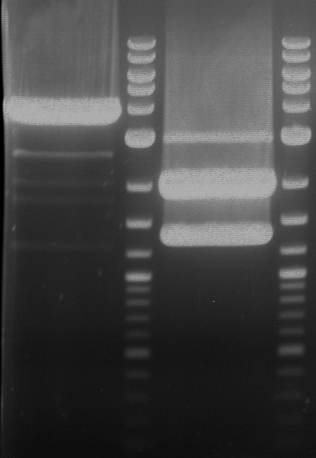
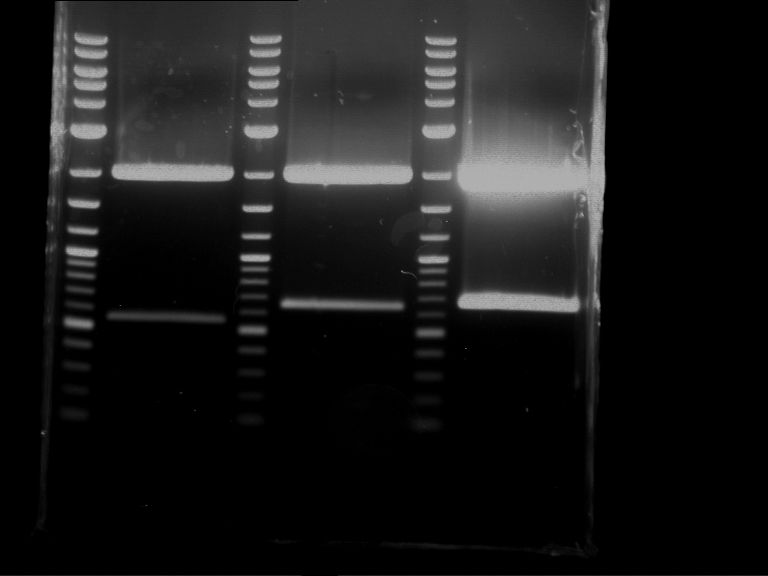
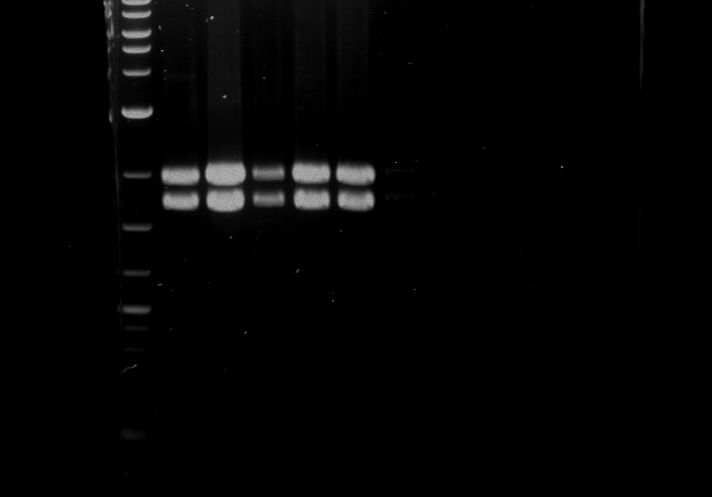
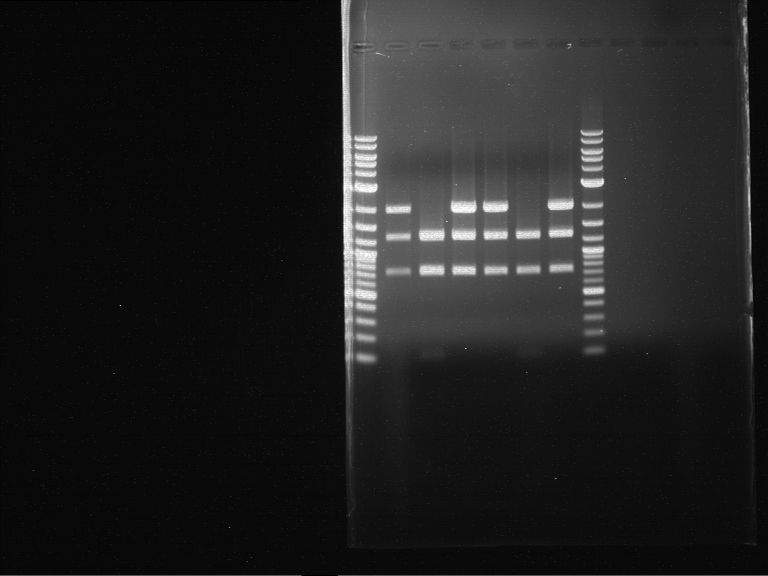
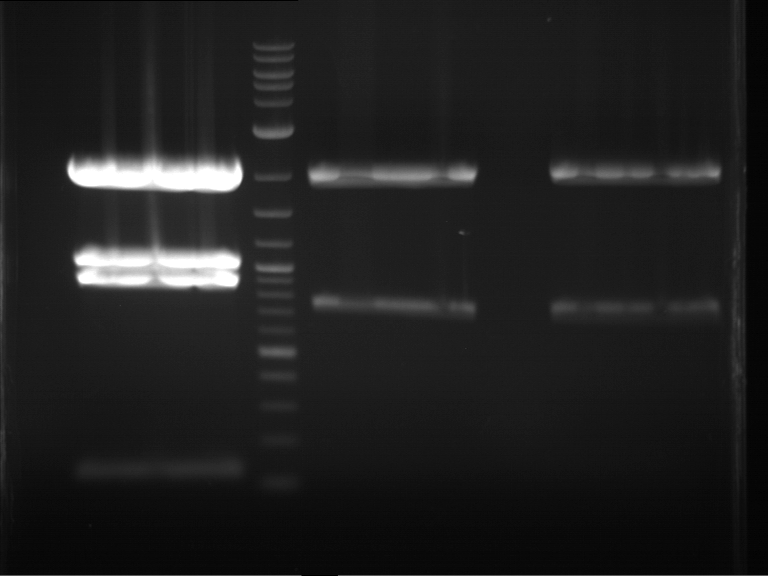
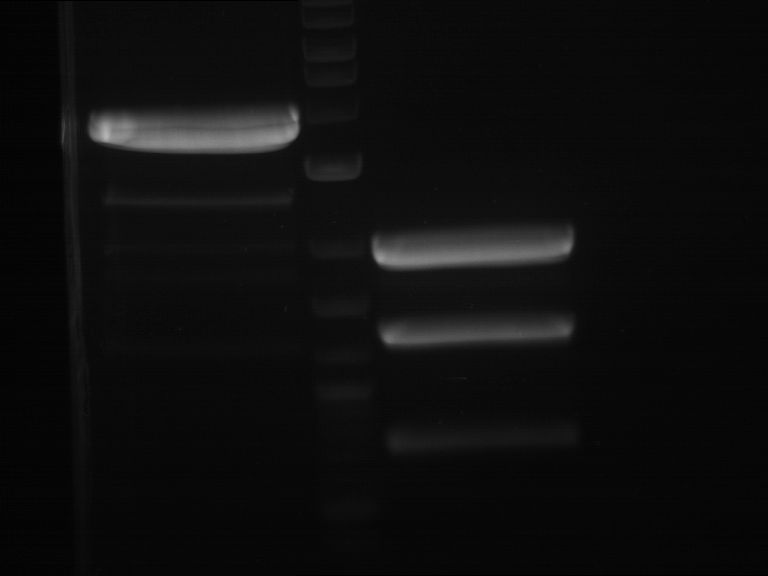
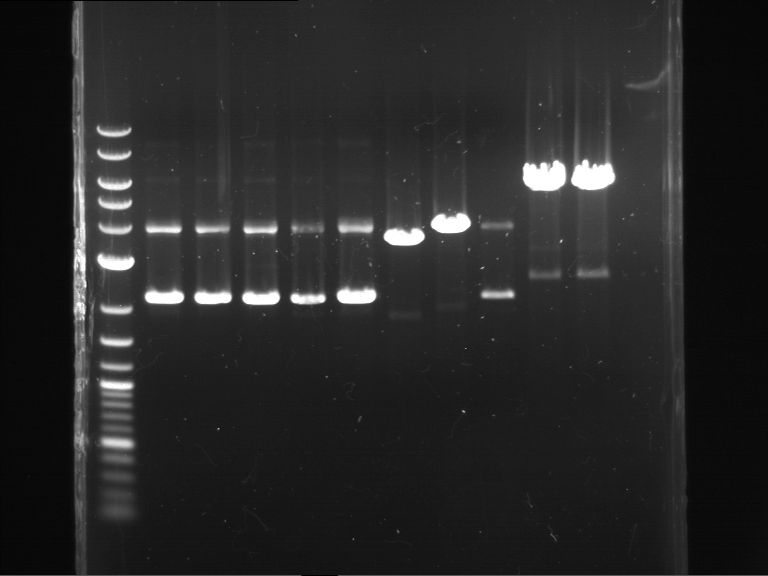
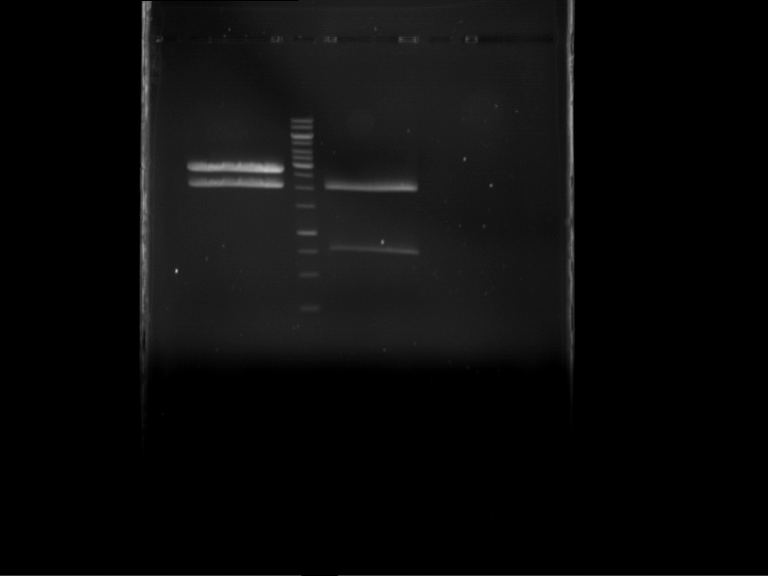
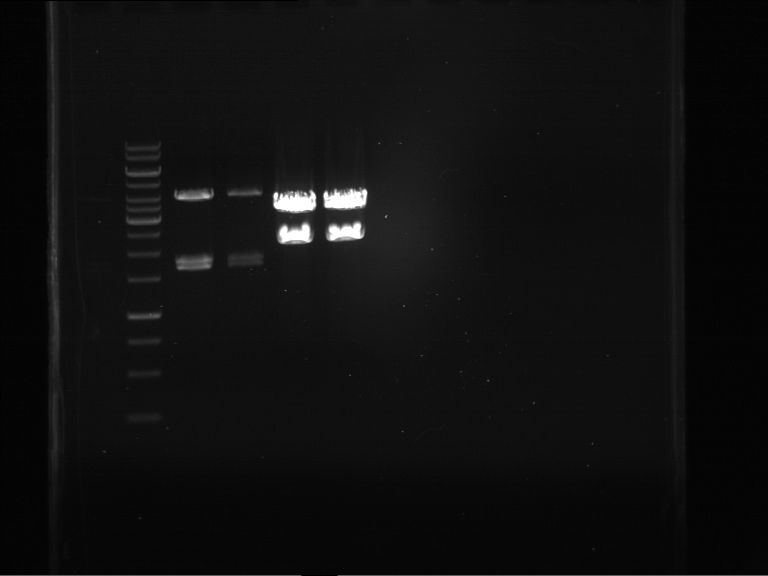
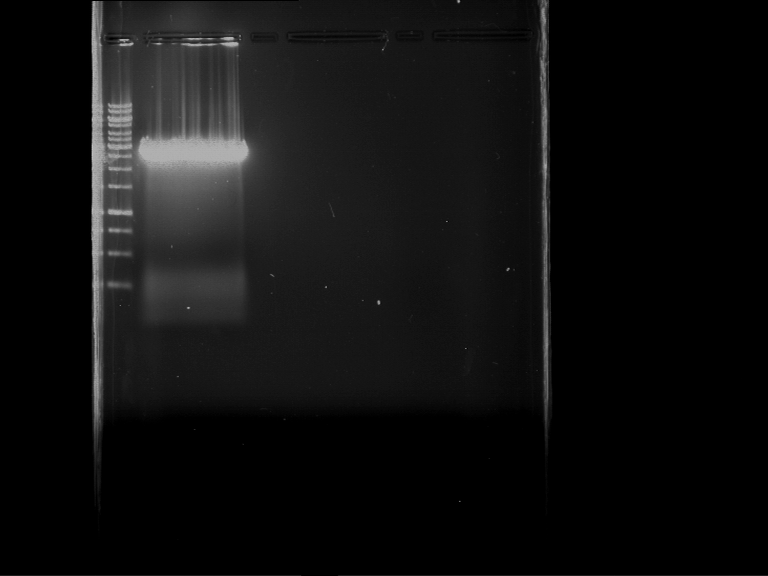
Analytical digestion and gelelectrophoresis of RFP-generator (RFC25, pSB1C3, P4 & P5)
Investigator: Jeff, Rosario, Florian
Aim of the experiment: Analytical digestion and gelelectrophoresis of RFP-generator (RFC25, pSB1C3, P4 & P5).
Procedure:
- Batch for analytical digestion for P4 with NgoMIV+AgeI-HF
| volume | reagent |
| 2.5 µl | Plasmid DNA P4 |
| 2 µl | NEBuffer 4 (10x) |
| 0.25 µl | NgoMIV (10 U/µl) |
| 0.25 µl | AgeI-HF (20 U/µl) |
| 15 µl | ddH2O |
| =20 µl | TOTAL |
- Batch for analytical digestion for P5 with NgoMIV+AgeI-HF
| volume | reagent |
| 2.5 µl | Plasmid DNA P5 |
| 2 µl | NEBuffer 4 (10x) |
| 0.25 µl | NgoMIV (10 U/µl) |
| 0.25 µl | AgeI-HF (20 U/µl) |
| 15 µl | ddH2O |
| =20 µl | TOTAL |
- Incubation for 90 min at 37 °C.
- Analytical gelelectrophoresis was performed at 90 V for 60 min.
Results:
| 1 kbp ladder DNA ladder | P4 | P5 |
| Mutation successful | Mutation successful! |
- Parts are compliant and do not contain RFC25 forbidden restriction sites.
Sequencing of pTUM vectors with pGAL, pADH, pTEF1, pTEF2
Investigator: Jeff, Rosario, Florian
Aim of the experiment: Sequencing of pTUM vectors with pGAL, pADH, pTEF1, pTEF2
Procedure:
Sequencing batch were prepared after manufacturer's protocol. (15 µl of plasmid DNA (50 - 100 ng) and 2 µl sequencing primer).
The different vectors we sequenced received the following barcodes:
- ADH in pTUM100: FR01002265
- TEF1 in pTUM100: FR01002266
- TEF2 in pTUM100: FR01002266
- GAL in pTUM100: FR01002268
Sequencing of TEF2 in pTUM100 was not interpretable. The other sequences were consistent with the sequences in the parts registry.
Wednesday, April 24th
Miniprep of Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3)
Investigator: Jeff, Florian
Aim of the experiment: Miniprep of Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3).
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
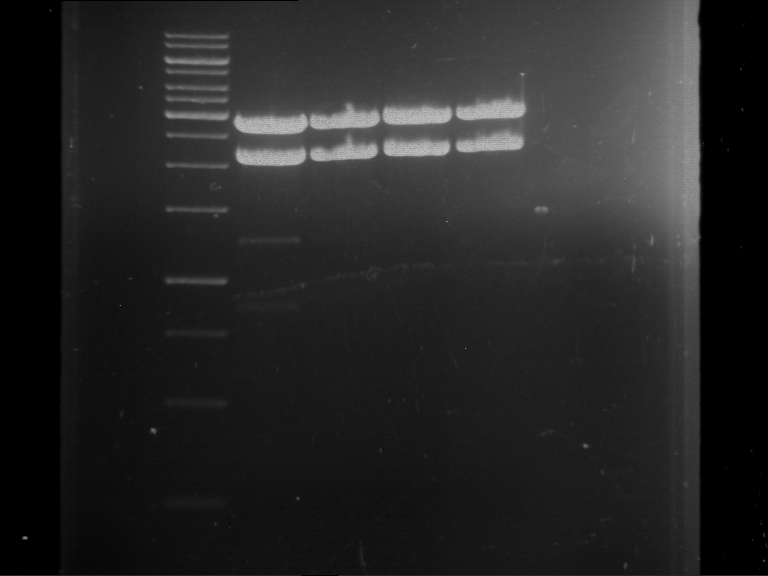
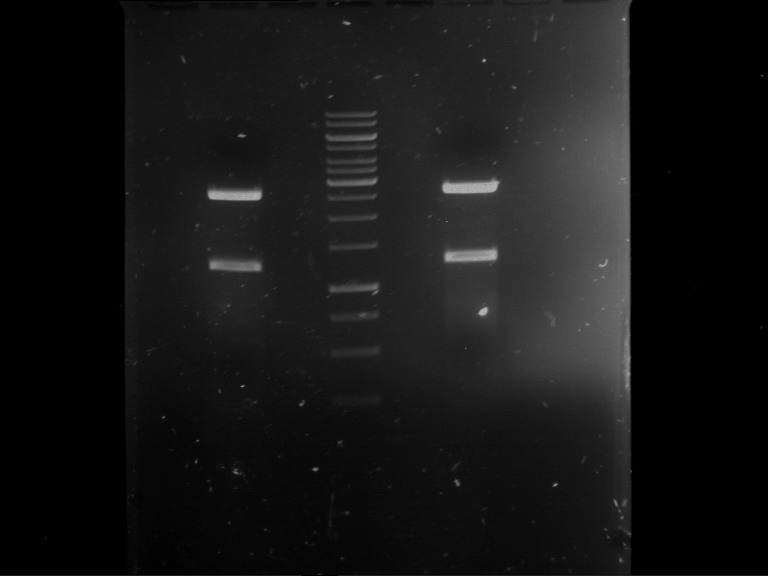
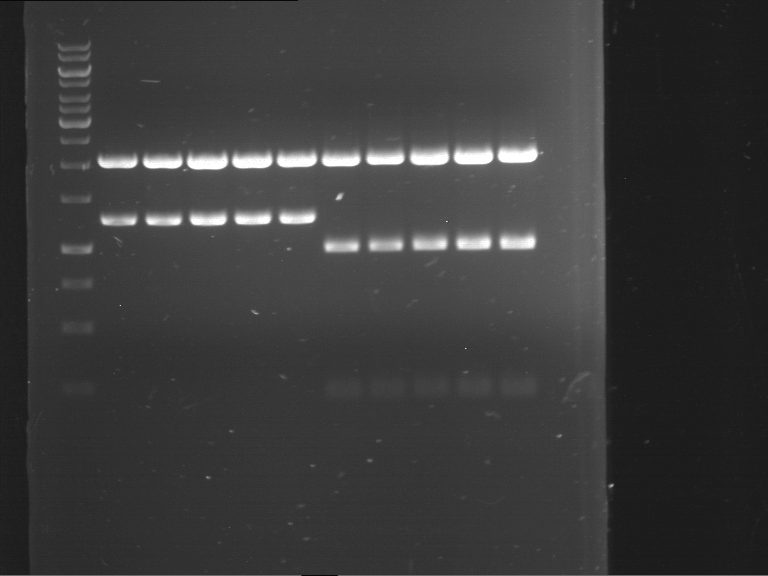
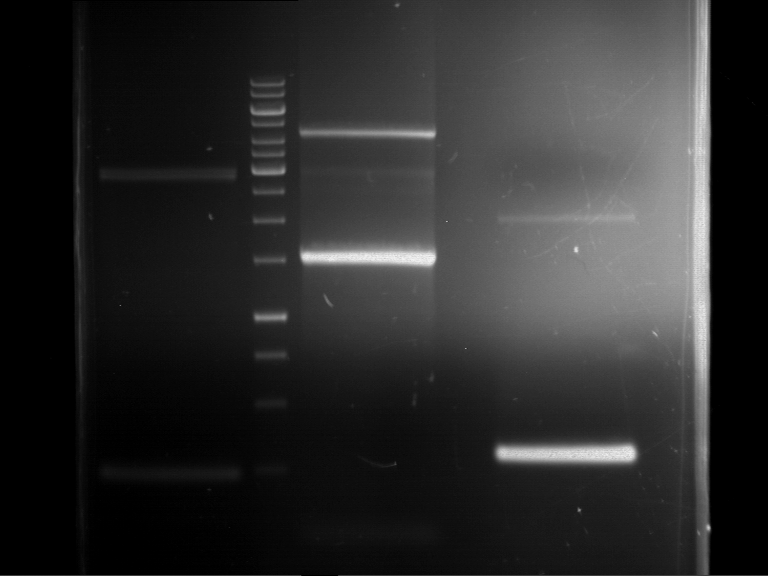
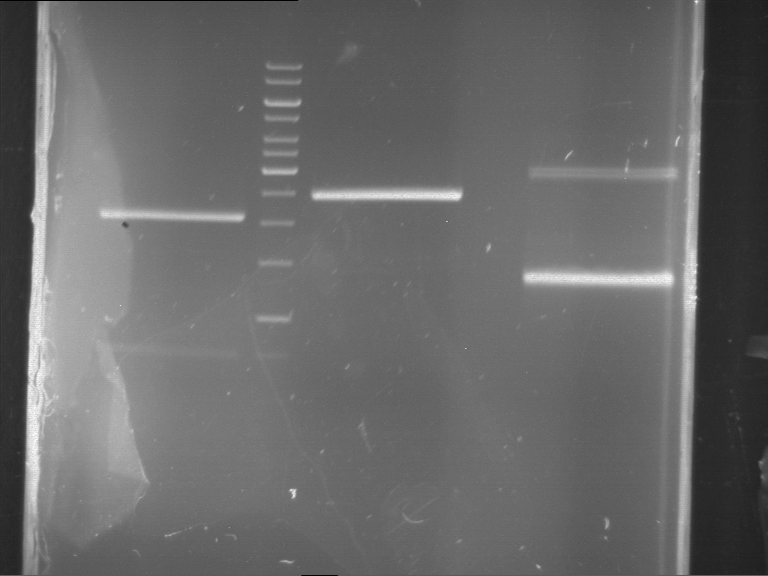
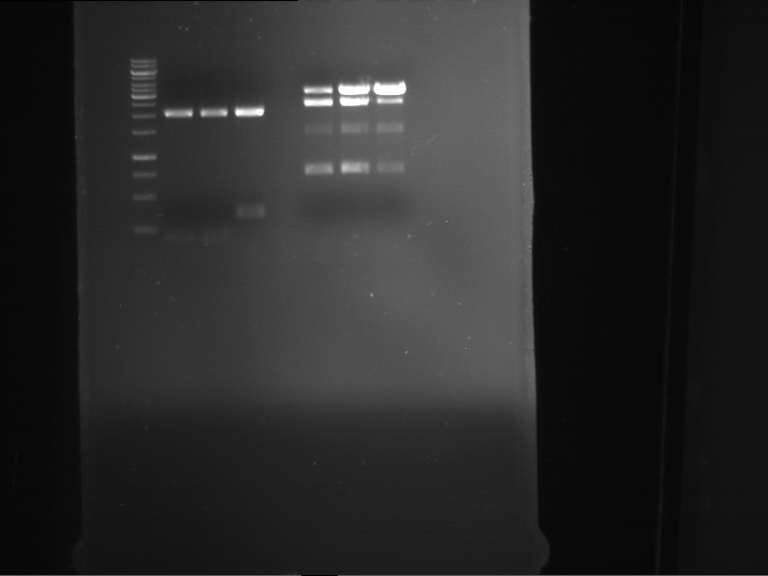
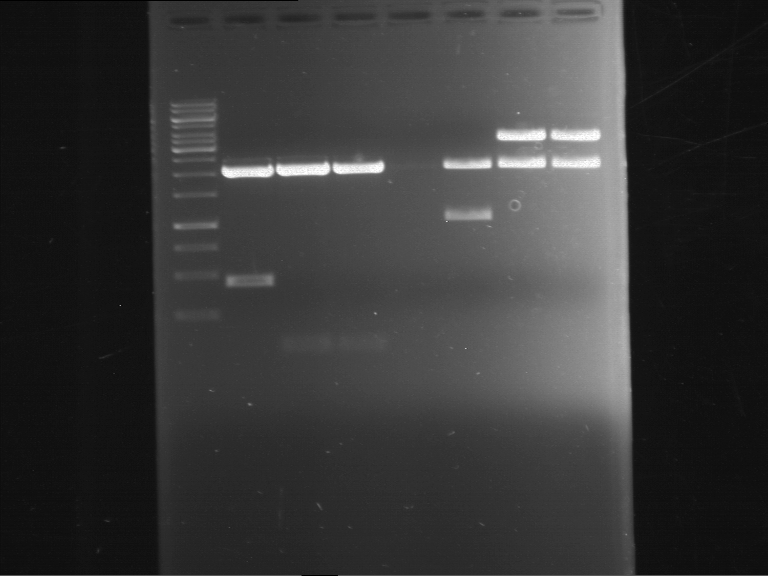
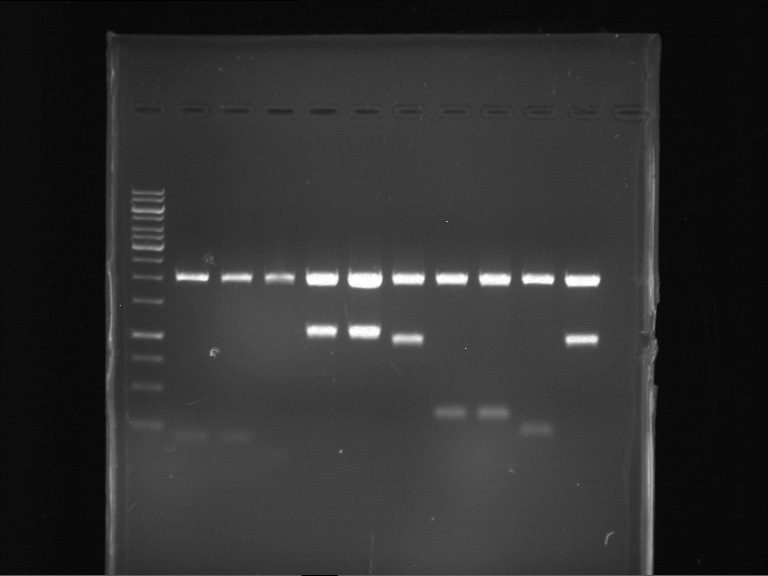
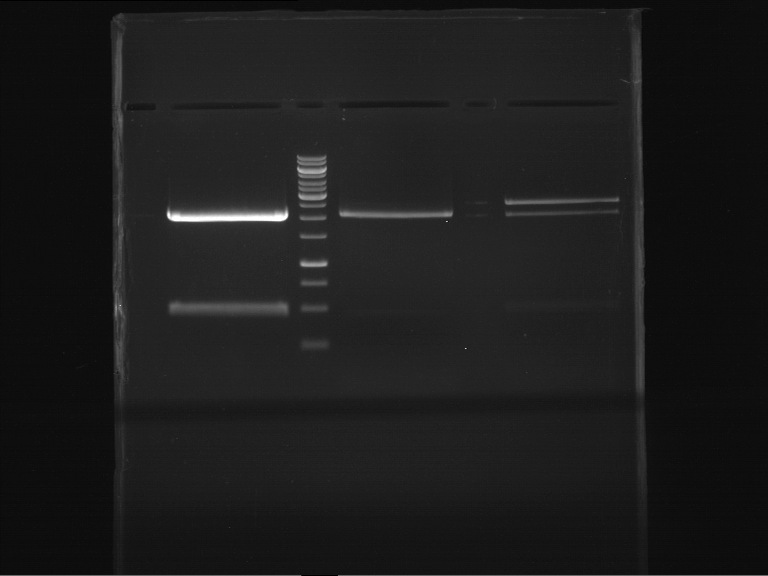
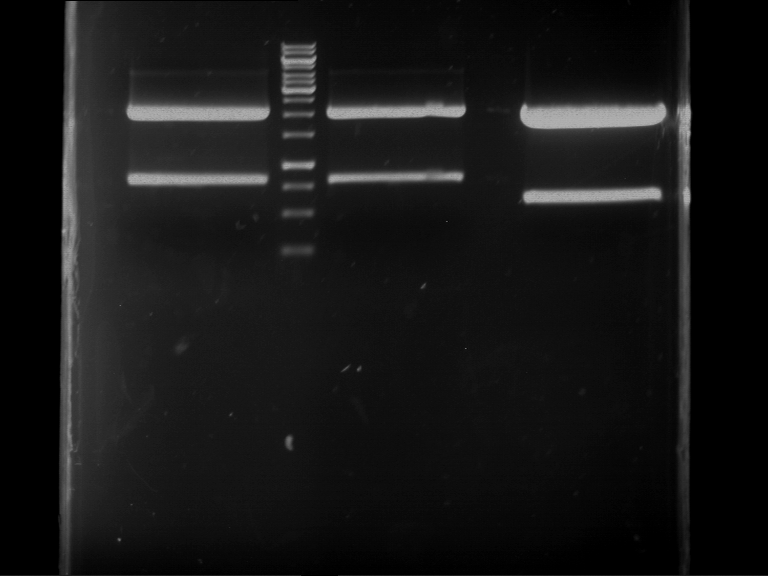
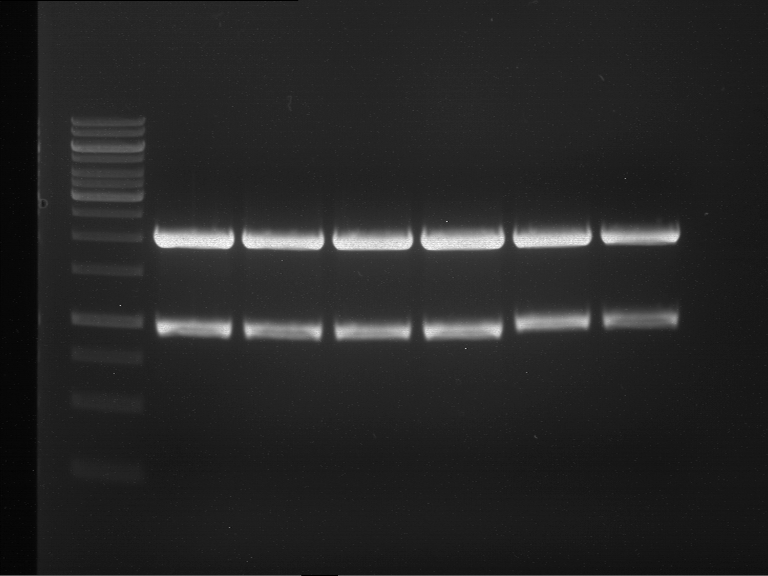
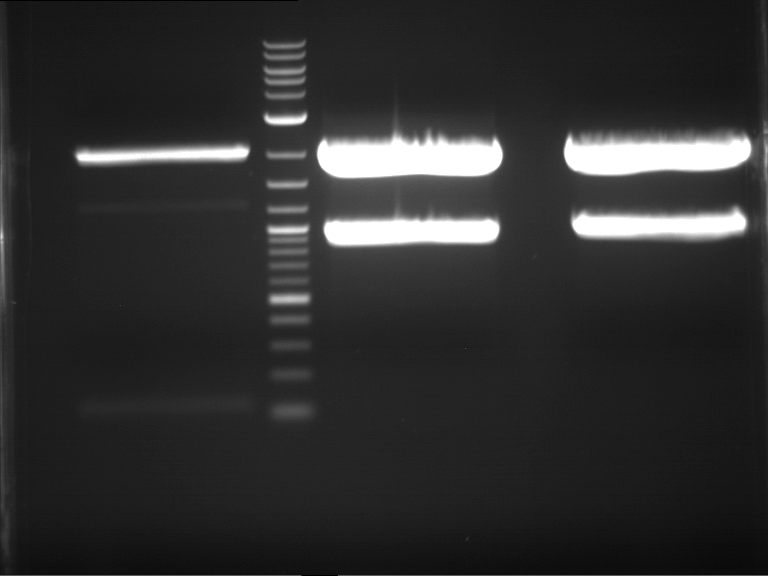
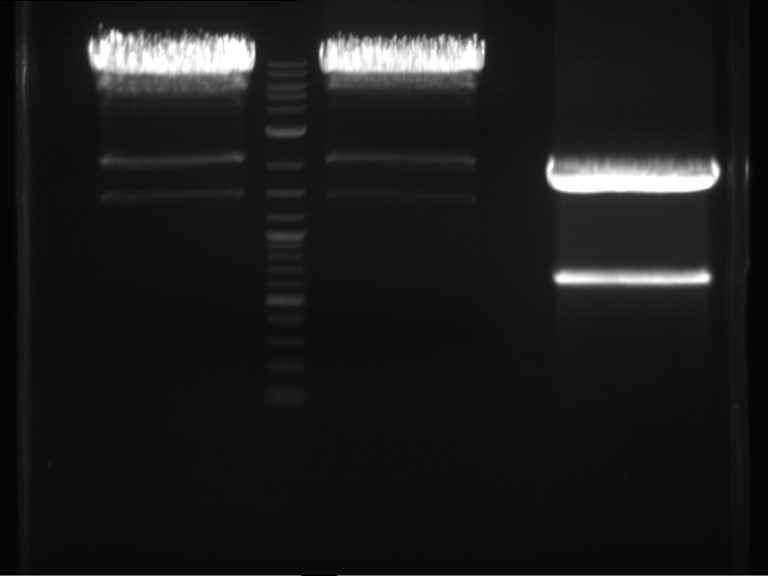
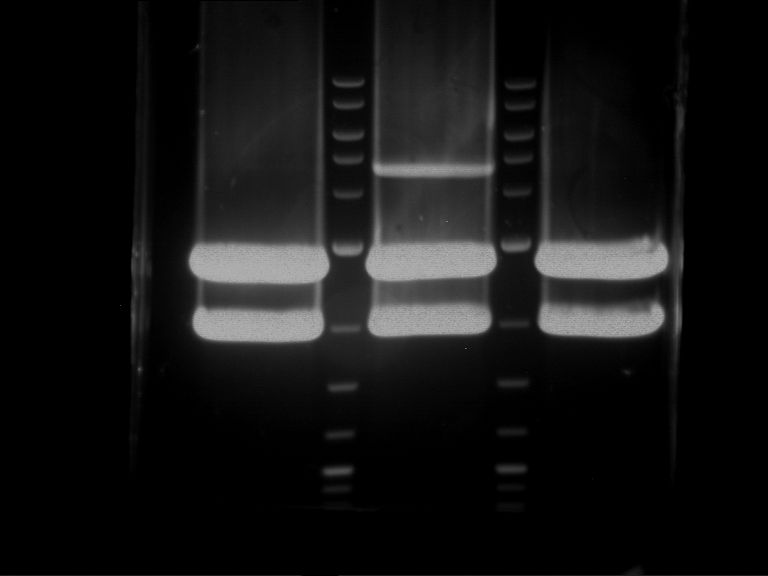
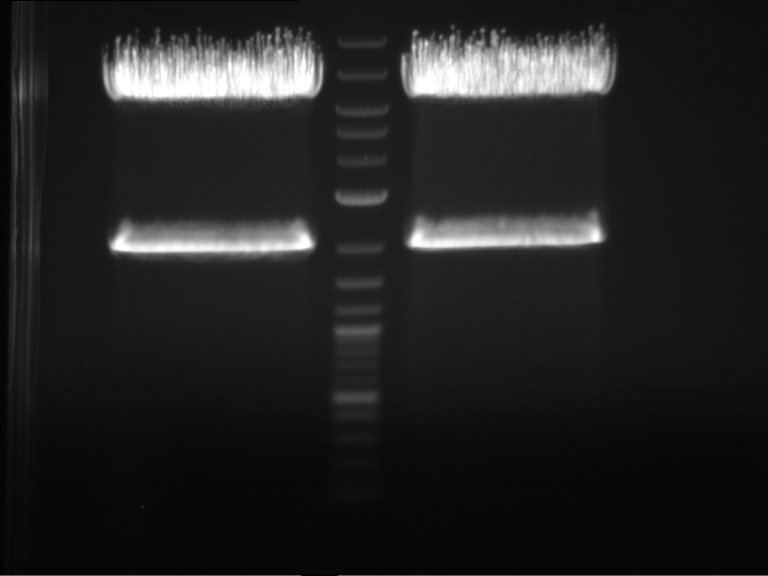
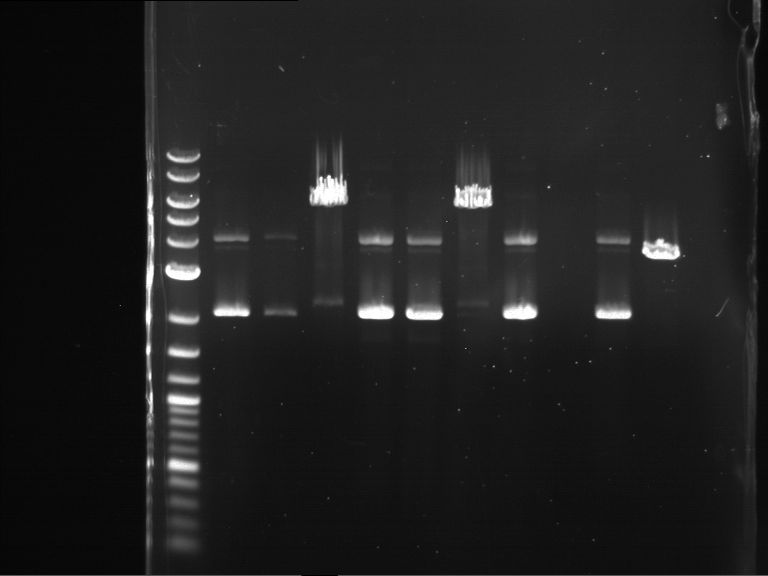
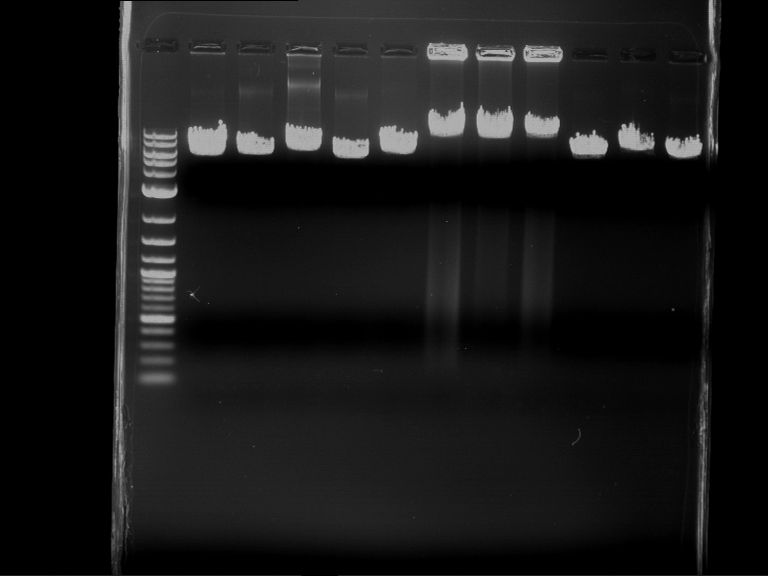
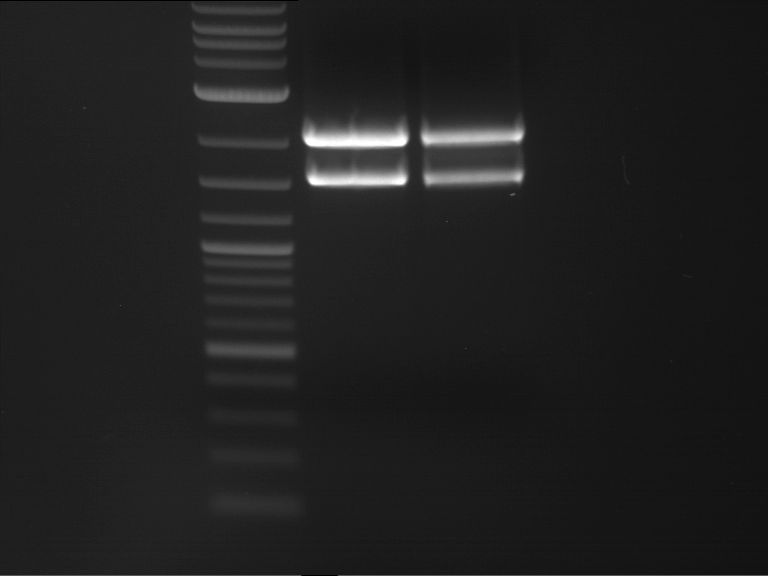
Analytical digestion and gelelectrophoresis of Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3), P7 - P10
Investigator: Jeff, Florian
Aim of the experiment: Analytical digestion and gelelectrophoresis of Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3), P7 - P10.
Procedure:
- Batch for analytical digestion for P7 with NgoMIV+AgeI-HF
| volume | reagent |
| 2.5 µl | Plasmid DNA P7 |
| 2 µl | NEBuffer 4 (10x) |
| 0.25 µl | NgoMIV (10 U/µl) |
| 0.25 µl | AgeI-HF (20 U/µl) |
| 15 µl | ddH2O |
| =20 µl | TOTAL |
- Batch for analytical digestion for P8 with NgoMIV+AgeI-HF
| volume | reagent |
| 2.5 µl | Plasmid DNA P8 |
| 2 µl | NEBuffer 4 (10x) |
| 0.25 µl | NgoMIV (10 U/µl) |
| 0.25 µl | AgeI-HF (20 U/µl) |
| 15 µl | ddH2O |
| =20 µl | TOTAL |
- Batch for analytical digestion for P9 with NgoMIV+AgeI-HF
| volume | reagent |
| 2.5 µl | Plasmid DNA P9 |
| 2 µl | NEBuffer 4 (10x) |
| 0.25 µl | NgoMIV (10 U/µl) |
| 0.25 µl | AgeI-HF (20 U/µl) |
| 15 µl | ddH2O |
| =20 µl | TOTAL |
- Batch for analytical digestion for P10 with NgoMIV+AgeI-HF
| volume | reagent |
| 2.5 µl | Plasmid DNA P10 |
| 2 µl | NEBuffer 4 (10x) |
| 0.25 µl | NgoMIV (10 U/µl) |
| 0.25 µl | AgeI-HF (20 U/µl) |
| 15 µl | ddH2O |
| =20 µl | TOTAL |
- Incubation for 90 min at 37 °C.
- Analytical gelelectrophoresis was performed at 90 V for 60 min.
Results:
| 1 kbp ladder DNA ladder | P7 | P8 | P9 | P10 |
| Part is correct | Part is correct | Part is correct | Part is correct |
Transformation of E. coli XL1 blue with EreA and EreB (pDEST14)
Investigator: Jeff, Florian
Aim of the experiment: Transformation of E. coli XL1 blue with EreA and EreB (pDEST14).
Procedure:
- Plasmid DNA was received dried in paper from McMaster University.
- DNA was resuspended in ddH2O
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 2 µl of DNA was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- The transformated cells were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated on chlorampenicol and ampicillin plates.
Week 2
Monday, April 29th
Miniprep of pRK792, pRK793, pRIL, ereA, ereB
Investigator: Rosario
Aim of the experiment: Miniprep of pRK792, pRK793, pRIL, ereA, ereB
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
- Concentrations were determined by measuring the absorption:
| Plasmid | c [ng/µl] |
| pRK792 | 214,5 |
| pRK793 | 154,3 |
| pRIL | 6,1 |
| ereA | 183,4 |
| ereB | 98,3 |
- The procedure will have to be repeated for pRIL
Midiprep of RFC25 compatible RFP generator in (pSB1C3)
Investigator: Andreas, Florian
Aim of the experiment: Midiprep of RFC25 compatible RFP generator in (pSB1C3)
Procedure:
- Midiprep was performed after manufacturer's protocol (QIAprep Midiprep, QIAGEN)
Tuesday, April 30th
Preparative digestion and gelelectrophoresis of mINF1 and Leptin with HindIII and EheI
Investigator: Louise, Johanna
Aim of the experiment: Testing the enzymes, since the earlier digestion with probably older enzymes did not work (no bands on gel) .
Procedure:
- Batch for preparative digestion for mINF1 with HindIII and EheI
| volume | reagent |
| 10 µl | Plasmid DNA mINF11 |
| 5 µl | Tango Buffer (10x) |
| 2 µl | HindIII (10 U/µl) |
| 3 µl | EheI (10 U/µl) |
| 30 µl | ddH2O |
| =50 µl | TOTAL |
- Batch for preparative digestion for Leptin with HindIII and EheI
| volume | reagent |
| 20 µl | Plasmid DNA Leptin |
| 5 µl | Tango Buffer (10x) |
| 2 µl | HindIII (10 U/µl) |
| 3 µl | EheI (10 U/µl) |
| 20 µl | ddH2O |
| =50 µl | TOTAL |
- Incubation for 3 h at 37 °C.
Transformation of E. coli XL1 blue with PSH21 (P17)
Investigator: Johanna, Louise
Aim of the experiment: Transformation of E. coli XL1 blue with PSH21.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice for 10 minutes.
- 5 µl of DNA was added to 250 µl of competent cells and gently mixed.
- 60 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding the DNA to 2 ml LB-medium.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- The transformated cells were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated on chlorampenicol and ampicillin plates.
Thursday, May 2nd
Miniprep of pSH21 (pAutoRex8) containing AlcR/AlcA promotor system
Investigator: Katrin
Aim of the experiment: Miniprep of pSH21 (pAutoRex8) containing AlcR/AlcA promotor system
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
- The concentration was measured (900 ng/µl)
Friday, May 3rd
Transformation of E. coli XL1 blue with pSB1C3-GFP, pSB1C3-mkate2, pSB1C3-mCherry
Investigator: Andreas
Aim of the experiment: Transformation of E. coli XL1 blue with pSB1C3-GFP, pSB1C3-mkate2, pSB1C3-mCherry.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice for 10 minutes.
- 5 µl of DNA was added to 250 µl of competent cells and gently mixed.
- 60 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding the DNA to 2 ml LB-medium.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- The transformated cells were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated on chlorampenicol and ampicillin plates.
Plating of received E. coli containing biobricks
Investigator: Jeff, Andreas
Aim of the experiment: The received biobricks were already transformed in E. coli and were in an agar stabs. These E. coli cells were transferred with an inoculation loop on antibiotic selection plates and were incubated over night.
Operational sequence:
- Bacterias containing plasmids with biobricks were transferred with a sterile inoculation loop on antibiotic plates and were incubated at 37 °C overnight.
- The biobricks were:
| Name | Function |
|---|---|
| [[http://partsregistry.org/Part:BBa_K157004 BBa_K157004]] | Fluoresceine A -binding derivative of a Lipocalin |
| [[http://partsregistry.org/Part:BBa_K863000 BBa_K863000]] | bpul (laccase from Bacillus pumilus) with T7 promoter, RBS and HIS tag |
| [[http://partsregistry.org/Part:BBa_K909009 BBa_K909009]] | cDNA of UV-B sensing protein UVR8 from Arabidopsis thaliana |
| [[http://partsregistry.org/Part:BBa_K337013 BBa_K337013]] | constitutive promoter SV40 |
| [[http://partsregistry.org/Part:BBa_K747096 BBa_K747096]] | constitutive promoter CMV |
| [[http://partsregistry.org/Part:BBa_K414002 BBa_K414002]] | constitutive promoter CaMV35S |
| [[http://partsregistry.org/Part:BBa_K147002 BBa_K147002]] | xylE gene coding for catechol-1,2-dioxygenase |
| [[http://partsregistry.org/Part:BBa_E2020 BBa_E2020]] | Cerulean |
| [[http://partsregistry.org/Part:BBa_K404319 BBa_K404319]] | mCFP |
| [[http://partsregistry.org/Part:BBa_E0040 BBa_E0040]] | GFPmut3b |
| [[http://partsregistry.org/Part:BBa_K592010 BBa_K592010]] | amilGFP |
| [[http://partsregistry.org/Part:BBa_E2050 BBa_E2050]] | mOrange |
| [[http://partsregistry.org/Part:BBa_K399001 BBa_K399001]] | mStrawberry |
| [[http://partsregistry.org/Part:BBa_E1010 BBa_E1010]] | mRFP1 |
| [[http://partsregistry.org/Part:BBa_E2060 BBa_E2060]] | mCherry |
| [[http://partsregistry.org/Part:BBa_K592012 BBa_K592012]] | eforRed |
| [[http://partsregistry.org/Part:BBa_K592011 BBa_K592011]] | cjBlue |
| [[http://partsregistry.org/Part:BBa_K864401 BBa_K864401]] | aeBlue |
| [[http://partsregistry.org/Part:BBa_K592009 BBa_K592009]] | amilCP, blue chromoprotein |
| [[http://partsregistry.org/wiki/index.php?title=Part:BBa_K414000 BBa_K414000]] | RD29A Promoter + Strong Plant Kozak (RBS) + RFP |
| [[http://partsregistry.org/wiki/index.php?title=Part:BBa_K414006 BBa_K414006]] | 35S Promoter + (Strong Plant RBS + GFP) From K414001 |
| [[http://partsregistry.org/wiki/index.php?title=Part:BBa_K414007 BBa_K414007]] | DREB1C promoter |
| [[http://partsregistry.org/wiki/index.php?title=Part:BBa_K678001 BBa_K678001]] | alcA promoter |
| [[http://partsregistry.org/Part:BBa_K243004 BBa_K243004]] | Short linker |
| [[http://partsregistry.org/Part:BBa_K243005 BBa_K243005]] | Middle linker |
| [[http://partsregistry.org/Part:BBa_K243006 BBa_K243006]] | Long Linker |
| [[http://partsregistry.org/Part:BBa_K243029 BBa_K243029]] | GSAT Linker |
| [[http://partsregistry.org/Part:BBa_K243030 BBa_K243030]] | SEG Linker |
Sunday, May 5th
Picking of the plated E. coli containing biobricks and the parts received from LMU
Investigator: Florian
Aim of the experiment: The colonies from the plates were transferred into liquid LB medium, so that minipreps can be performed the next day.
Operational sequence:
- Tubes with 5 ml LB medium and the corresponding antibiotic were prepared.
- A single colony from each plate was transferred into a tube with a pipet tip.
- The biobricks were:
| Label | Name | Function |
|---|---|---|
| p19 | mCherry in pSB1C3 | |
| p20 | [[http://partsregistry.org/Part:BBa_K823039 BBa_K823039]] | GFP in pSB1C3 |
| p21 | [[http://partsregistry.org/Part:BBa_K823029 BBa_K823029]] | mKate2 in pSB1C3 |
| p22 | [[http://partsregistry.org/wiki/index.php?title=Part:BBa_K414006 BBa_K414006]] | 35S Promoter + (Strong Plant RBS + GFP) From K414001 |
| p23 | [[http://partsregistry.org/Part:BBa_K414002 BBa_K414002]] | constitutive promoter CaMV35S |
| p24 | [[http://partsregistry.org/Part:BBa_K909009 BBa_K909009]] | cDNA of UV-B sensing protein UVR8 from Arabidopsis thaliana |
| p25 | [[http://partsregistry.org/Part:BBa_K399001 BBa_K399001]] | mStrawberry |
| p26 | [[http://partsregistry.org/wiki/index.php?title=Part:BBa_K414007 BBa_K414007]] | DREB1C promoter |
| p27 | [[http://partsregistry.org/Part:BBa_K863000 BBa_K863000]] | bpul (laccase from Bacillus pumilus) with T7 promoter, RBS and HIS tag |
| p28 | [[http://partsregistry.org/Part:BBa_K337013 BBa_K337013]] | constitutive promoter SV40 |
| p29 | [[http://partsregistry.org/Part:BBa_K592011 BBa_K592011]] | cjBlue |
| p30 | [[http://partsregistry.org/wiki/index.php?title=Part:BBa_K678001 BBa_K678001]] | alcA promoter |
| p31 | [[http://partsregistry.org/Part:BBa_K592012 BBa_K592012]] | eforRed |
| p32 | [[http://partsregistry.org/Part:BBa_K592010 BBa_K592010]] | amilGFP |
| p33 | [[http://partsregistry.org/Part:BBa_K864401 BBa_K864401]] | aeBlue |
| p34 | [[http://partsregistry.org/Part:BBa_K404319 BBa_K404319]] | mCFP |
| p35 | [[http://partsregistry.org/wiki/index.php?title=Part:BBa_K414000 BBa_K414000]] | RD29A Promoter + Strong Plant Kozak (RBS) + RFP |
| p36 | [[http://partsregistry.org/Part:BBa_K592009 BBa_K592009]] | amilCP, blue chromoprotein |
| p37 | [[http://partsregistry.org/Part:BBa_K747096 BBa_K747096]] | constitutive promoter CMV |
| p38 | [[http://partsregistry.org/Part:BBa_K147002 BBa_K147002]] | XylE dioxygenase (catechol dioxygenase) |
| p39 | [[http://partsregistry.org/Part:BBa_K243005 BBa_K243005]] | Middle linker |
| p40 | [[http://partsregistry.org/Part:BBa_K243029 BBa_K243029]] | GSAT Linker |
| p41 | [[http://partsregistry.org/Part:BBa_E0040 BBa_E0040]] | GFPmut3b |
| p42 | [[http://partsregistry.org/Part:BBa_K243006 BBa_K243006]] | Long Linker |
| p43 | [[http://partsregistry.org/Part:BBa_K243030 BBa_K243030]] | SEG Linker |
| p44 | [[http://partsregistry.org/Part:BBa_K243004 BBa_K243004]] | Short linker |
| p45 | [[http://partsregistry.org/Part:BBa_K157004 BBa_K157004]] | Fluoresceine A -binding derivative of a Lipocalin |
| p46 | [[http://partsregistry.org/Part:BBa_E2060 BBa_E2060]] | mCherry |
| p47 | [[http://partsregistry.org/Part:BBa_E2020 BBa_E2020]] | Cerulean |
| p48 | [[http://partsregistry.org/Part:BBa_E1010 BBa_E1010]] | mRFP1 |
| p49 | [[http://partsregistry.org/Part:BBa_E2050 BBa_E2050]] | mOrange |
- There were no colonies on the plate with K864401 (p33), therefore the LB medium could not be infected
- The tubes were placed into the incubator at 37 °C
Week 3
Monday, May 6th
Miniprep of the prepared E. coli containing biobricks and the parts from LMU
Investigators: Florian, Johanna, Jefferey
Aim of the experiment: A miniprep of the overnight culture of our E. coli containig our biobricks and LMU parts was performed using the Qiagen miniprep kit.
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
- Resulting concentrations were determined by measuring the absorption:
| Label | Name | Concentration [ng/µl] |
|---|---|---|
| p19 | mCherry in pSB1C3 | 40,2 |
| p20 | GFP in pSB1C3 | 58,5 |
| p21 | mKate2 in pSB1C3 | 131,3 |
| p22 | 35S Promoter + (Strong Plant RBS + GFP) From K414001 | 42,8 |
| p23 | constitutive promoter CaMV35S | 39,3 |
| p24 | cDNA of UV-B sensing protein UVR8 from Arabidopsis thaliana | 30,0 |
| p25 | mStrawberry | 46,6 |
| p26 | DREB1C promoter | 34,8 |
| p27 | bpul (laccase from Bacillus pumilus) with T7 promoter, RBS and HIS tag | 52,5 |
| p28 | constitutive promoter SV40 | 58,7 |
| p29 | cjBlue | 44,4 |
| p30 | alcA promoter | 26,9 |
| p31 | eforRed | 64,8 |
| p32 | amilGFP | 59,3 |
| p34 | mCFP | 78,4 |
| p35 | RD29A Promoter + Strong Plant Kozak (RBS) + RFP | 61,2 |
| p36 | amilCP, blue chromoprotein | 89,2 |
| p37 | constitutive promoter CMV | 83,6 |
| p38 | XylE dioxygenase (catechol dioxygenase) | 57,4 |
| p39 | Middle linker | 64,3 |
| p40 | GSAT Linker | 109,6 |
| p41 | GFPmut3b | 118,1 |
| p42 | Long Linker | 133,0 |
| p43 | SEG Linker | 107,5 |
| p44 | Short linker | 60,2 |
| p45 | Fluoresceine A -binding derivative of a Lipocalin | 82,7 |
| p46 | mCherry | 230,2 |
| p47 | Cerulean | 25,7 |
| p48 | mRFP1 | 299,3 |
| p49 | mOrange | 181,2 |
Sequencing of FluA, SV40, CaMV35S, xylE, LMU mKate2, LMU GFP, LMU mCherry, DREB1C, RD29A
Investigators: Andreas, Florian, Johanna, Jefferey, Rosario
Aim of the experiment: Sequencing of genes for which no prior sequencing results were available.
Procedure: The DNA was prepared for sequencing according to the Eurofins SmartSeq Kit protocol (15 µl of plasmid DNA (50 - 100 ng) and 2 µl sequencing primer).
The different genes we sequenced received the following barcodes:
| Label | Name | Barcode |
|---|---|---|
| p45 | FluA | FR01002355 |
| p28 | SV40 | FR01002356 |
| p23 | CaMV35S | FR01002357 |
| p38 | xylE | FR01002358 |
| p21 | LMU mKate2 | FR01002359 |
| p20 | LMU GFP | FR01002360 |
| p19 | LMU mCherry | FR01002345 |
| p46 | mCherry | FR01002346 |
| p26 | DREB1C | FR01002347 |
| p35 | RD29A | FR01002348 |
Preparation of ordered primers
Investigators: Andreas, Jefferey, Rosario
Aim of the experiment: Dissolving of primer pallets to provide a concentration of 100 pmol/µl
Procedure: We added an amount of water which gave us a final primer concentration of 100 pmol/µl.
Primers were labeled as follows:
| Label | Oligonr. |
|---|---|
| 6 | EreA_for |
| 7 | EreA_rev |
| 8 | EreA_SDM1_for |
| 9 | EreA_SDM1_rev |
| 10 | EreA_SDM2_for |
| 11 | EreA_SDM2_rev |
| 12 | EreA_SDM3_for |
| 13 | EreA_SDM3_rev |
| 14 | EreB_for |
| 15 | EreB_rev |
| 16 | EreB_SDM1_for |
| 17 | EreB_SDM1_rev |
| 18 | EreB_SDM2_for |
| 19 | EreB_SDM2_rev |
| 20 | EreB_SDM3_for |
| 21 | EreB_SDM3_rev |
Tuesday, May 7th
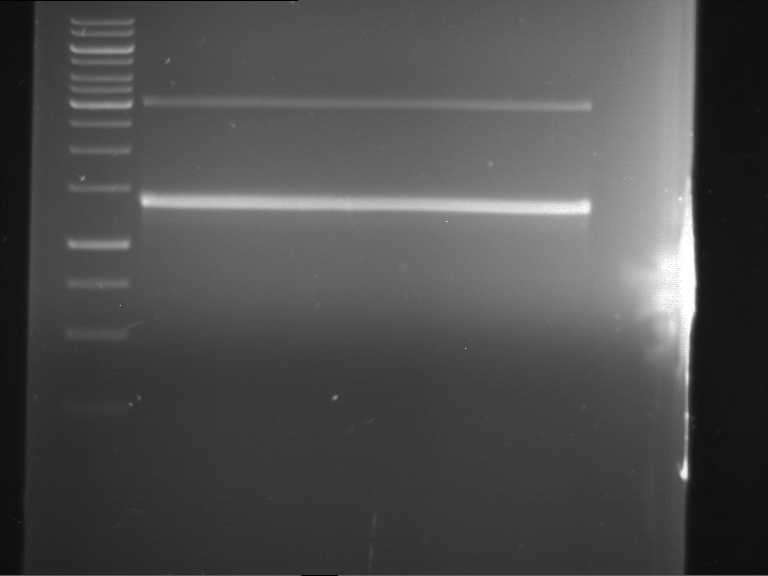
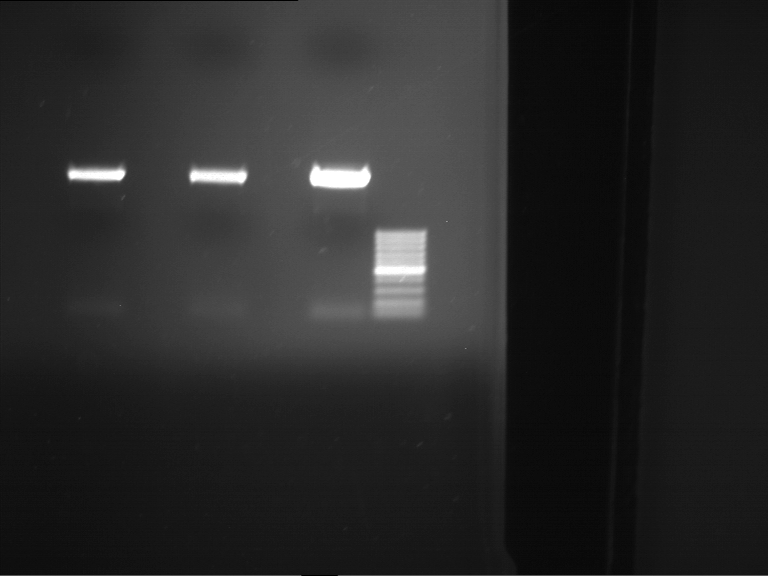
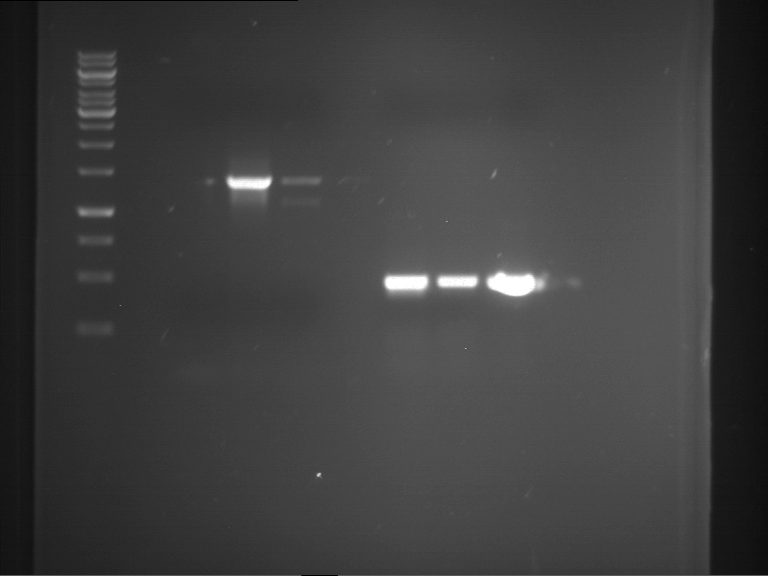
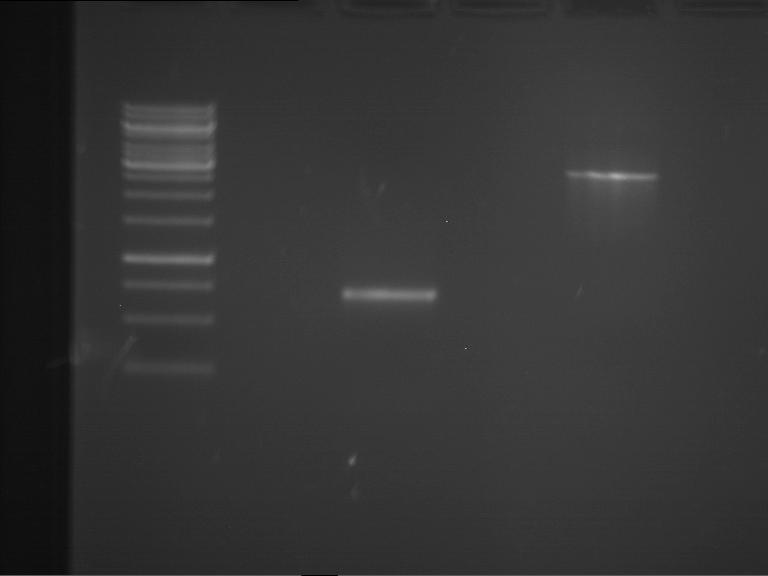
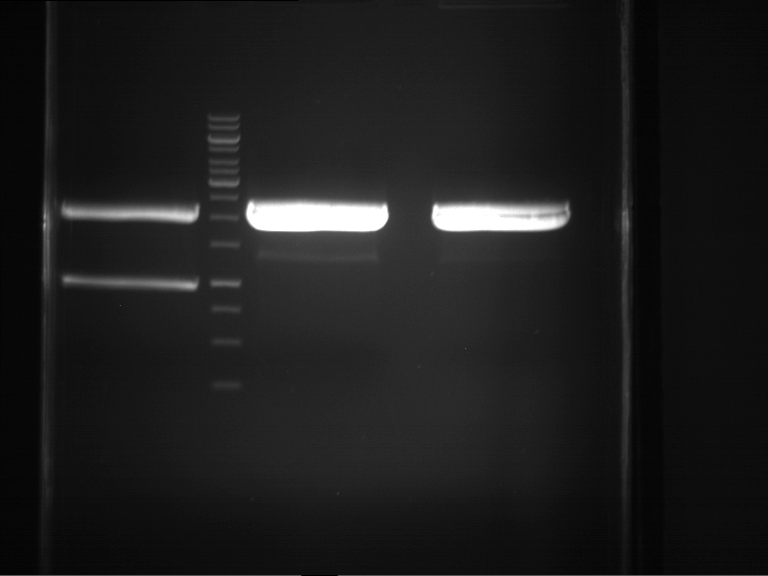
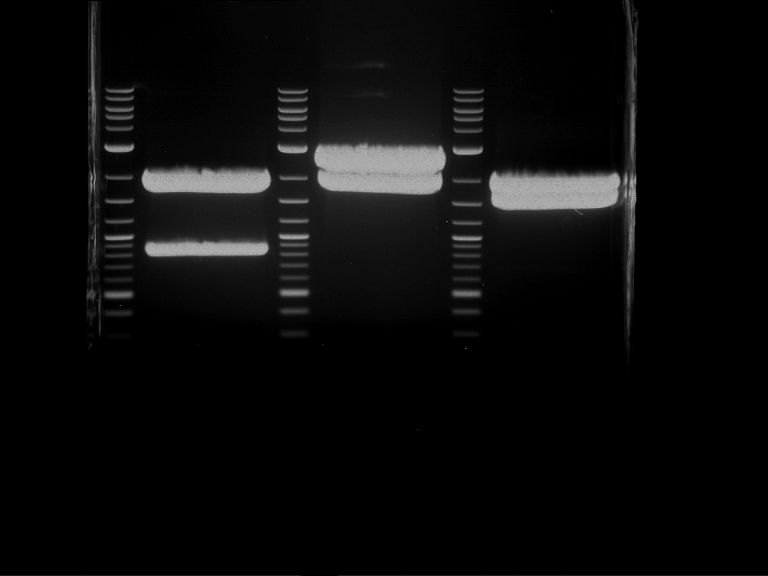
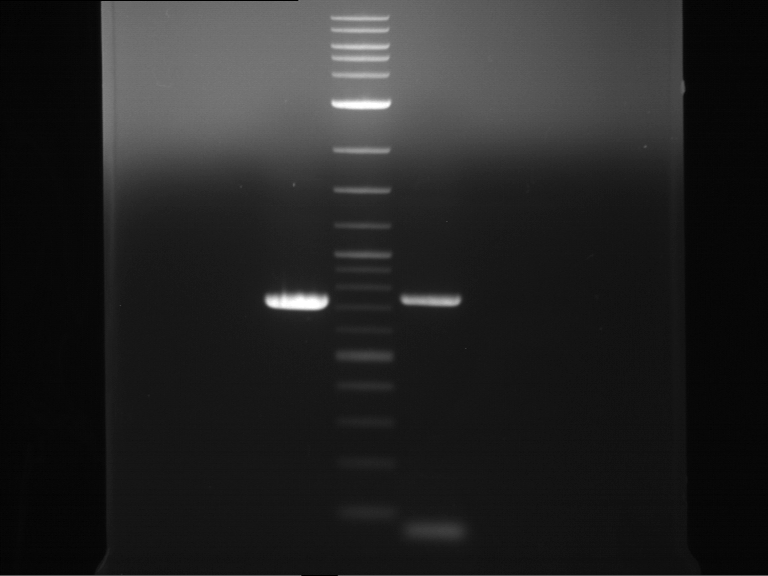
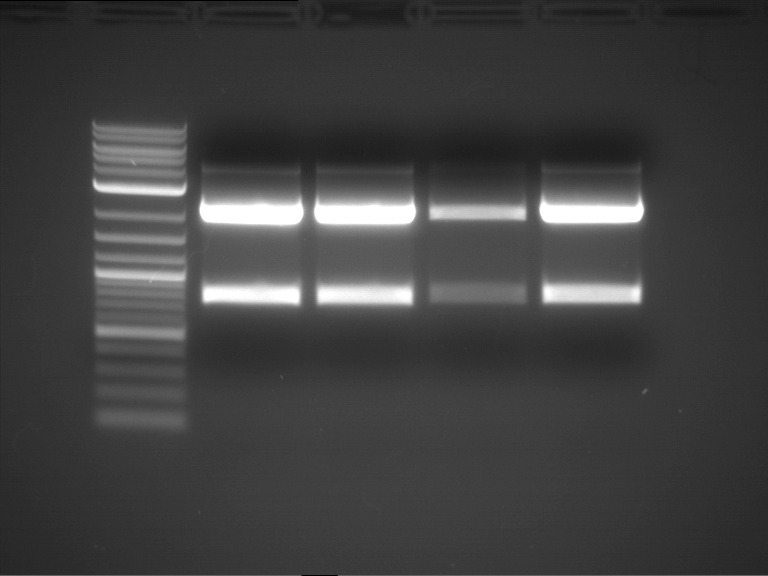
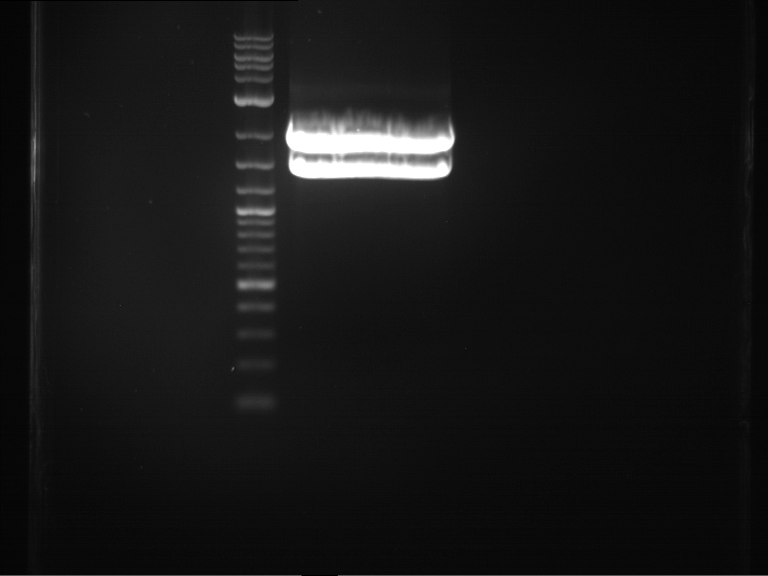
PCR, purification and analytical geleletrophoresis of EreA (P15) and EreB (P16)
Investigator: Jeff, Louise, Florian, Rosario, Andi, Johanna
Aim of the experiment: PCR of EreA (P15) and EreB (P16).
Procedure:
Operational sequence:
- PCR reaction mixture
| volume | reagent |
| 10 µl | 5x OneTaq Standard Reaction Buffer |
| 1 µl | 10 mM dNTPs |
| 1 µl | 10 µM Forward Primer (P15: O46 (EreA_for); P16: O31 (EreB_for)) |
| 1 µl | 10 µM Reverse Primer (P15: O47 (EreA_rev); P16: O32 (EreB_rev)) |
| 0.25 µL | OneTaq Hot Start DNA Polymerase (Finally: 1.25 units/50 µL) |
| 1 µl | Plasmid DNA (P15; P16) |
| 35.75 µL | ddH2O Water |
| =50 µL | TOTAL |
- Mix with pipette
- The gradient PCR program was performed after following scheme with following conditions (Tm=54 °C; ΔG=2 °C; P15 in row 11(=52 °C); P16 in row 1 (=54.0 °C)):
| Initial denaturation | 94 °C | 30 s |
| 30 cycles | 94 °C | 30 s |
| Tm=54 °C; ΔG=2 °C | 60 s | |
| 68 °C | 80 s | |
| Final extension | 68 °C | 5 min |
| Hold | 4 °C | infinite |
- After PCR, the product was purified with QIAquick PCR Purification Kit, Qiagen.
- 9 µl of the purified PCR products were mixed with 1 µl DNA loading buffer (10x) for analytical gelelectrophoresis
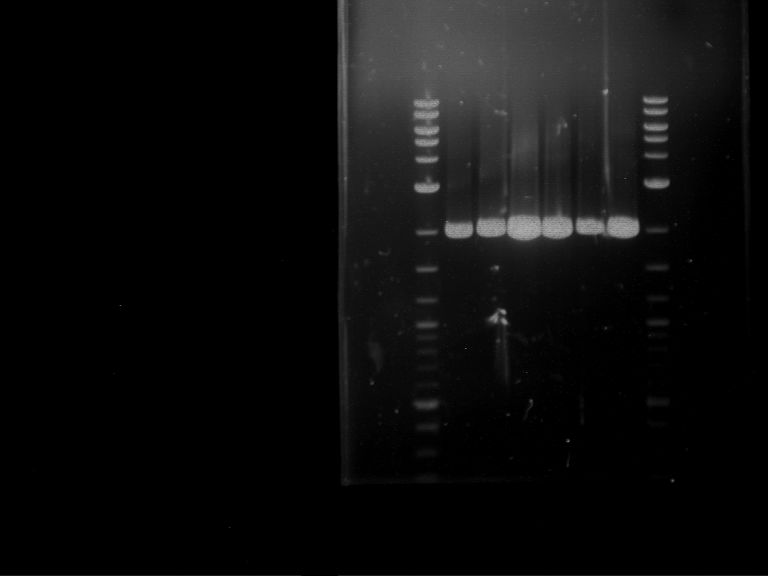
- Analytical gelelectrophoresis was performed at 90 V for 60 min in 1% agarose gel.
| 1 kbp ladder | F1 | F2 |
| Fragment-size like expected | Fragment-size like expected |
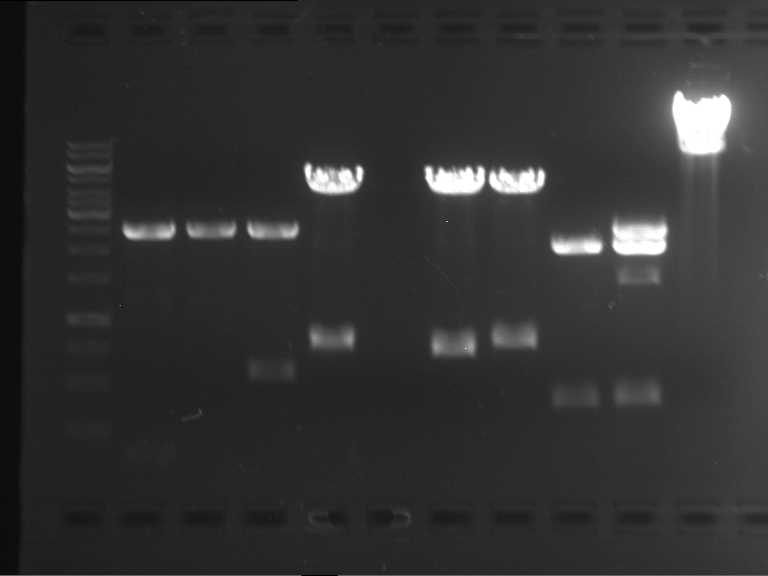
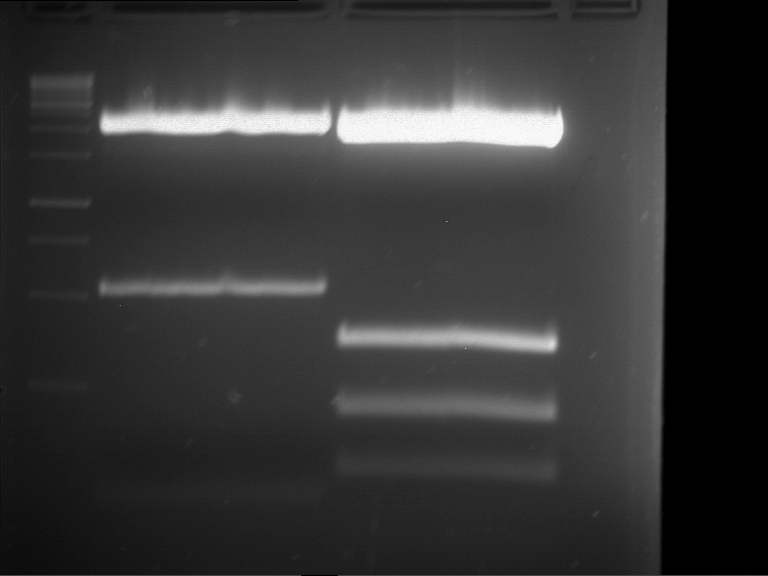
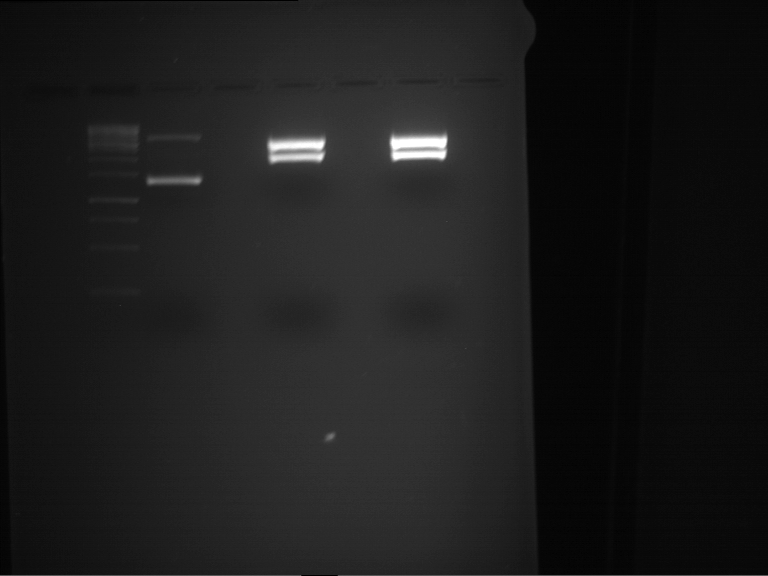
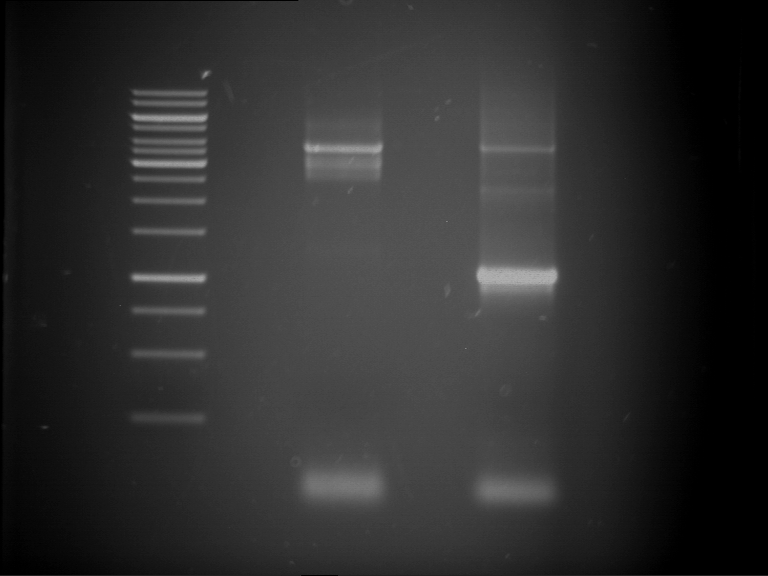
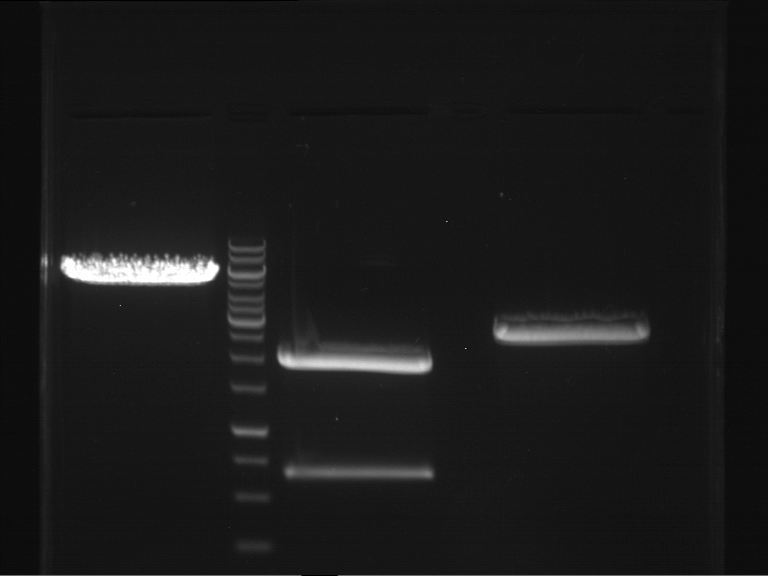
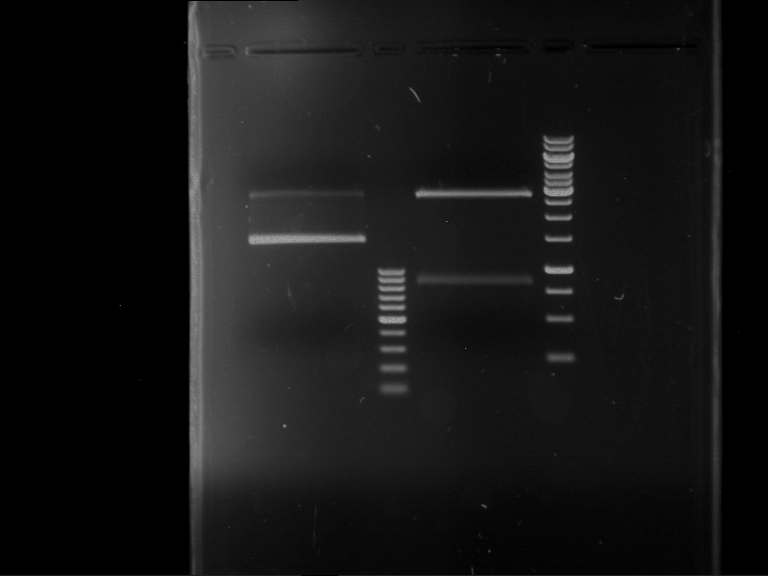
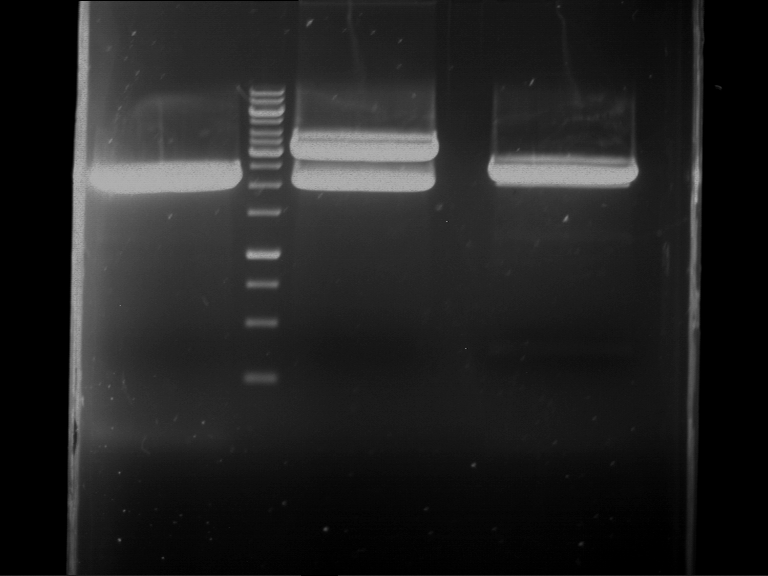
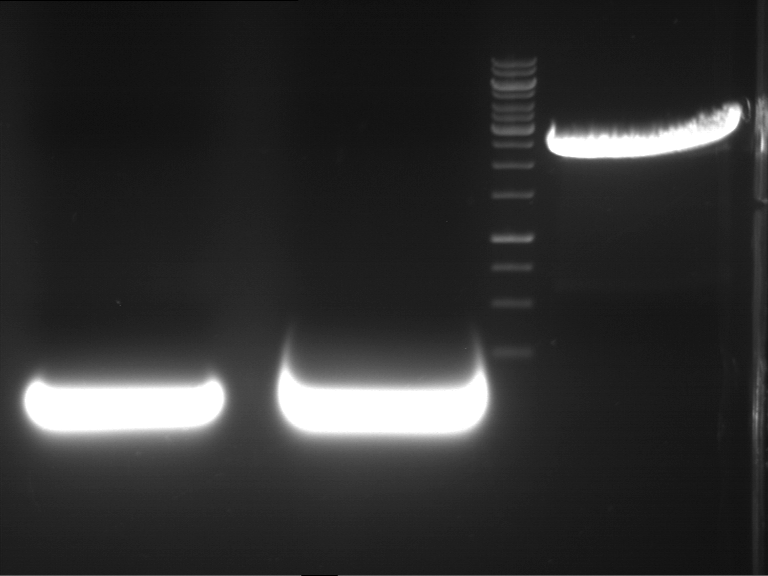
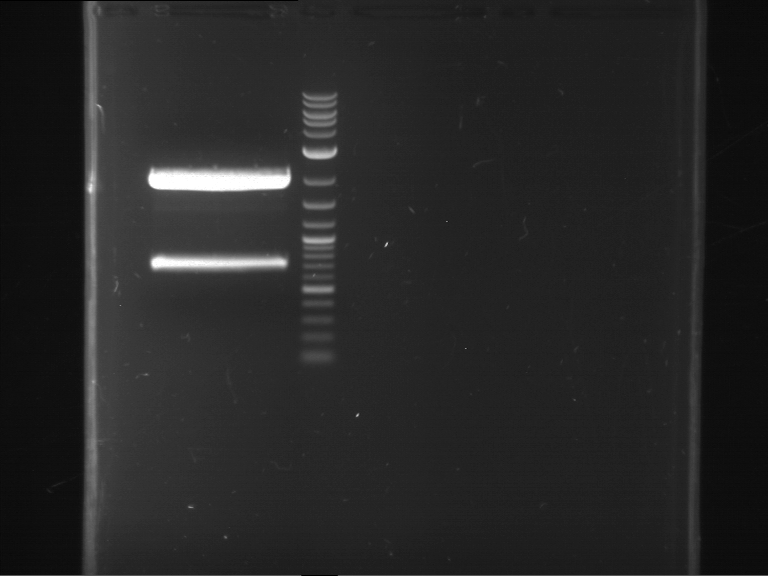
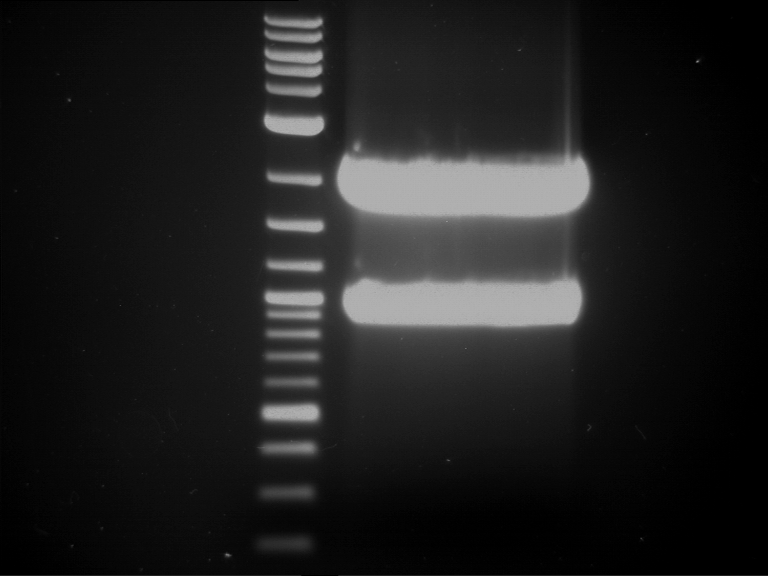
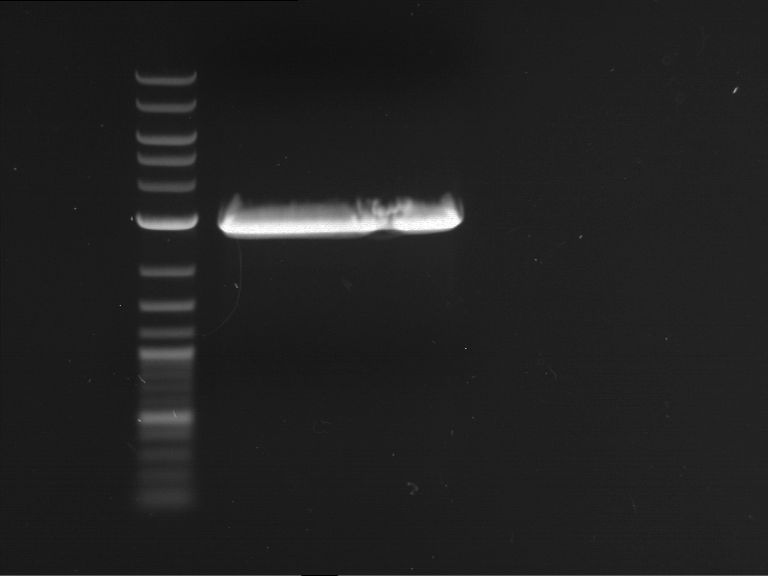
Preparative digestion and gelelectrophoresis of PhyB (P9) and PIF3-20aa linker (P50) with AgeI and PstI as well as 20aalinker-PIF3 (P51) with EcoRI and NgoMIV
Investigator: Jeff, Andi, Florian, Rosario, Louise, Johanna
Aim of the experiment: Preparative digestion and gelelectrophoresis of PhyB (P9) and PIF3-20aa linker (P50) with AgeI and PstI as well as 20aalinker-PIF3 (P51) with EcoRI and NgoMIV .
Procedure:
- Batch for preparative digestion of PhyB (P9) and PIF3-20aa linker (P50) with AgeI and PstI
| volume | reagent |
| 20 µl | Plasmid DNA P9/P50 |
| 4 µl | NEBuffer 4 |
| 0.4 µl | BSA (100x) |
| 1 µl | AgeI-HF |
| 1 µl | PstI-HF |
| 13.6 µl | ddH2O |
| =40 µl | TOTAL |
- Batch for preparative digestion of 20aalinker-PIF3 (P51) with EcoRI and NgoMIV
| volume | reagent |
| 20 µl | Plasmid DNA P51 |
| 4 µl | NEBuffer 4 |
| 1 µl | EcoRI-HF |
| 1 µl | NgoMIV |
| 14 µl | ddH2O |
| =40 µl | TOTAL |
- Incubation for 3 h at 37 °C.
- 4 µl of DNA loading buffer (10x) were added to the reaction batches after digestion and were loaded on an 1% low-melting agarose gel for preparative gelelectrophoresis.
- Preparative gelelectrophoresis was performed at 70 V for 90 min.
| P9 digestion = F3 | 1 kbp ladder | P50 digestion = F4 | P51 digestion = F5 |
| Fragment-size like expected; band was cut out | Fragment-size like expected; band was cut out | Fragment-size like expected; band was cut out |
- Bands were extracted by QIAquick Gel Extraction Kit, QIAGEN.
Wednesday, May 8th
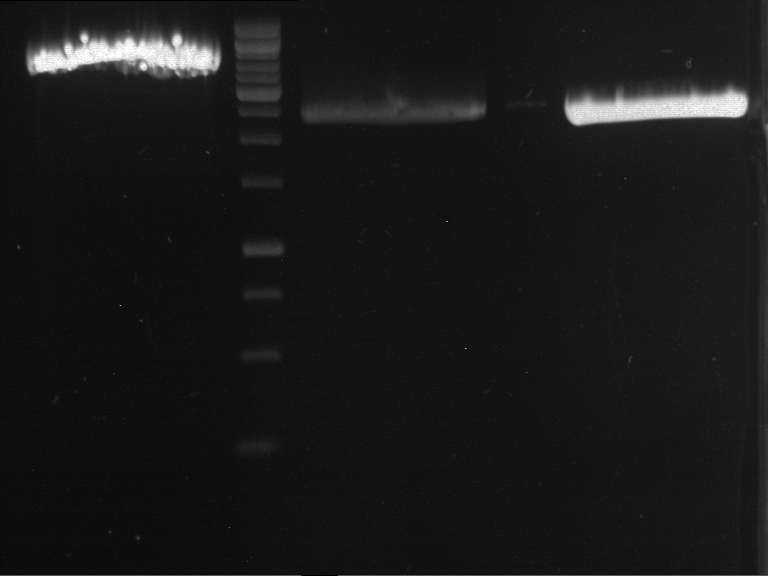
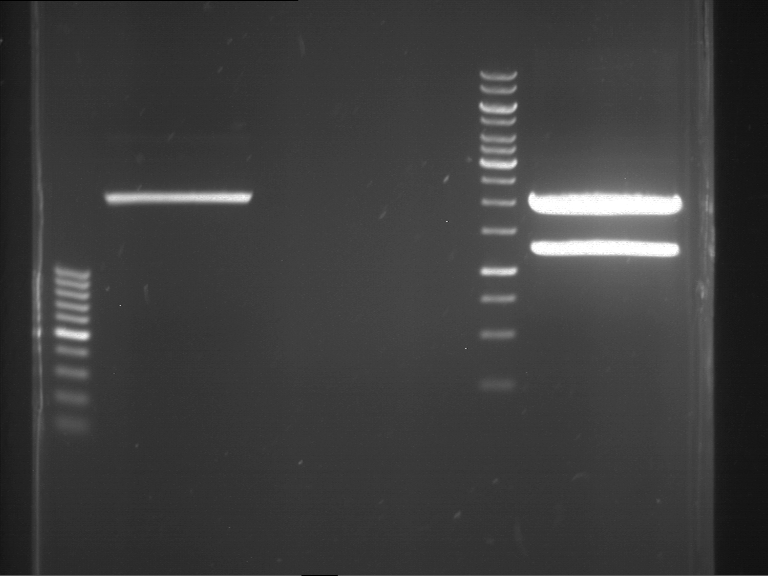
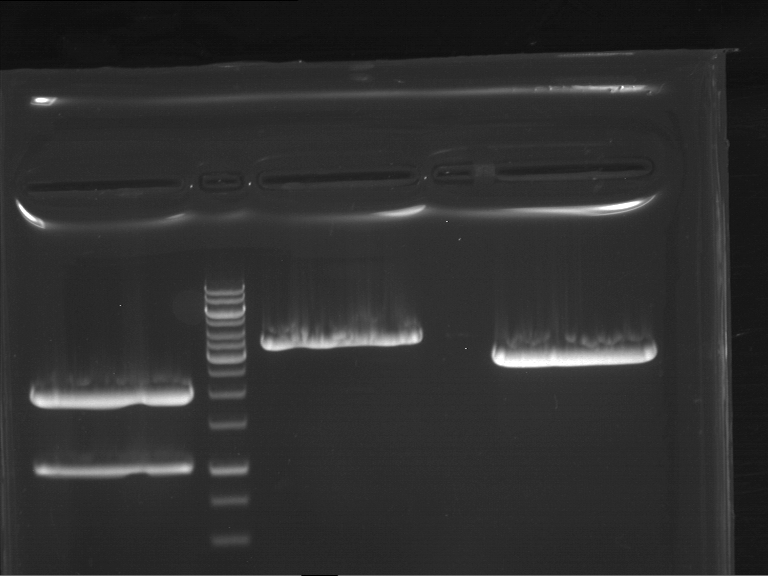
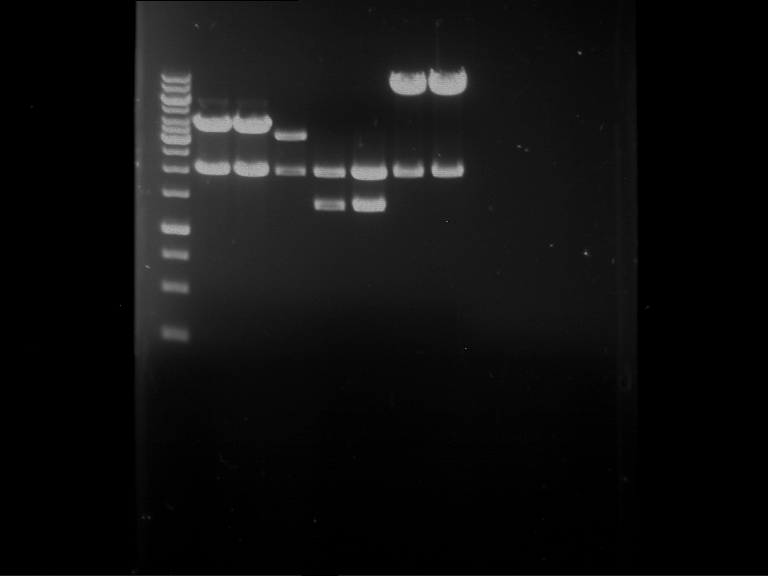
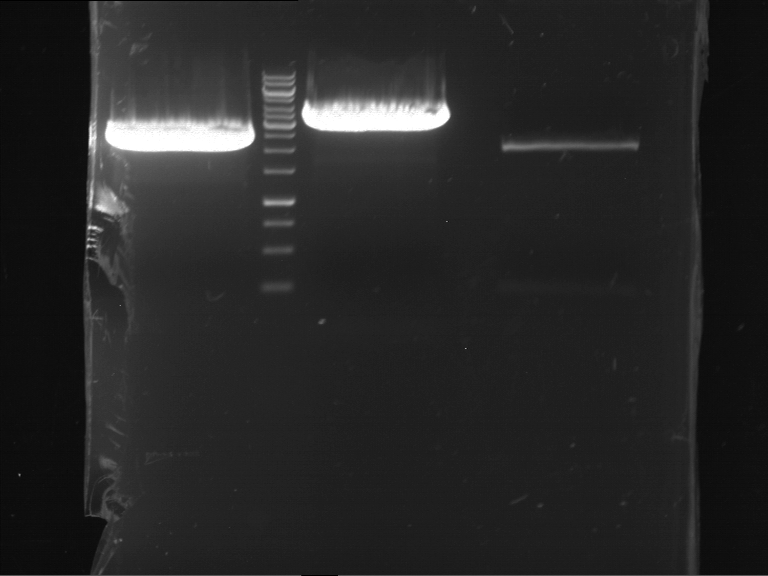
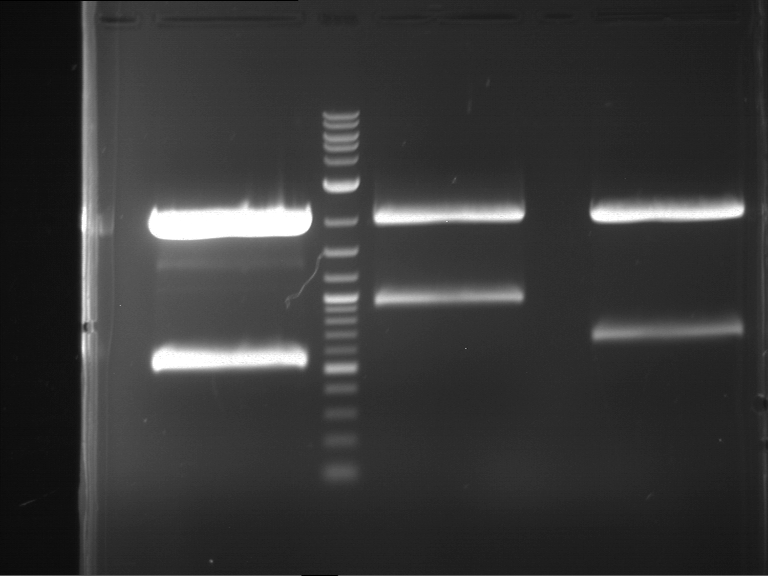
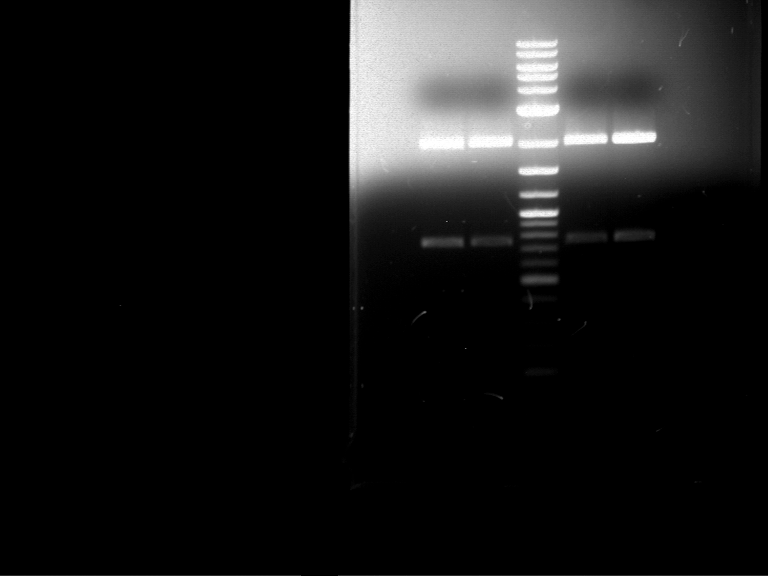
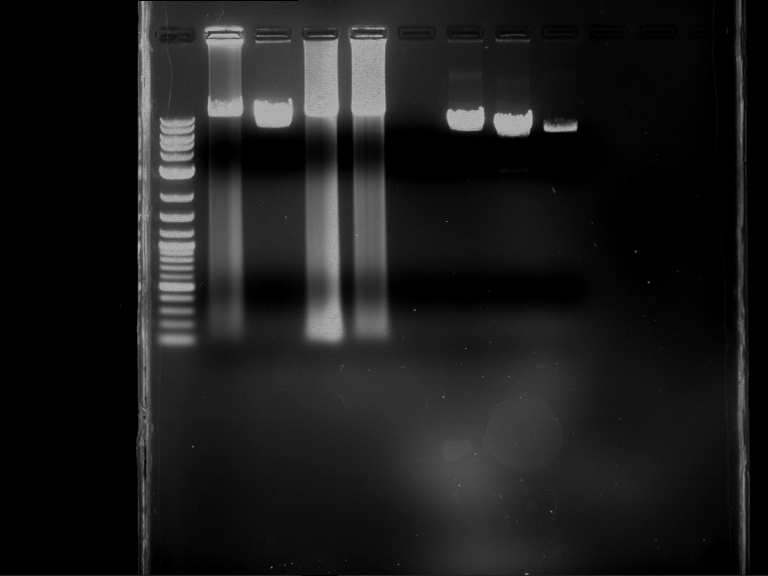
Preparative digestion and gelelectrophoresis of PCR product of EreA (F1) and EreB (F2) with XbaI & AgeI
Investigator: Jeff, Andi, Florian, Rosario, Louise
Aim of the experiment: Preparative digestion and gelelectrophoresis of PCR product of EreA (F1) and EreB (F2) with XbaI & AgeI.
Procedure:
- Batch for preparative digestion of PCR product of EreA (F1) and EreB (F2) with XbaI & AgeI.
| volume | reagent |
| 25 µl | PCR product F1/F2 |
| 5 µl | NEBuffer 4 |
| 0.5 µl | BSA (100x) |
| 1 µl | AgeI-HF |
| 1 µl | XbaI |
| 17.5 µl | ddH2O |
| =50 µl | TOTAL |
- Batch for preparative digestion of P52 for preperation of pSB1C3 vector:
| volume | reagent |
| 20 µl | Plasmid DNA P52 |
| 4 µl | NEBuffer 4 |
| 0.4 µl | BSA (100x) |
| 1 µl | AgeI-HF |
| 1 µl | XbaI |
| 13.6 µl | ddH2O |
| =40 µl | TOTAL |
- Incubation for 3 h at 37 °C.
- 5 µl of DNA loading buffer (10x) were added to the reaction batches for F1 & F2 and 4 µl of DNA loading buffer (10x) were added to the reaction batches for Pxx after digestion and were loaded on an 1% agarose gel for preparative gelelectrophoresis.
- Preparative gelelectrophoresis was performed at 90 V for 60 min.
| F1 digestion = F6 | F2 digestion = F7 | 1 kbp ladder | P52 digestion = F8 |
| Length was like expected | Length was like expected | Length was like expected; the lower band was extracted |
- Bands were extracted by QIAquick Gel Extraction Kit, QIAGEN.
Ligation of F8+F6 (EreA in pSB1C3), F8+F7 (EreB in pSB1C3)
Investigator: Jeff, Andi, Florian, Rosario, Louise
Aim of the experiment: Ligation of F8+F6 (EreA in pSB1C3), F8+F7 (EreB in pSB1C3). Quickchanges have still to be performed.
Procedure:
Ligation batch for F8+F6 (EreA in pSB1C3), F8+F7 (EreB in pSB1C3):
- Ligation batch for F8+F6
| volume | reagent |
| 1.35 µl | F8 (74.3 ng/µl, 2086 bp) |
| 6.76 µl | F6 (25.9 ng/µl, 1218 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 8.89 µl | ddH2O |
| =20 µl | TOTAL |
- Ligation batch for F8+F7
| volume | reagent |
| 1.35 µl | F8 (74.3 ng/µl, 2086 bp) |
| 8.78 µl | F7 (20.6 ng/µl, 1257 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 6.87 µl | ddH2O |
| =20 µl | TOTAL |
- Negative control was als prepared. Instead of the insert, the same volume of ddH2O was added to the reaction batches.
- Cycled ligation has been performed after following protocol in a thermocycler:
| 100 cycles | 12 °C | 60 s |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| Hold | 16 °C | infinite |
Lid temperature = 37 °C
Midiprep of cloning vector RFC25 RFP generating device
Investigators: Andreas, Rosario
Aim of the experiment: Midiprep of our cloning vector RFC25 RFP generating device.
Procedure: Midiprep was performed after manufacturer's protocol (QIAprep Midiprep, QIAGEN)
The yield of our vector was 1854 ng.
Thursday, May 9th
Transformation of E. coli XL1 blue with F8(pSB1C3)+F6(EreA) and F8(pSB1C3)+F7(EreB)
Investigator: Florian, Jeff, Louise, Katrin, Ingmar
Aim of the experiment: Transformation of E. coli XL1 blue with F8(pSB1C3)+F6(EreA) and F8(pSB1C3)+F7(EreB). Quickchanges has now to be performed to remove forbidden restriction sites.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 10 µl of ligation products F8+F6, F8+F7, F8 negative control were added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of the cell suspension was plated on one chloramphenicol plate.
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on a new chlorampenicol plate.
Friday, May 10th
Quick Change mutagenesis to remove mutations found by sequencing in our gene constructs
Investigator: Andi, Jeff, Rosario
Aim of the experiment: Deletion of found mutations.
Operational sequence
1. PCR general setup
Reaction batch 1
| volume | reagent |
| 2.5 µl | 10x Pfu Ultra II buffer |
| 4 µl | DNA template |
| 1 µl | 1:10 dilution of appropriate Oligo (= 5 pmol) |
| 16.5 µl | ddH2O |
| 0.5 µl | dNTP mix |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
Reaction batch 2
| volume | reagent |
| 2.5 µl | 10x Pfu Ultra II buffer |
| 4 µl | DNA template |
| 1 µl | 1:10 dilution of appropriate Oligo (= 5 pmol) |
| 16.5 µl | ddH2O |
| 0.5 µl | dNTP mix |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
PCR cycling parameters
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 30 sec |
| 2 | 12 | 95 °C | 30 s |
| 55 °C | 1 min | ||
| 68 °C | 6 min |
- Having completed the PCR cycling parameters listed above both PCR reaction batches were mixed together and the cycling parameters listed above were one time more applied.
- After the PCR was finished 1 µl of the DpnI restriction enzyme (10 U/µl)was added
- Now the reaction was mixed gently and thoroughly, spinned down in a microcentrifuge for 1 minute, and immediately incubated at 37 °C for 1 hour to digest the parental supercoiled dsDNA
2. Used constructs and primers
| Quickchange number | Plasmid number | Geneconstruct | Oligo number |
| QC 1 | P27 | BPul_Laccase+ pSB1C3 | O26 & O27 |
Picking of transformed Plasmids F8 + F6
Investigator: Andi
Aim of the study: Picking from the overnight transformation of the ligated F8+F6, to see whether ligation is successful or not.
Operational sequence:
- Picking and overnight culture after standard laboratory's protocol. (CamR LB-medium)
PCR of TEV (P14, pRK793)
Investigator: Katrin, Jeff
Aim of the experiment: PCR, purification and analytical geleletrophoresis of TEV (P14, pRK793) to produce TEV with RFC25 pre- and suffx (still contains forbidden SpeI-site).
Procedure:
Operational sequence:
- PCR reaction mixture
| volume | reagent |
| 10 µl | 5x OneTaq Standard Reaction Buffer |
| 1 µl | 10 mM dNTPs |
| 1 µl | 10 µM Forward Primer (O34 (TEV_fw)) |
| 1 µl | 10 µM Reverse Primer (O35 (TEV_rv)) |
| 0.25 µL | OneTaq Hot Start DNA Polymerase (Finally: 1.25 units/50 µL) |
| 1 µl | Plasmid DNA (P14) |
| 35.75 µL | ddH2O Water |
| =50 µL | TOTAL |
- Mix with pipette
- The PCR program was performed after following scheme with following conditions:
| Initial denaturation | 94 °C | 30 s |
| 30 cycles | 94 °C | 30 s |
| 55 °C | 60 s | |
| 68 °C | 150 s | |
| Final extension | 68 °C | 5 min |
| Hold | 4 °C | infinite |
- After PCR, the product was purified with QIAquick PCR Purification Kit, Qiagen.
- Resulting product was labeled as F12.
Digestion of P61 with S1 Nuclease
Investigator: Katrin
Oligohybridization of O22 (MinMCS_for) and O23 (MinMCS_rev)
Investigator: Andi
Aim of the experiment: Oligohybridization of MinMCS_for (O22) and MinMCS_rev (O23
Procedure:
- 25 µL of 100 pmol/µl of MiniMCS fw and 25 µL of 100 pmol/µl MiniMCS rev
- Heating up to 95 °C for 5 min
- Cooling at RT in a styropor box overnight
Preparation of ordered primers
Investigators: Andreas, Rosario, Katrin, Ingmar
Aim of the experiment: Dissolving of primer pallets to provide a concentration of 100 pmol/µl
Procedure: We added an amount of water which gave us a final primer concentration of 100 pmol/µl.
Primers were labeled as follows:
| Label | Oligonr. |
|---|---|
| 24 | BPU_for |
| 25 | BPU_rev |
| 26 | BPU_SDM1_for |
| 27 | BPU_SDM1_rev |
| 28 | BPU_SDM2_for |
| 29 | BPU_SDM2_rev |
| 30 | BPU_SDM3_for |
| 31 | BPU_SDM3_rev |
| 32 | BPU_SDM4_for |
| 33 | BPU_SDM4_rev |
| 34 | TEV_for |
| 35 | TEV_rev |
| 36 | TEV_t387c_for |
| 37 | TEV_t387c_rev |
| 38 | N_Split_TEV_for |
| 39 | N_Split_TEV_rev |
| 40 | C_Split_TEV_for |
| 41 | C_Split_TEV_rev |
| 42 | PIF6_2-100_for |
| 43 | PIF6_2-100_rev |
| 44 | p104AgeI a71g fw |
| 45 | p104AgeI a71g rev |
| 46 | xylE_for |
| 47 | xylE_rev |
| 48 | xylE_c312a_for |
| 49 | xylE_c312a_rev |
| 50 | xylE_g483a_for |
| 51 | xylE_g483a_rev |
| 52 | xylE_a834g_for |
| 53 | xylE_a834g_rev |
Saturday, May 11th
Preparative digestion of PCR product of S219V pRK793 (F12) with XbaI & AgeI
Investigator: Louise
Aim of the experiment: Preparative digestion of PCR product of S219V pRK793 (F12) with XbaI & AgeI. Quickchange has still to be performed (forbidden SpeI restriction site).
Procedure:
- Batch for preparative digestion of PCR product S219V pRK793 (F12) with XbaI & AgeI.
| volume | reagent |
| 20 µl | PCR product F12 |
| 4 µl | NEBuffer 4 |
| 0.4 µl | BSA (100x) |
| 1 µl | AgeI-HF |
| 1 µl | XbaI |
| 13.6 µl | ddH2O |
| =40 µl | TOTAL |
- Incubation for 3 h at 37 °C
- Digested products were purified to remove nucleotides and enzymes with QIAquick PCR Purification Kit, QIAGEN.
- The digested fragment was labelled F15
Miniprep of overnight culture of transformated E.~coli XL1-Blue with F8 (pSB1C3) + F6 (EreA)
Investigator: Louise
Aim of the experiment: Miniprep of overnight culture of transformated E.~coli XL1-Blue with F8 (pSB1C3) + F6 (EreA) of 2 clones picked the day before
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
- The plasmids were labelled as P63 and P64
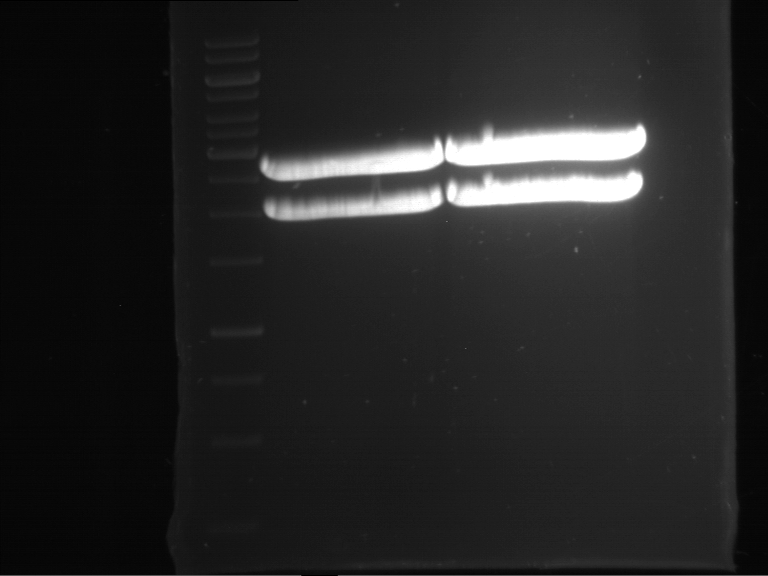
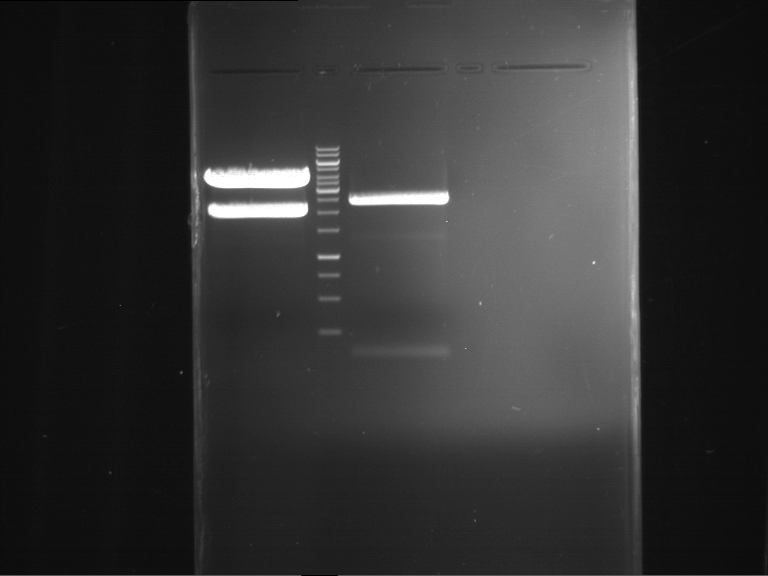
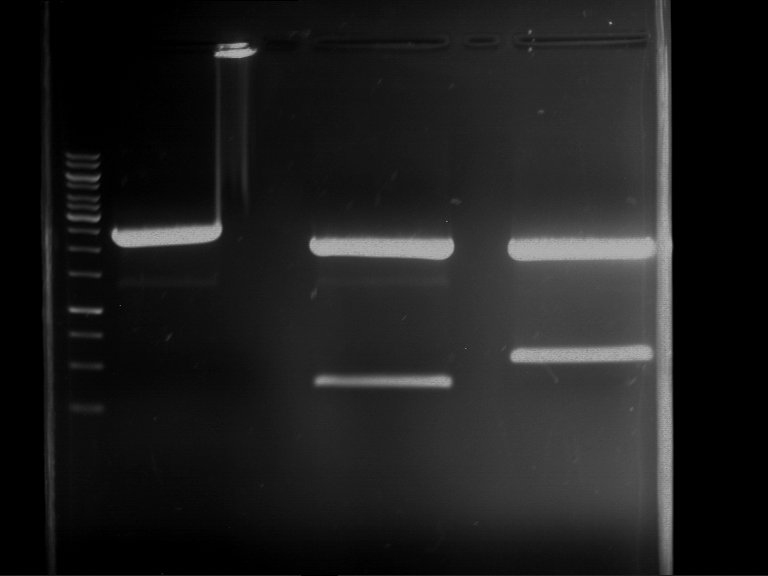
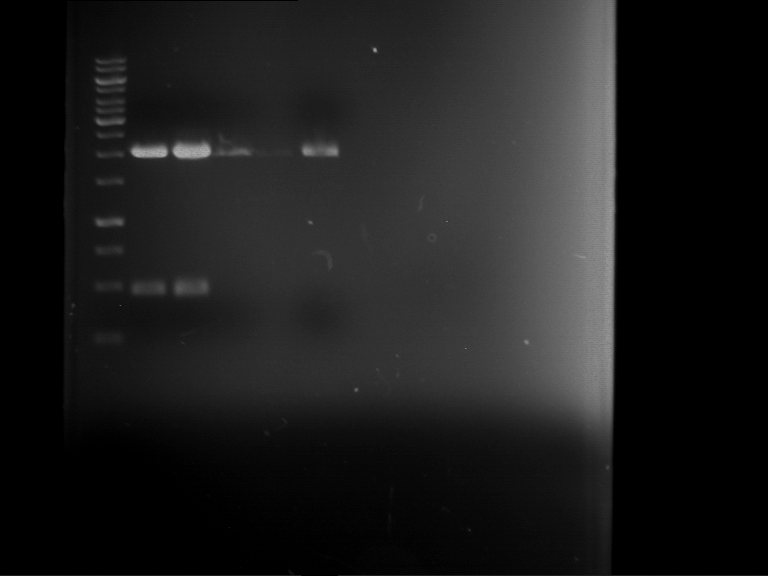
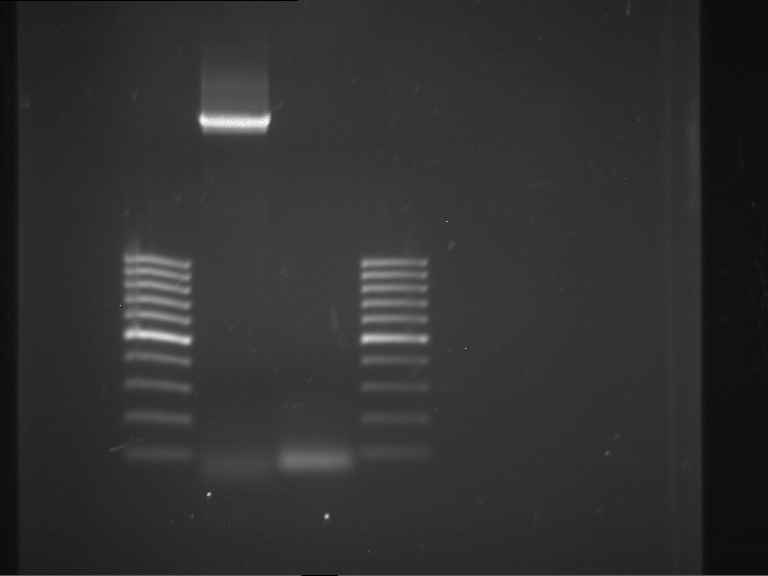
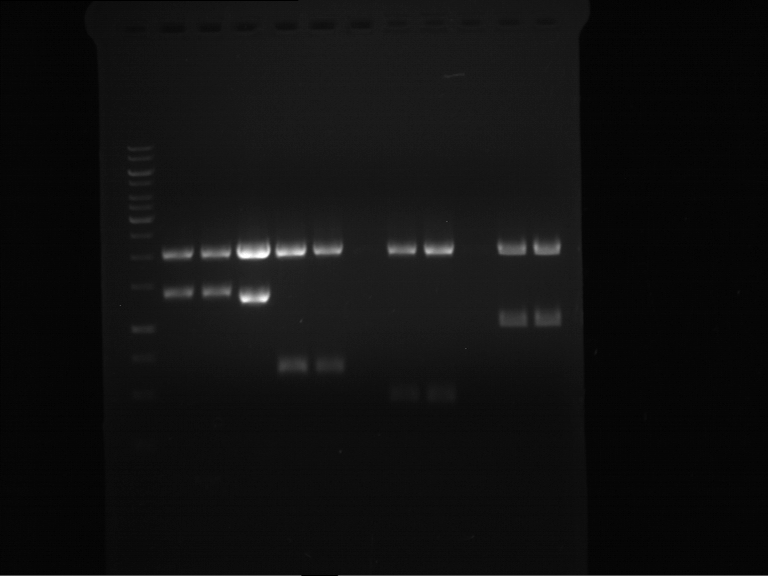
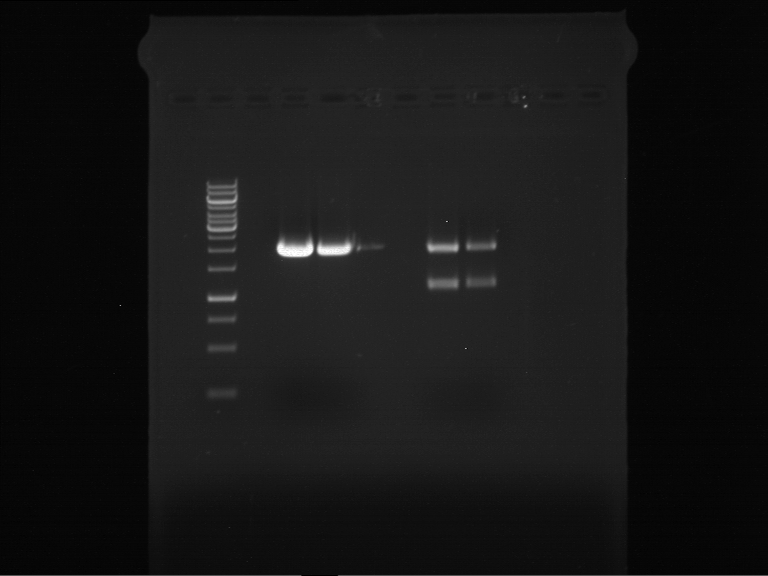
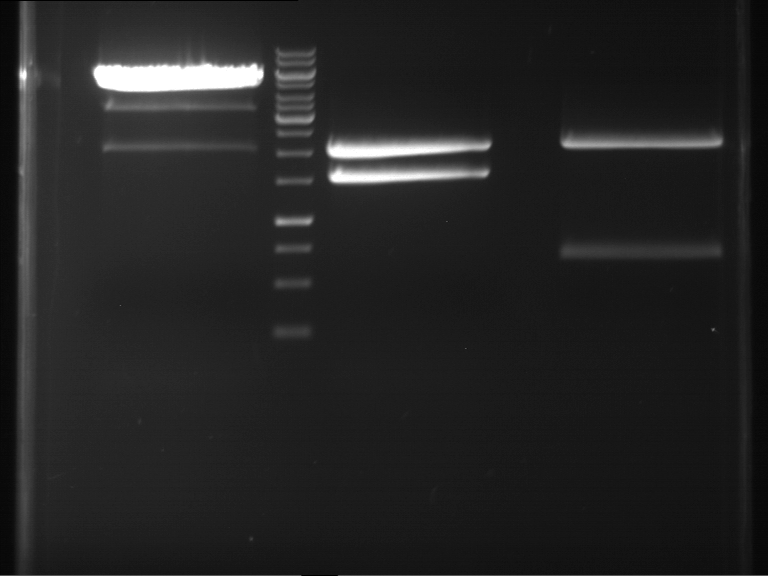
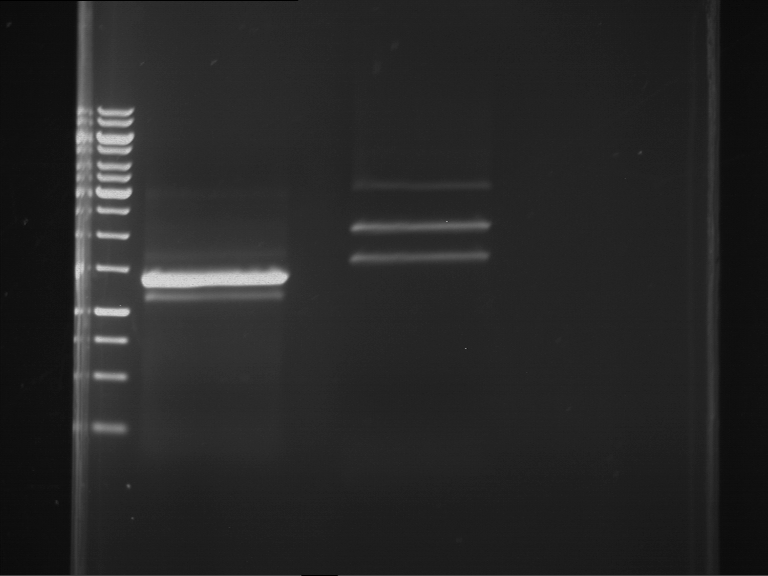
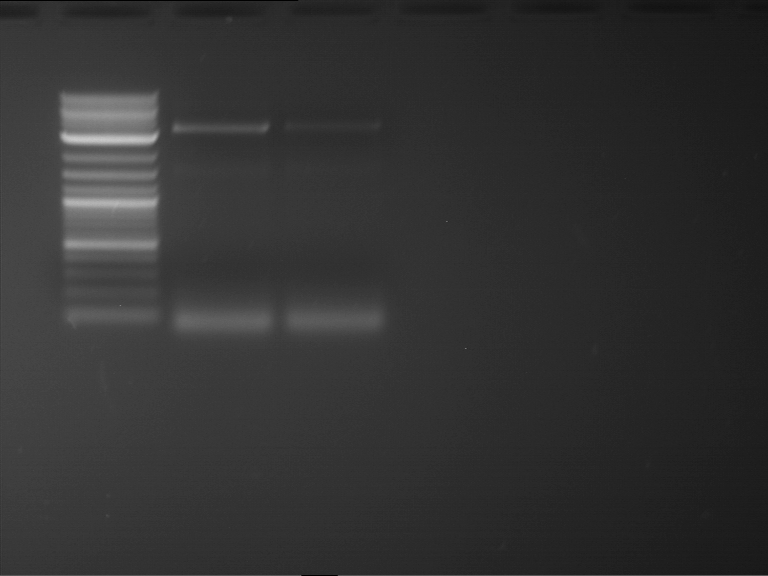
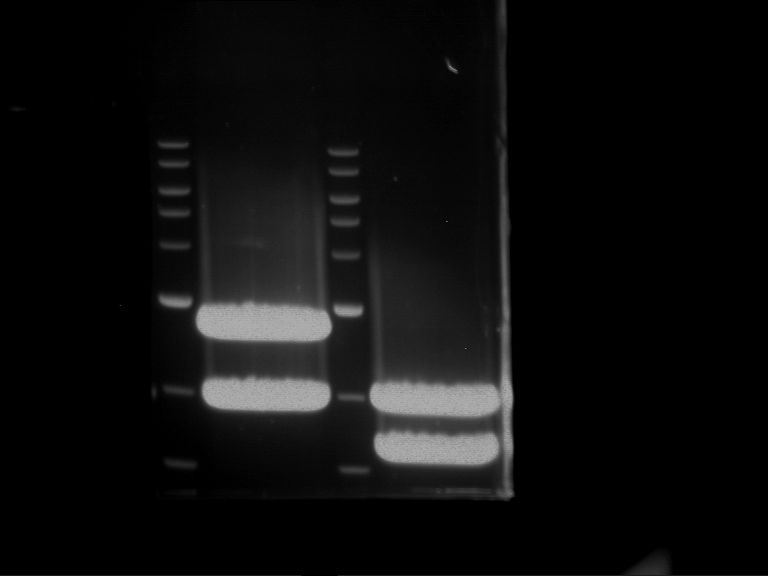
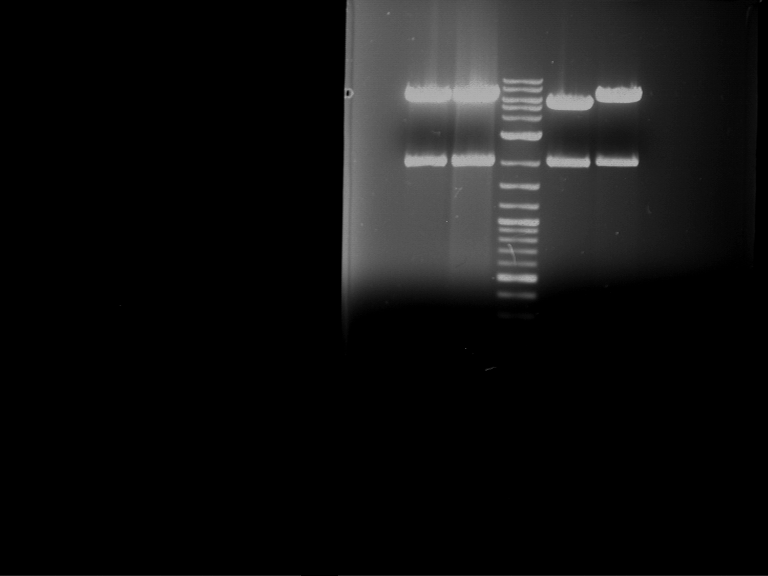
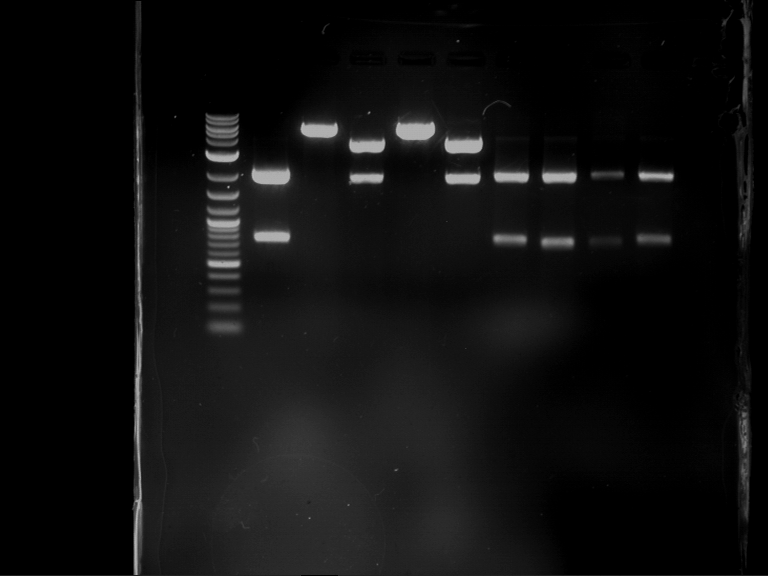
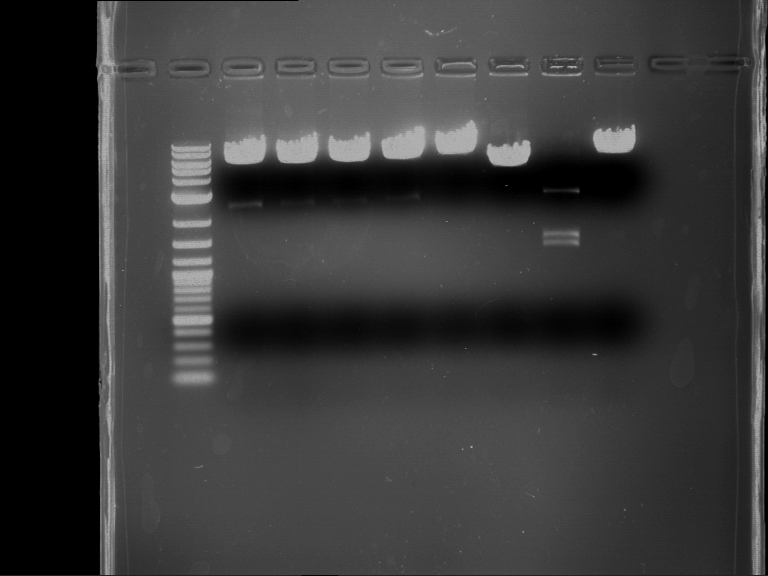
Analytical digestion and gelelectrophoresis of P63 and P64 with AgeI-HF and XbaI
Investigator: Louise
Aim of the experiment: Analytical digestion and gelelectrophoresis of P63 and P64 with AgeI-HF and XbaI
Procedure:
- Batch for analytical digestion of P63 and P64 with AgeI-HF and XbaI
| volume | reagent |
| 2.5 µl | Plasmid DNA P63/P64 |
| 2 µl | NEBuffer 4 (10x) |
| 0.2 µl | BSA (100x) |
| 0.25 µl | XbaI(10 U/µl) |
| 0.25 µl | AgeI-HF (20 U/µl) |
| 14.8 µl | ddH2O |
| =20 µl | TOTAL |
- Incubation for 90 min at 37 °C.
- Analytical gelelectrophoresis was performed at 90 V for 60 min.
| Analytical digestion of P63 with AgeI-HF and XbaI | 1 kbp ladder | Analytical digestion of P64 with AgeI-HF and XbaI |
| Insertion successful | Insertion successful |
Preparative digestion of PCR product of EreB (F2) with XbaI & AgeI
Investigator: Louise, Johanna
Aim of the experiment: Preparative digestion of PCR product EreB (F2) with XbaI & AgeI (second try since there did not grow any transformed bacteria in the first time).
Procedure:
- Batch for preparative digestion of PCR product and EreB (F2) with XbaI & AgeI.
| volume | reagent |
| 20 µl | PCR product F2 |
| 5 µl | NEBuffer 4 |
| 0.5 µl | BSA (100x) |
| 1 µl | AgeI-HF |
| 1 µl | XbaI |
| 22.5 µl | ddH2O |
| =50 µl | TOTAL |
- Incubation for 3 h at 37 °C
- The digested fragment was labelled F14
Purification and ligation of digested PCR product EreB (F14)
Investigator: Louise, Rosario
Aim of the experiment: PCR Purification and ligation of digested PCR product EreB (F14).
Procedure: PCR Purification of EreB PCR (F14)
- To purify the digested Fragment(F14) the QIAquick PCR Purification Kit was used. The concentration of the purified product was found to be 5 ng/µl via Nano drop.
Ligation of F14 + F8(pSB1C3)
- Ligation batch for F8+F14 (positive control)
| volume | reagent |
| 1.35 µl | F8 (74.3 ng/µl, 2086 bp) |
| 15.65 µl | F14 (5 ng/µl, 1257 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- Negative control was also prepared. Instead of the insert, the same volume of ddH2O was added to the reaction batch.
- Cycled ligation has been performed after following protocol in a thermocycler:
| 99 cycles | 12 °C | 60 s |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| Hold | 16 °C | infinite |
Lid temperature = 37 °C
Transformation of ligation products of F10 + F11 into E.coli Xl1-Blue
Investigator: Andi
Aim of the experiment:Transformation of the ligation products of F10 + F11 and the negative control in pTUM104 in XL1 Blue.
Operation Sequence
- melting of 2x 200 µl Ca-competent E.coli XL1-Blue cells on ice
- Preparing 4 Tubes with 100 µl of Ca-competent E.coli XL1-Blue cells
- addition of 5 µl of the ligation products
- incubation for 30 min on ice
- heat shock for 5 min at 37 °C
- transfer of cells to 1 ml LB-medium without antibiotics and incubate at 37°C and 180 rpm for 30 min
- plate 100 µl on an Amp-LB-plate
- sediment the leftover in a centrifuge (30 - 60 sec, 13 000 rpm) and resuspend the sediment in 100 µl LB-medium and plate it as well on an Amp-LB-plate
Transformation of Quickchange Product P62 into E.coli Xl1-Blue
Investigator: Andi
Aim of the experiment:Transformation of the Quickchange Product P62 and the negative control in XL1 Blue.
Operation Sequence
- melting of 1x 200 µl Ca-competent E.coli XL1-Blue cells on ice
- Preparing 4 Tubes with 50 µl of Ca-competent E.coli XL1-Blue cells
- addition of 1 µl of the Quickchange Product
- incubation for 30 min on ice
- heat shock for 5 min at 37 °C
- transfer of cells to 1 ml LB-medium without antibiotics and incubate at 37°C and 180 rpm for 30 min
- plate 100 µl on an Amp-LB-plate
- sediment the leftover in a centrifuge (30 - 60 sec, 13 000 rpm) and resuspend the sediment in 100 µl LB-medium and plate it as well on an Amp-LB-plate
Quickchange mutagenesis of pTUM104
Investigator: Ingmar
Aim of the experiment: Sequencing results showed a forbiden AgeI restriction site in this vector. The sdm will delete this restricition site
Procedure: PCR
Reaction batch 1
| volume | reagent |
| 2.5 µl | 10x Pfu Ultra II buffer |
| 4 µl | Plasmid P6 template |
| 0.5 µl | 1:10 dilution of O44 (10 pmol/µL) |
| 16.5 µl | ddH2O |
| 0.5 µl | dNTP mix |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
Reaction batch 2
| volume | reagent |
| 2.5 µl | 10x Pfu Ultra II buffer |
| 4 µl | Plasmid P6 template |
| 0.5 µl | 1:10 dilution of O45 (10 pmol/µL) |
| 16.5 µl | ddH2O |
| 0.5 µl | dNTP mix |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
PCR cycling parameters
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 30 sec |
| 2 | 10 | 95°C | 30 sec |
| 55°C | 1 min | ||
| 67°C | 6 min |
- Having completed the PCR cycling parameters listed above both PCR reaction batches were mixed together and the cycling parameters listed above were one time more applied.
- Digestion of the parental DNA with DpnI: Addition of 1 µl DpnI to the PCR batch and incubate for 1 h at 37 °C. The resulting product was named fragmet 14.
Transformation into E.coli Xl1-Blue Operation Sequence
- melting of 100 µl Ca-competent E.coli XL1-Blue cells on ice
- addition of 1 µl of the PCR product
- incubation for 30 min on ice
- heat shock for 5 min at 37 °C
- transfer of cells to 1 ml LB-medium without antibiotics and incubate at 37°C and 180 rpm for 30 min
- plate 100 µl on an Amp-LB-plate
- sediment the leftover in a centrifuge (30 - 60 sec, 13 000 rpm) and resuspend the sediment in 100 µl LB-medium and plate it as well on an Amp-LB-plate
Sunday, May 12th
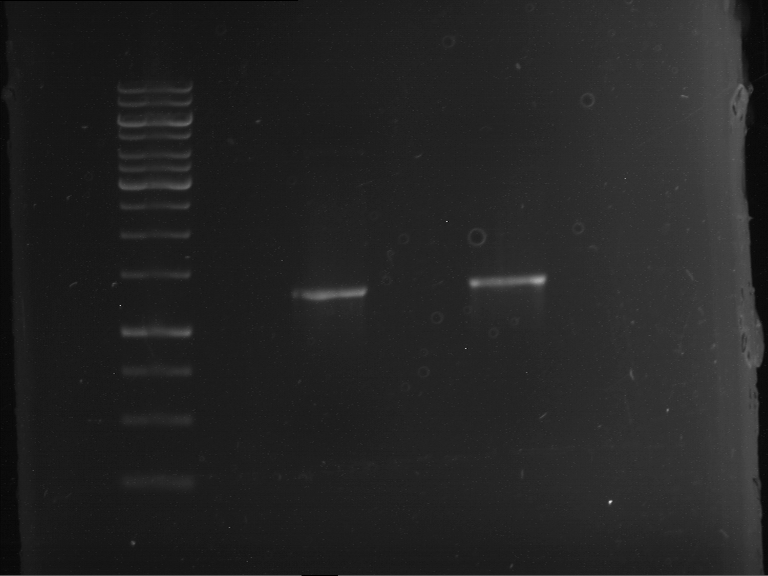
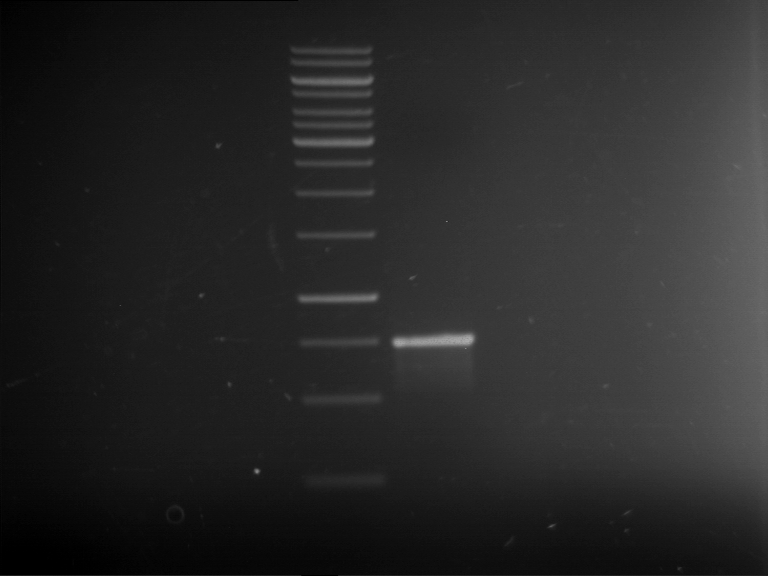
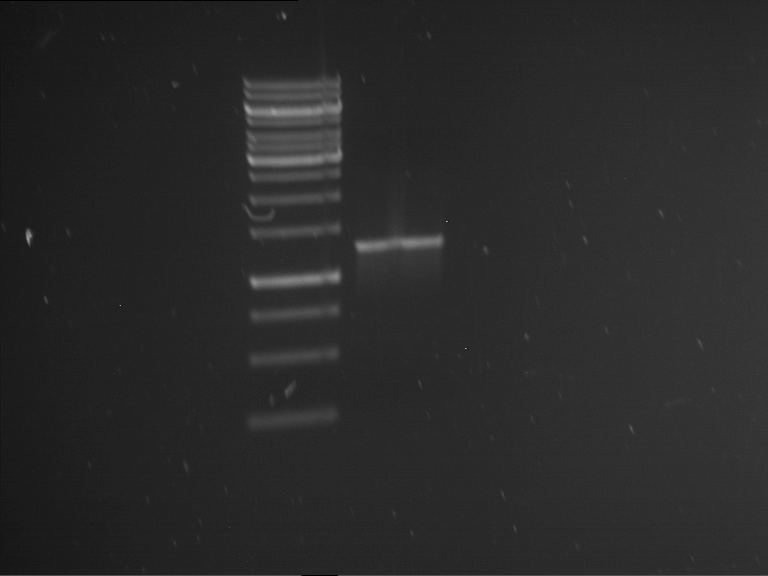
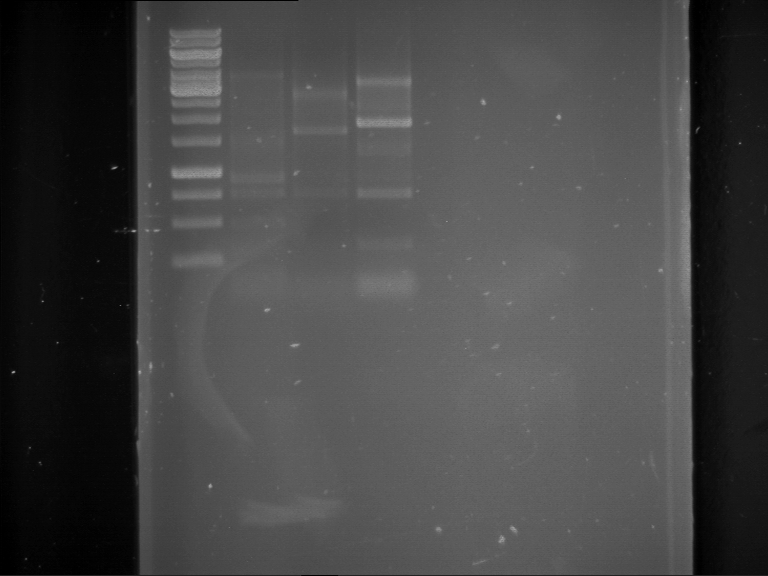
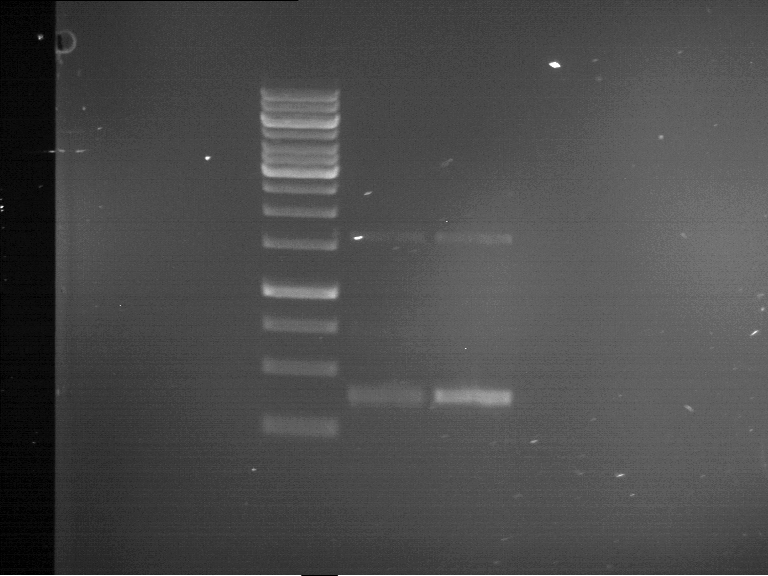
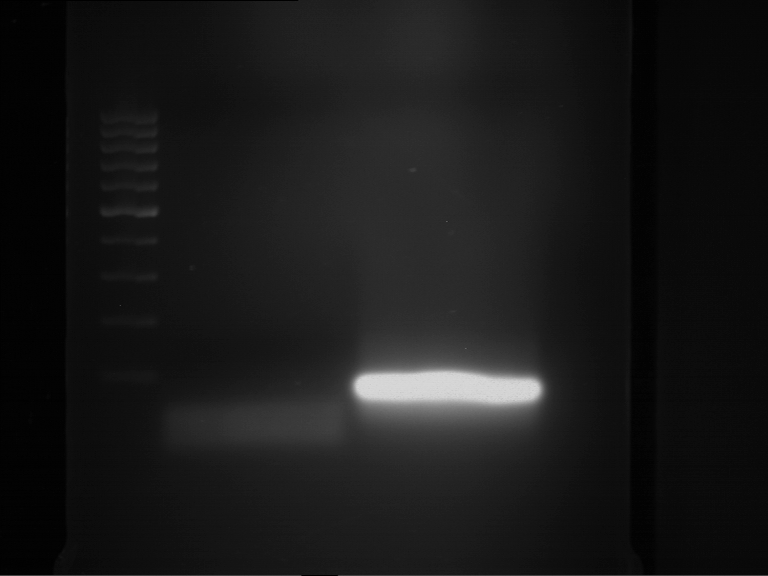
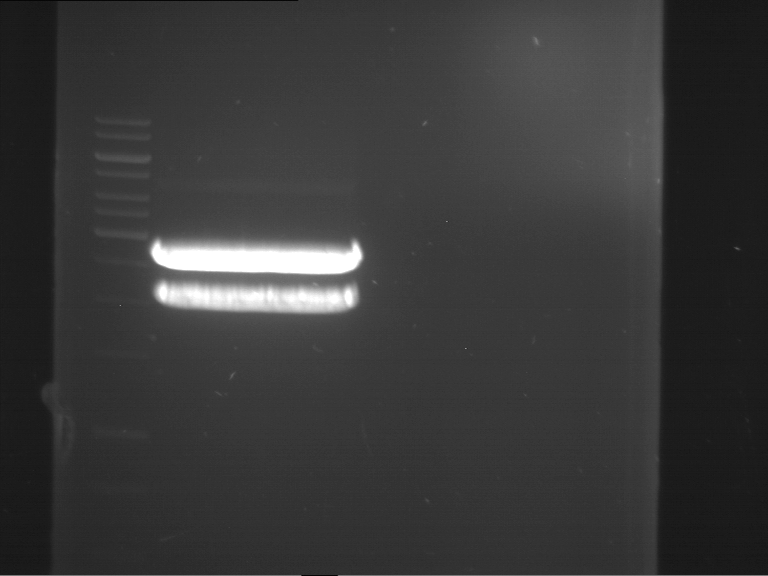
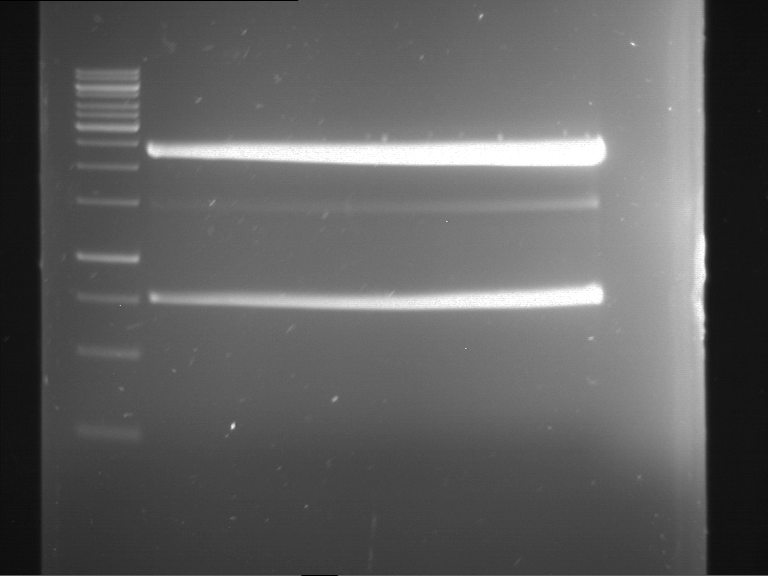
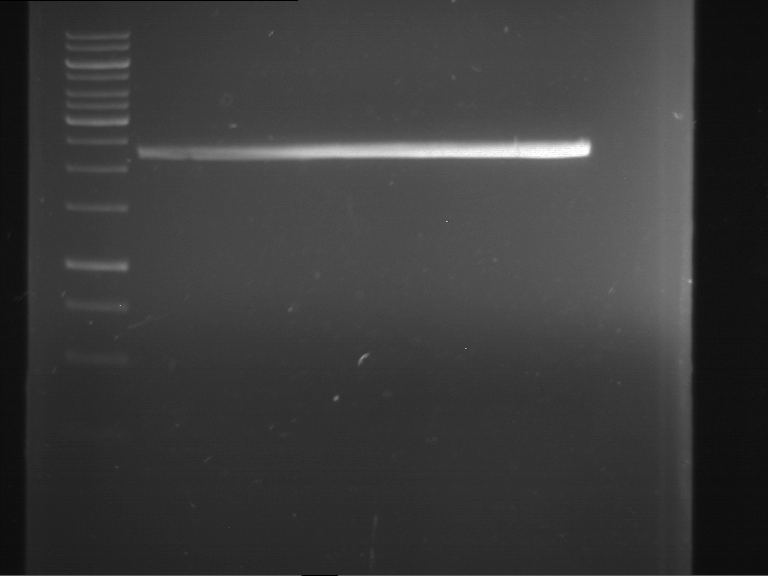
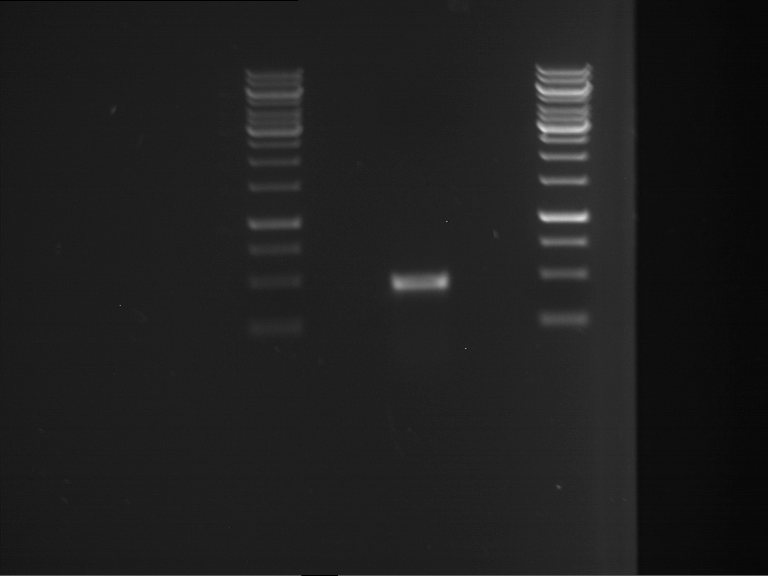
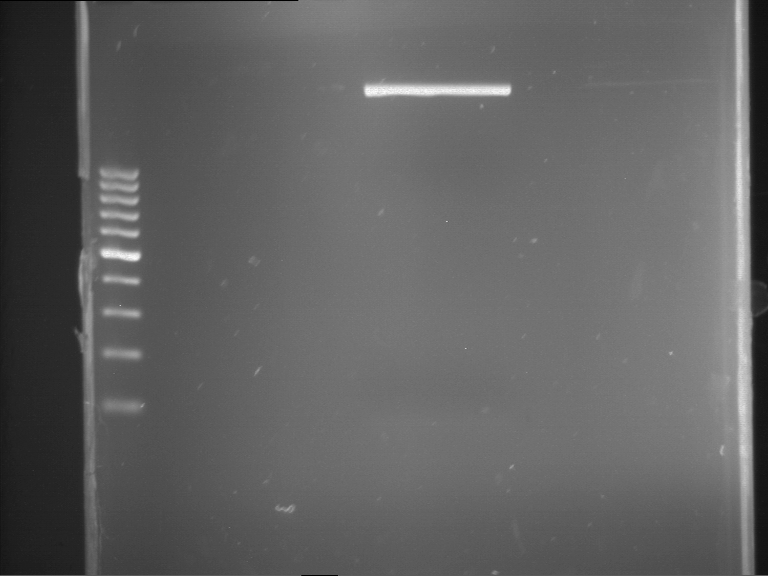
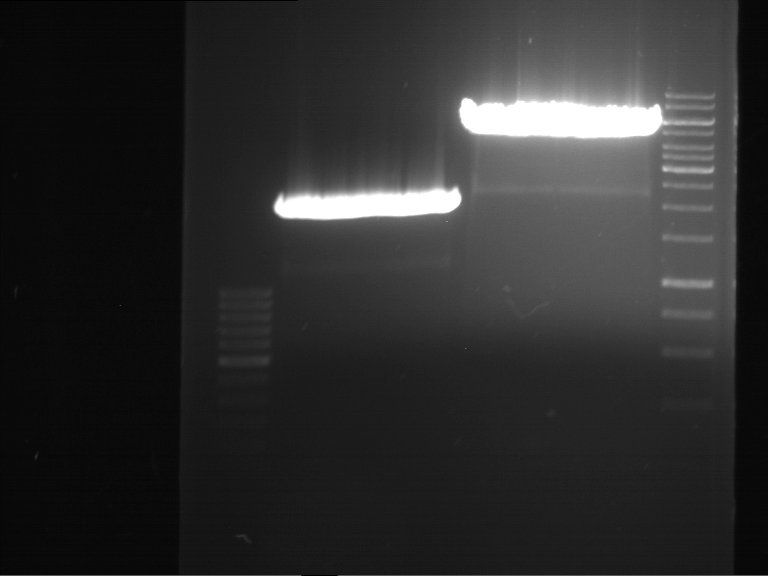
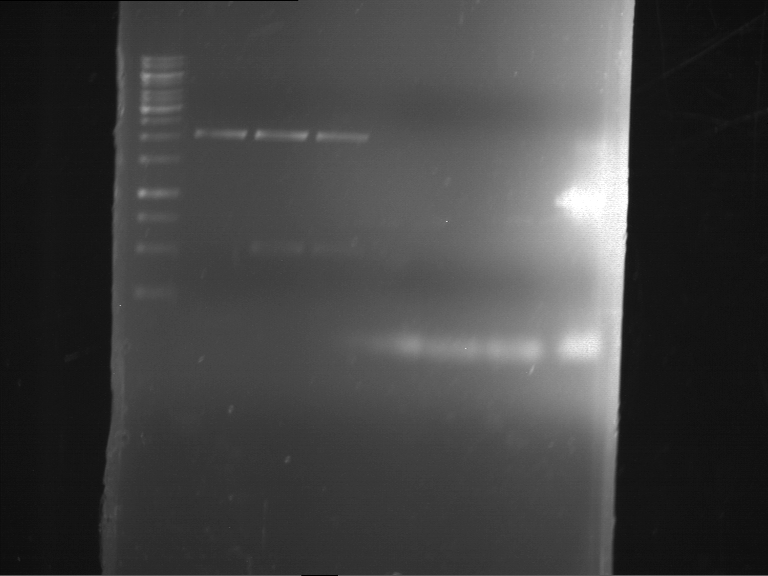
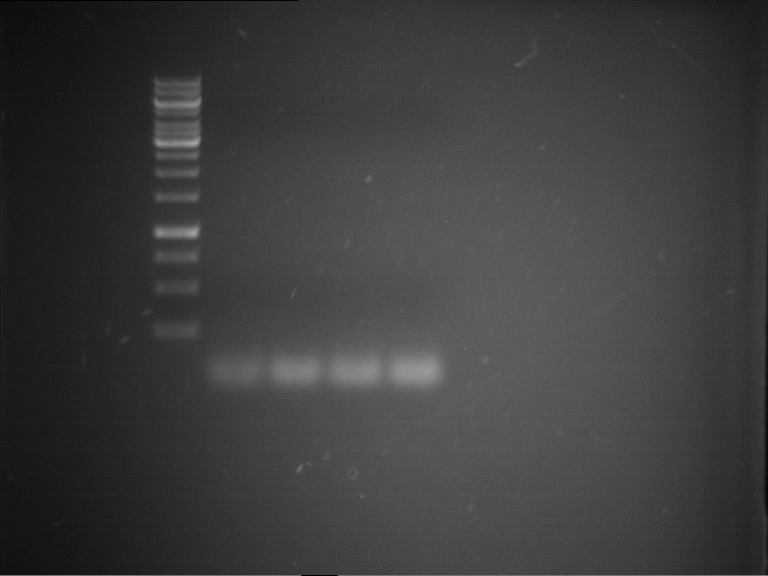
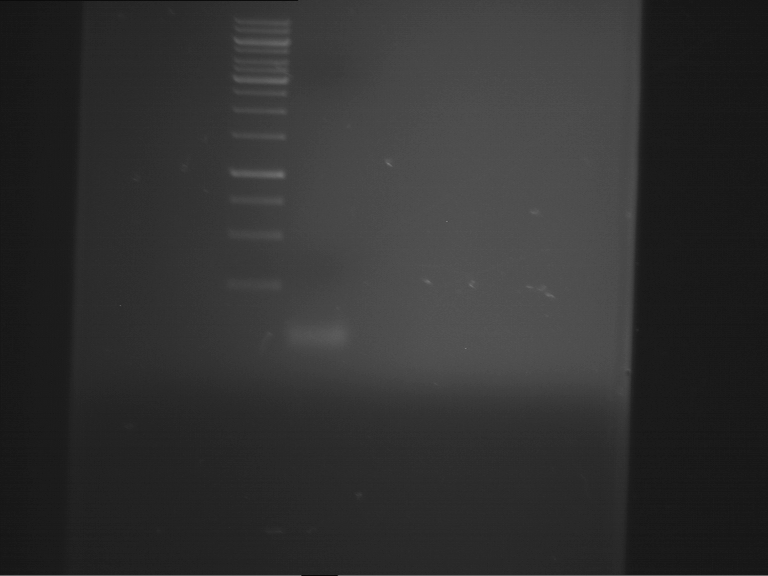
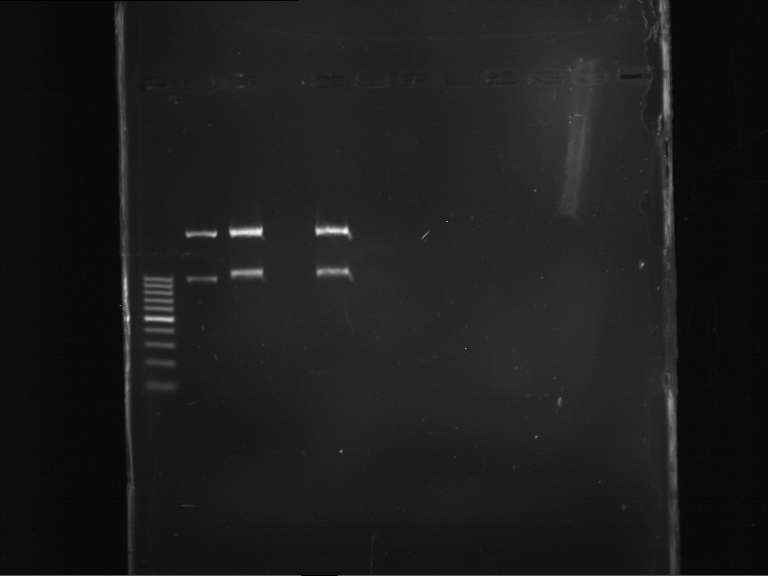
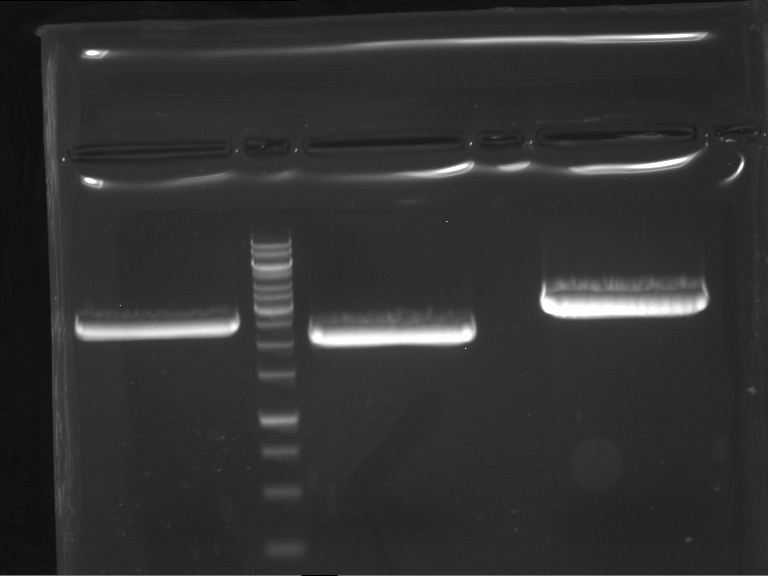
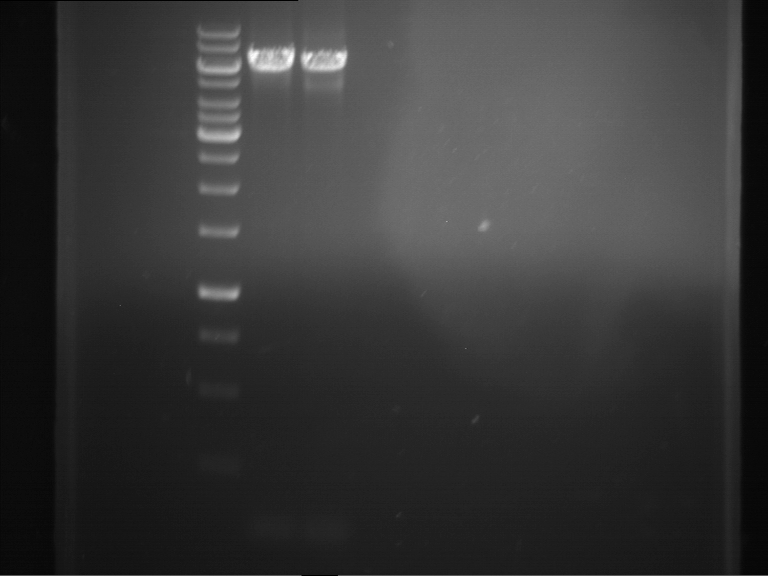
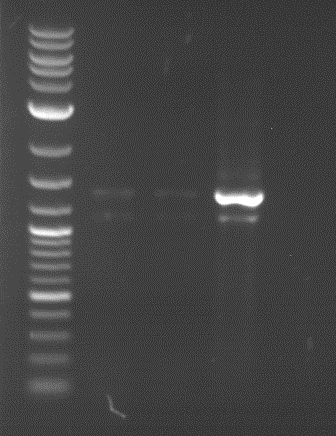
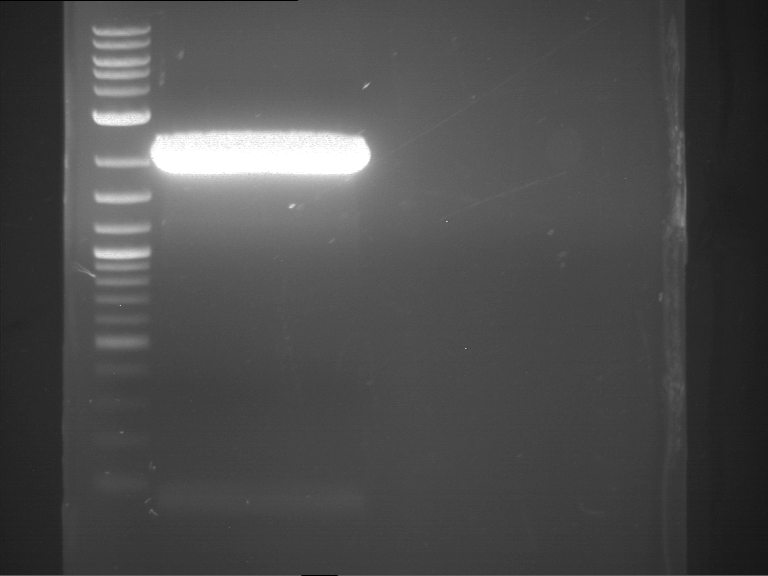
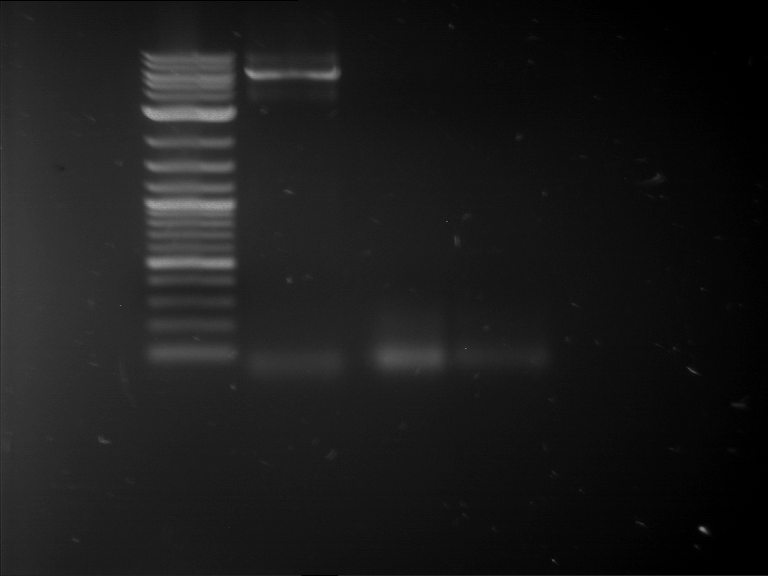
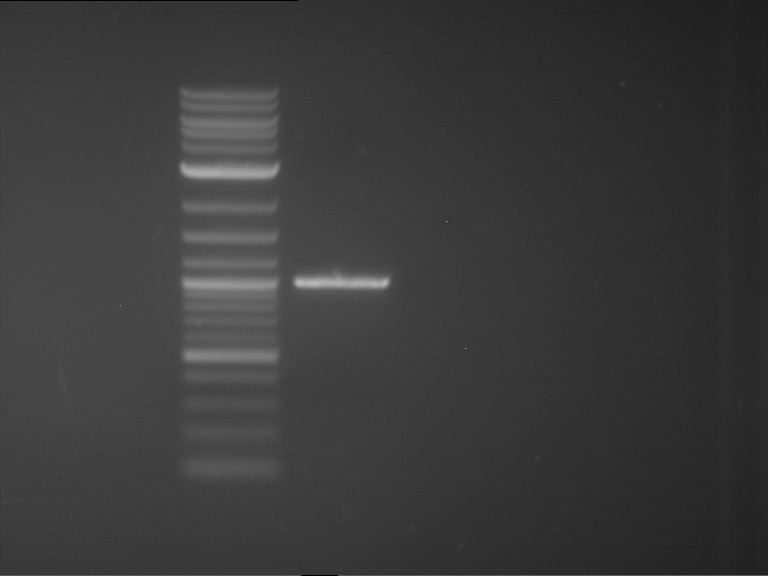
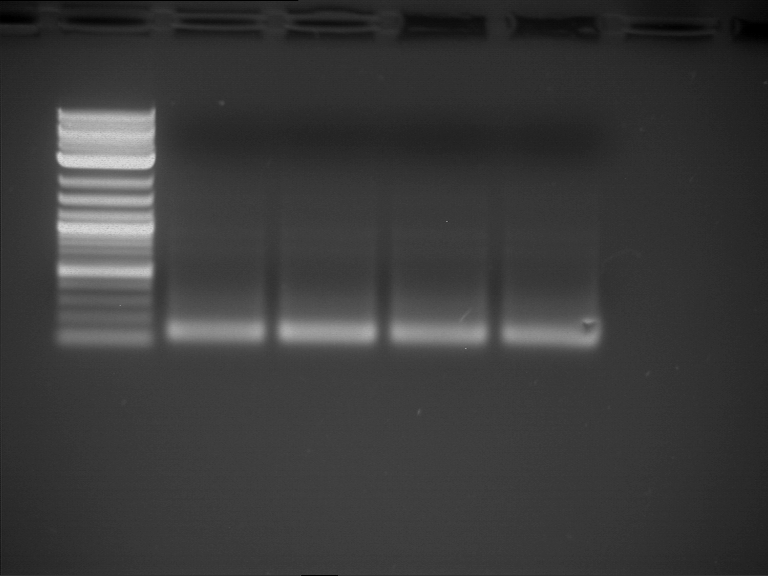
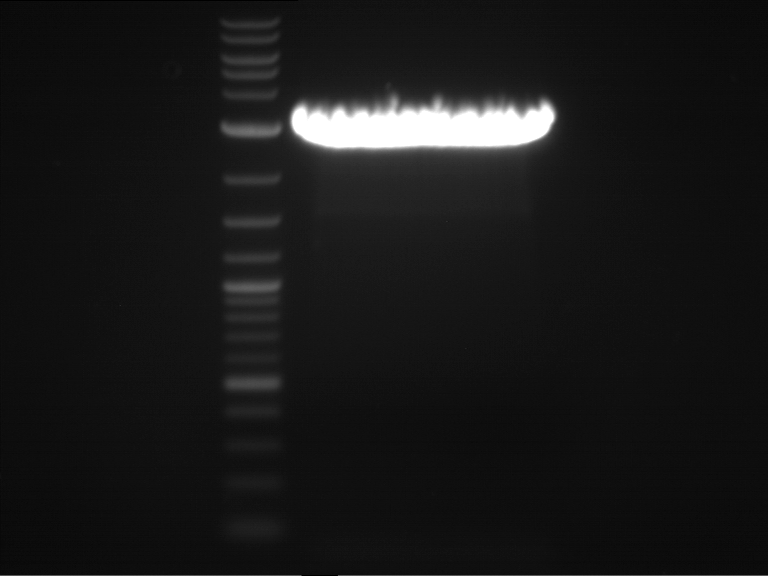
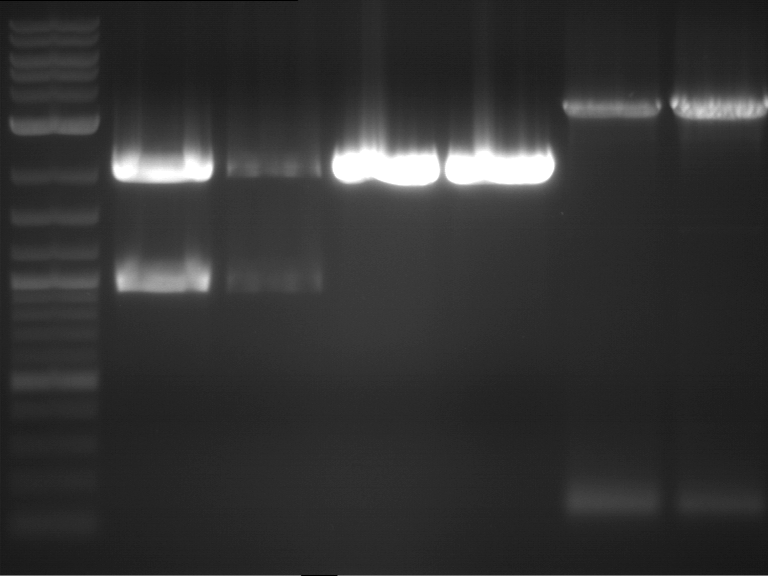
Analytical gelelectrophoresis of F15 (digested TEV PCR product)
Investigator: Jeff, Rosario
Aim of the experiment: Analytical gelelectrophoresis of F15 (digested TEV PCR product) to check whether PCR has worked.
Procedure:
- 5 µl of F15 was mixed with 0.55 µl of DNA loading buffer (10x) and analytical gelelectrophoresis was performed at 1% agarose gel for 60 min at 90 V.
- PCR product has expected length
Transformation of E. coli XL1 blue with ligation product F8+F15 (TEV in pSB1C3, RFC25)
Investigator: Jeff, Rosario
Aim of the experiment: Transformation of E. coli XL1 blue with ligation product F8+F15 (TEV in pSB1C3, RFC25). Further quickchanges still have to be done.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 10 µl of the ligation products were added to 200 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of the cell suspension was plated on one chloramphenicol plate.
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on a new chlorampenicol plate.
Transformation of E. coli XL1 blue with ligation products F8+F14 (EreB in pSB1C3, RFC25)
Investigator: Jeff, Rosario
Aim of the experiment: Transformation of E. coli XL1 blue with ligation product F8+F14 (EreB in pSB1C3, RFC25). Further quickchanges still have to be done.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 10 µl of the ligation products were added to 200 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of the cell suspension was plated on one chloramphenicol plate.
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on a new chlorampenicol plate.
Transformation of E. coli XL1 blue with P23
Investigator: Jeff, Rosario
Aim of the experiment: Transformation of E. coli XL1 blue with P23 for insertion of miniMCS.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 2 µl of DNA was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of the cell suspension was plated on one chloramphenicol plate.
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on a new chlorampenicol plate.
Picking of transformed Plasmid F10+F11 (IRES from polio virus, blunt ligated with pSB1C3 for sequencing)
Investigator: Andi
Aim of the study: Picking of transformed Plasmid F10+F11 (AlcR blunt ligated with pSB1C3 for sequencing).
Operational sequence:
- Picking and overnight culture after standard laboratory's protocol. (CamR LB-medium)
- 10 colonies were picked.
Week 4
Monday, May 13th
Miniprep of overnight culture of transformated E.~coli XL1-Blue with F10+F11
Investigator: Rosario, Jeffery, Johanna, Flo
Aim of the experiment: Miniprep of overnight culture of transformated E.~coli XL1-Blue F10+F11 of 10 clones picked the day before
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
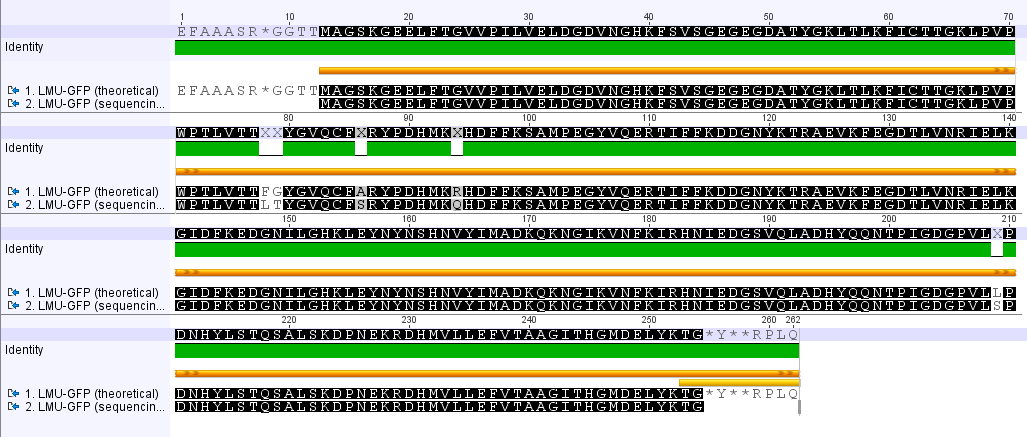
Analyzing sequencing results
Investigator: Johanna
Aim of the experiment: Comparison of the sequencing results with the Biobrick (BB) sequences.
Procedure: Using Geneious to align the sequencing results with the BB sequences.
LMU GFP in RFC 25(p20)
- The provided sequence in the parts registry is totally wrong.
- The comparison of the amino acid seqeunces reveals several point mutations.
FluA in RFC 25 (p45)
- Did not yield sequencing results (does the primer bind ?)
- No presence of RCF 25 standard restriction enzymes
mCherry in RFC 25 (p46)
- Did not yield sequencing results (does the primer bind ?)
- No presence of RCF 25 standard restriction enzymes
CaMV35S in RFC 25 (p23)
- high identity
- high match with HQ699544 (Blast)
LMU mKate2in RFC 25 (p21)
- no continous ORF
- Did not yield sequencing results
DREB1C promoter in RFC 25 (p26)
- Present of fluorecent protein gene
xylE in RCF 25 (p38)
- Did not yield sequencing results
SV40 in RCF 25 (p28)
- sequencing result agrees with the laccases
- the sequencing result can't be the one of SV40
Quickchange mutagenesis SDMI of p27
Investigator: Rosario, Jeff, Flo, Andi
Aim of the experiment: Sequencing results showed a forbidden restriction site in this vector. The sdm will delete this restricition site. The resulting Plasmid was labeled 65.
Procedure: PCR
Reaction batch 1
| volume | reagent |
| 2.5 µl | 10x Pfu Ultra II buffer |
| 0.5 µl | Plasmid P27 template |
| 0.5 µl | 1:10 dilution of O26 (10 pmol/µL) |
| 20. 5 µl | ddH2O |
| 0.5 µl | dNTP mix |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
Reaction batch 2
| volume | reagent |
| 2.5 µl | 10x Pfu Ultra II buffer |
| 0.5 µl | Plasmid P27 template |
| 0.5 µl | 1:10 dilution of O27 (10 pmol/µL) |
| 20.5 µl | ddH2O |
| 0.5 µl | dNTP mix |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
PCR cycling parameters
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 30 sec |
| 2 | 10 | 95°C | 30 sec |
| 55°C | 1 min | ||
| 67°C | 4 min |
- Having completed the PCR cycling parameters listed above both PCR reaction batches were mixed together and the cycling parameters listed above were one time more applied, but the amount of cycles was increased to 20
- Digestion of the parental DNA with DpnI: Addition of 1 µl DpnI to the PCR batch and incubate for 1 h at 37 °C.
Quickchange mutagenesis SDMI of p38
Investigator: Jeff, Flo, Andi, Rosario
Aim of the experiment: Sequencing results showed a forbidden restriction site in this vector. The sdm will delete this restricition site. The resulting Plasmid was labeled 67.
Procedure: PCR
Reaction batch 1
| volume | reagent |
| 2.5 µl | 10x Pfu Ultra II buffer |
| 0,5 µl | Plasmid P38 template |
| 0.5 µl | 1:10 dilution of O48 (10 pmol/µL) |
| 20.5 µl | ddH2O |
| 0.5 µl | dNTP mix |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
Reaction batch 2
| volume | reagent |
| 2.5 µl | 10x Pfu Ultra II buffer |
| 0.5 µl | Plasmid P38 template |
| 0.5 µl | 1:10 dilution of O49 (10 pmol/µL) |
| 20.5 µl | ddH2O |
| 0.5 µl | dNTP mix |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
PCR cycling parameters
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 30 sec |
| 2 | 10 | 95°C | 30 sec |
| 55°C | 1 min | ||
| 67°C | 4 min |
- Having completed the PCR cycling parameters listed above both PCR reaction batches were mixed together and the cycling parameters listed above were one time more applied, but the amount of cycles have been increased to 20
- Digestion of the parental DNA with DpnI: Addition of 1 µl DpnI to the PCR batch and incubate for 1 h at 37 °C.
Quickchange mutagenesis SDMI of P63
Investigator: Jeff, Rosario, Flo, Andi
Aim of the experiment: Sequencing results showed a forbidden restriction site in this vector. The sdm will delete this restricition site. The resulting Plasmid was labeled P66.
Procedure: PCR
Reaction batch 1
| volume | reagent |
| 2.5 µl | 10x Pfu Ultra II buffer |
| 0.25 µl | Plasmid P63 template; 1:2 dilution |
| 0.5 µl | 1:10 dilution of O8 (10 pmol/µL) |
| 20.75 µl | ddH2O |
| 0.5 µl | dNTP mix |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
Reaction batch 2
| volume | reagent |
| 2.5 µl | 10x Pfu Ultra II buffer |
| 0.25 µl | Plasmid P63 template; 1:2 dilution |
| 0.5 µl | 1:10 dilution of O9 (10 pmol/µL) |
| 20.75 µl | ddH2O |
| 0.5 µl | dNTP mix |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
PCR cycling parameters
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 30 sec |
| 2 | 10 | 95°C | 30 sec |
| 55°C | 1 min | ||
| 67°C | 6 min |
- Having completed the PCR cycling parameters listed above both PCR reaction batches were mixed together and the cycling parameters listed above were one time more applied, the amount of cycles was raised to 20
- Digestion of the parental DNA with DpnI: Addition of 1 µl DpnI to the PCR batch and incubate for 1 h at 37 °C.
Transformation of E. coli XL1 blue with P65
Investigator: Jeff, Rosario, Johanna, Andi, Flo
Aim of the experiment: Transformation of E. coli XL1 blue with P65 .
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 2 µl of DNA was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of the cell suspension was plated on one chloramphenicol plate.
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on a new chlorampenicol plate.
Transformation of E. coli XL1 blue with P66
Investigator: Jeff, Rosario, Flo, Johanna, Andi
Aim of the experiment: Transformation of E. coli XL1 blue with P66.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 2 µl of DNA was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of the cell suspension was plated on one chloramphenicol plate.
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on a new chlorampenicol plate.
Transformation of E. coli XL1 blue with P67
Investigator: Jeff, Rosario, Johanna, Andi, Flo
Aim of the experiment: Transformation of E. coli XL1 blue with P67.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 2 µl of DNA was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of the cell suspension was plated on one chloramphenicol plate.
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on a new chlorampenicol plate.
Tuesday, May 14th
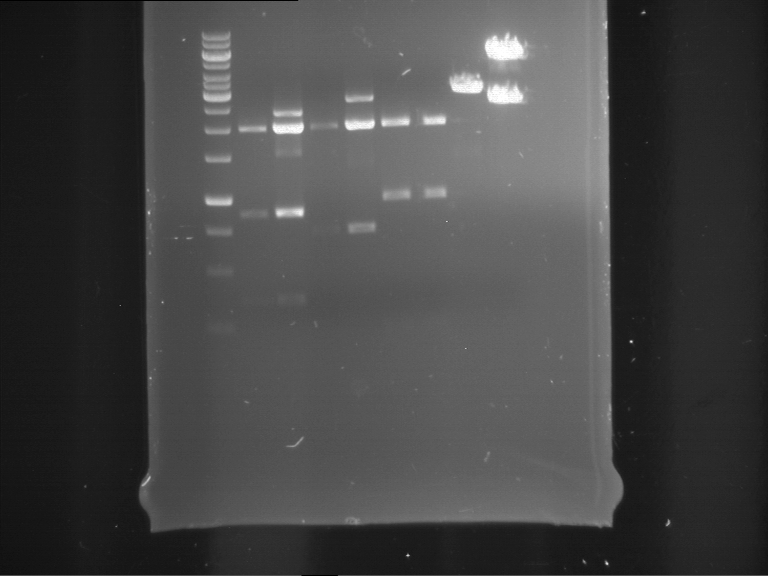
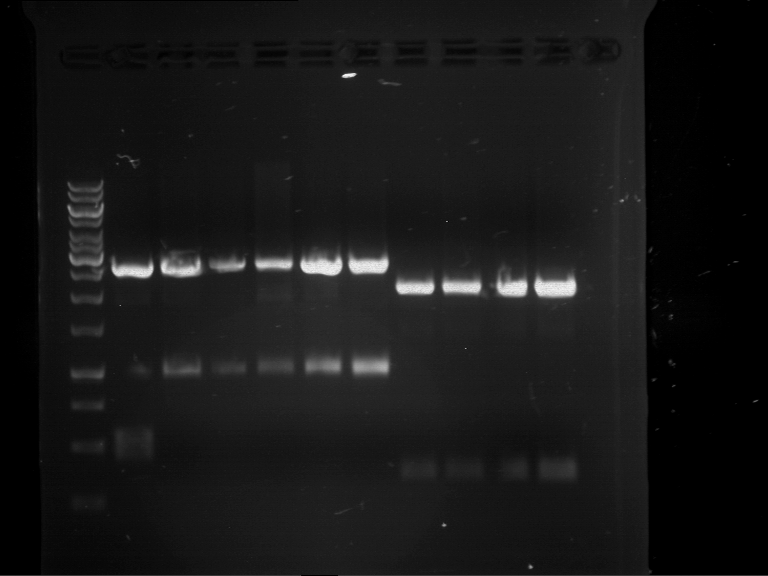
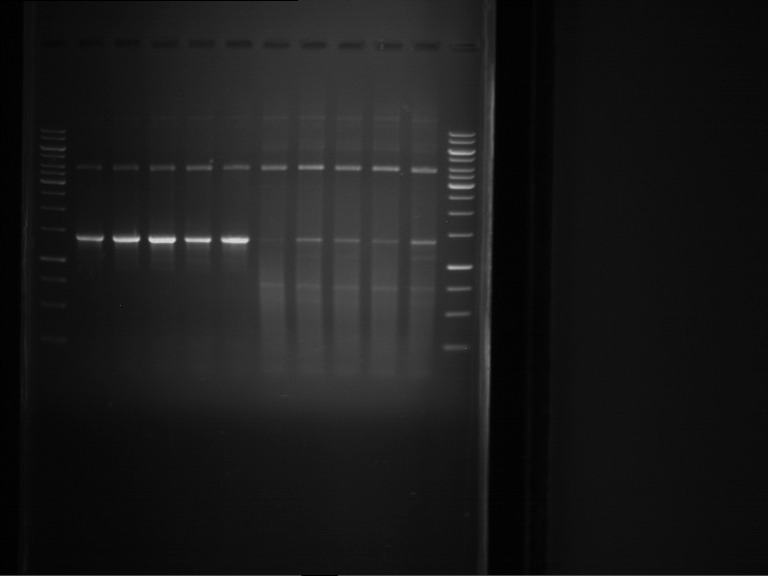
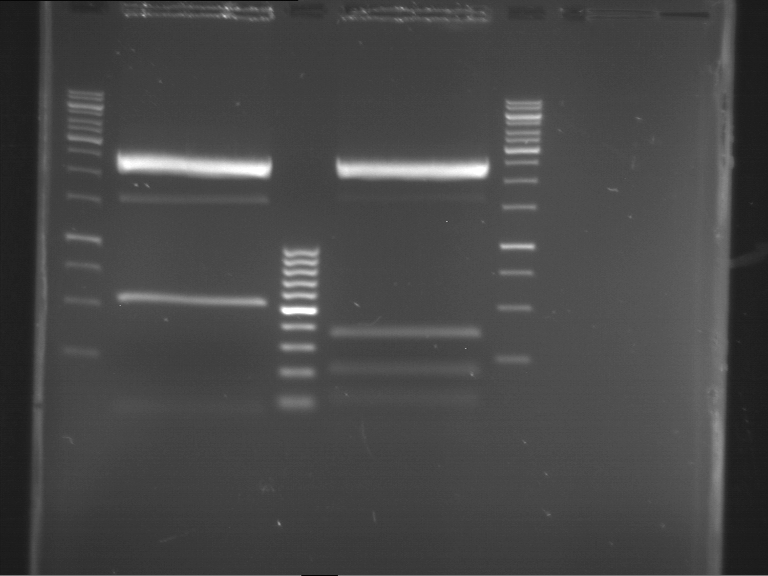
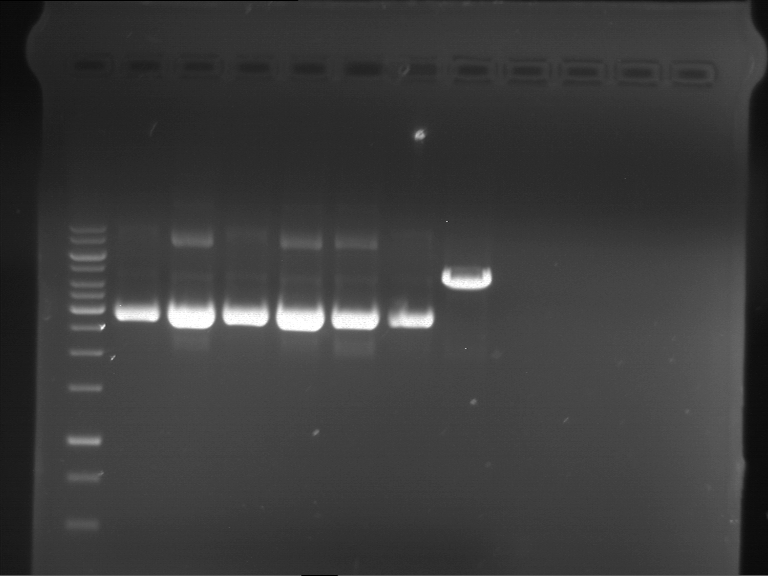
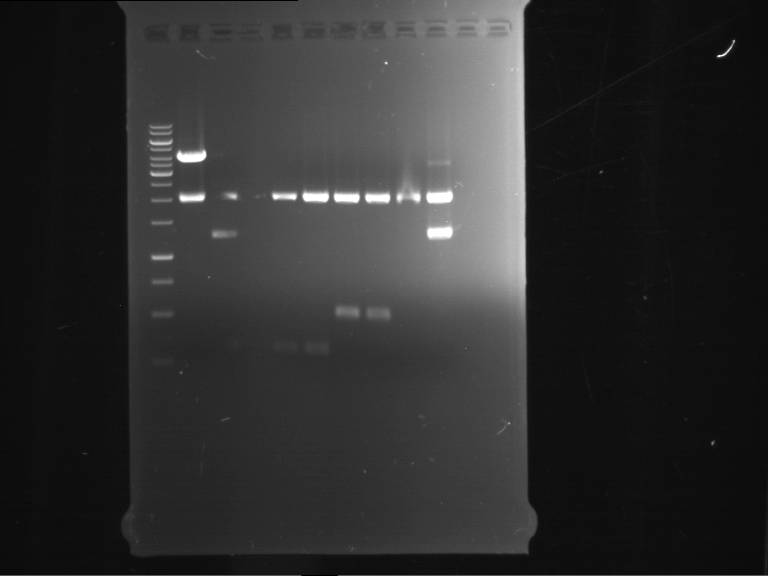
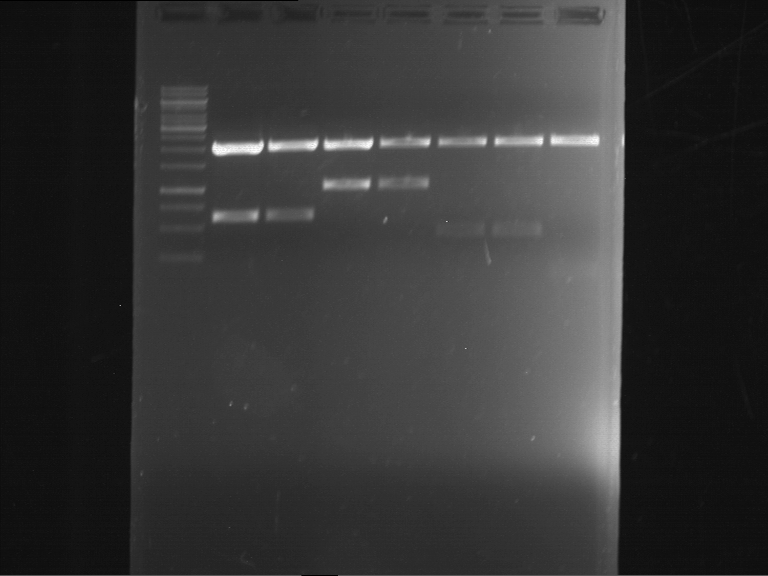
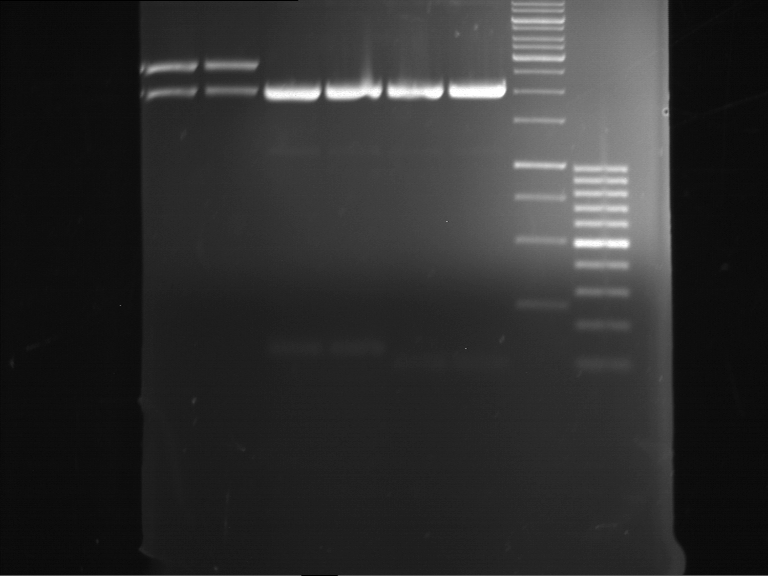
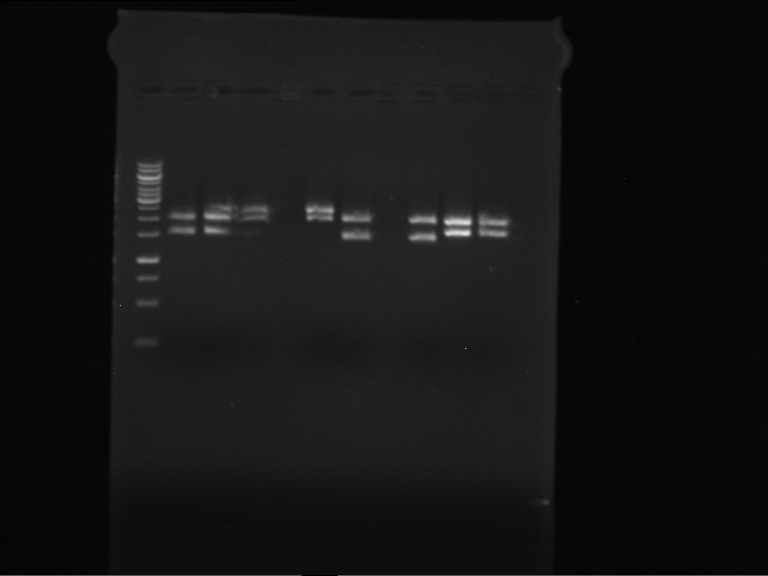
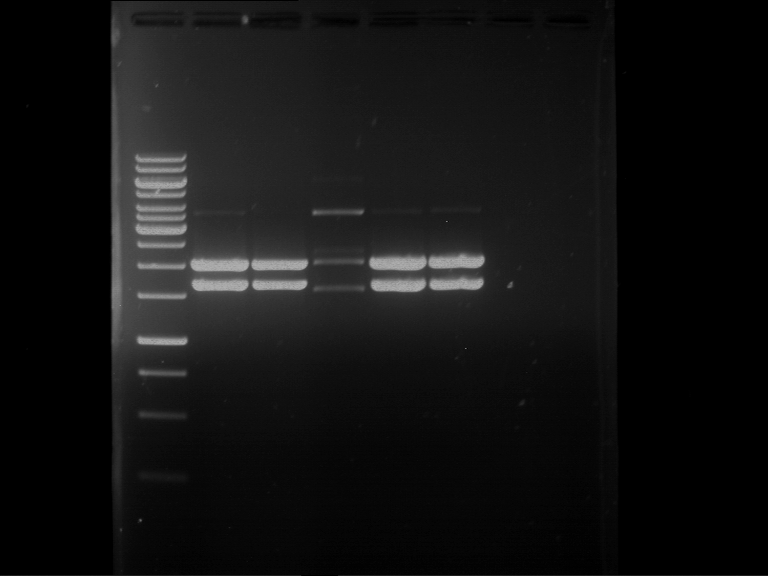
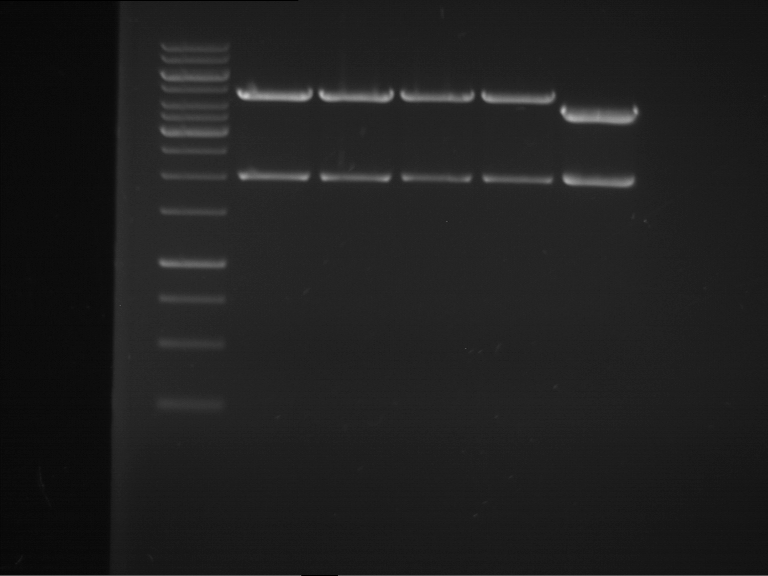
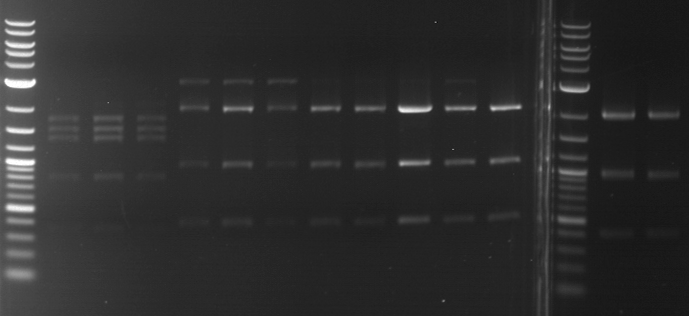
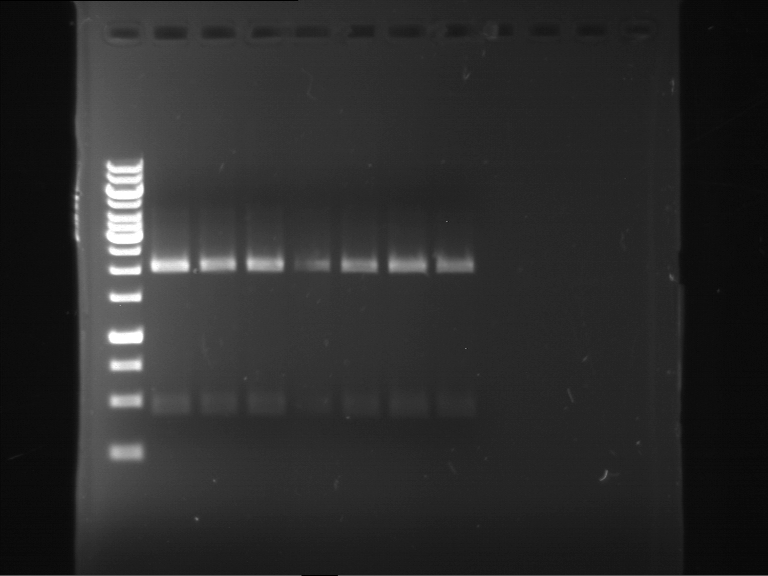
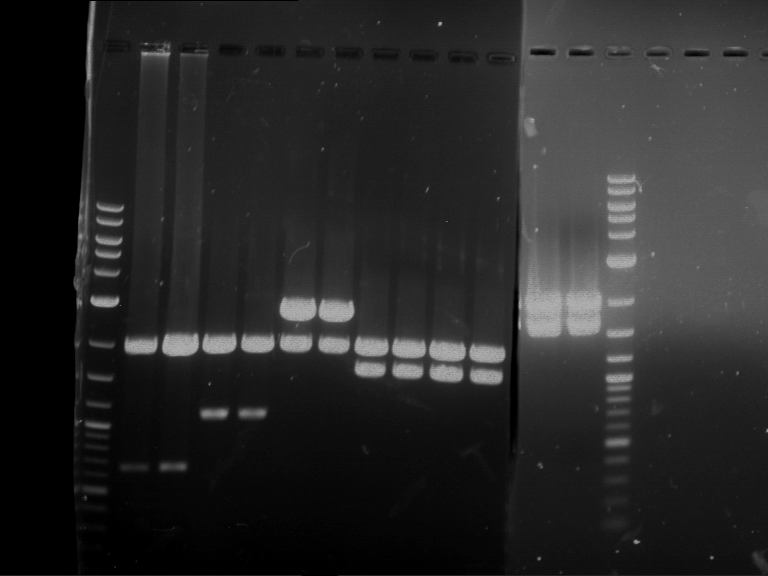
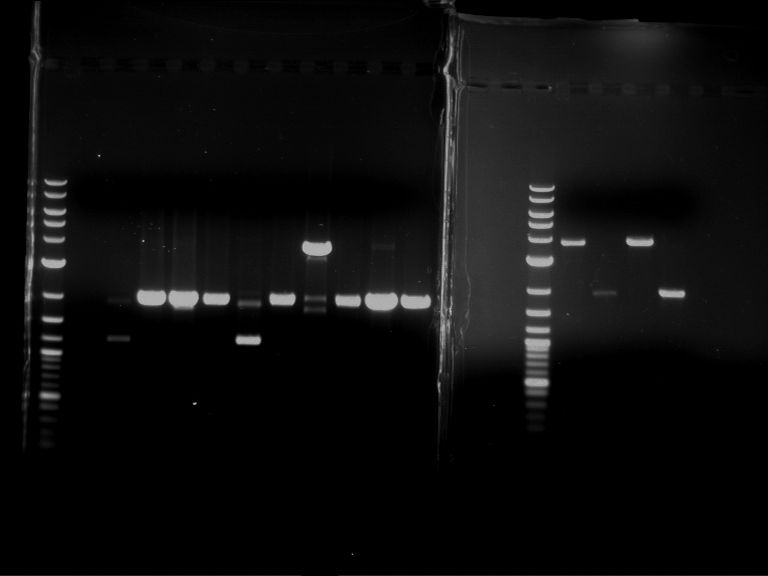
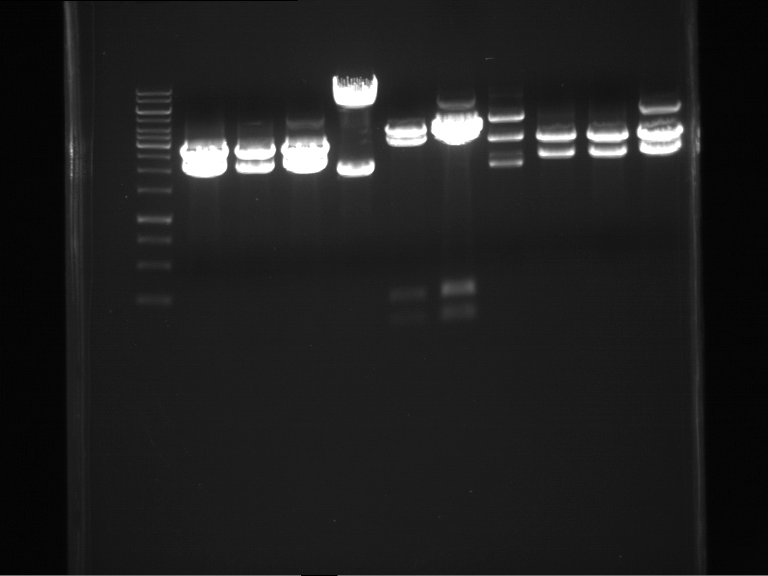
Analytical digestion of P15 and P19 to P52
Investigator: Florian, Johanna
Aim of the experiment: Analytical digestion and gelelectrophoresis of P15 and P19 to P52 to check the sequencing results.
Procedure:
- Batch for analytical digestion of P15 and P19 to P52
| volume | reagent |
| 5 µl | Plasmid DNA |
| 5 µl | Mastermix |
| =10 µl | TOTAL |
Batch of Mastermix
| volume | reagent |
| 40 µl | NEBuffer 4 (10x) |
| 4 µl | BSA (100x) |
| 0.2 µl | NotI(10 U/µl) |
| 148 µl | ddH2O |
| =200 µl | TOTAL |
- Incubation for 90 min at 37 °C.
- Analytical gelelectrophoresis was performed at 90 V for 60 min.
- Discussion:
- Gel 1, Row 1:
| lane | part | plasmid | band 1 | band 2 | result |
| 2 | EreA | pDEST14 | 3,25 kb | no cut, no restriction sites in pDEST14 | |
| 3 | mCherry(LMU) | pSB1C3 | 2,0 kb | 750 bp | probably right |
| 4 | GFP(LMU)(845 bp) | pSB1C3 | 2,0 kb | 750 bp | maybe switched with mKate2(LMU) |
| 5 | mKate2(LMU)(702 bp) | pSB1C3 | 2,0 kb | 900 bp | maybe switched with GFP(LMU) |
| 6 | 35S + Strong Plant + GFP(1775 bp) | pSB1C3 | 2,0 kb | 750 bp | ? |
| 7 | UVR8(1332 bp) | pSB1C3 | 2,0 kb | 1,1 kb | ? |
| 8 | mStrawberry(708 bp) | pSB1C3 | 2,0 kb | 1,4 kb | ? |
| 9 | DREB1C promoter(463 bp) | pSB1C3 | 2,5 kb | probably only one cut -> one NotI site mutated | |
| 10 | Laccase(1587 bp) | pSB1C3 | 2,0 kb | 1,3 kb | ? |
| 11 | SV40(419 bp) | pSB1C3 | 2,0 kb | 1,6 kb | this could be the laccase |
| 12 | cjBlue(696 bp) | pSB1C3 | 2,0 kb | 0,5 kb | ? |
- Gel 1, Row 2:
| lane | part | plasmid | band 1 | band 2 | result |
| 2 | alcA (843 bp) | pSB1C3 | 2,0 kb | 0,7 kb | ? |
| 3 | eforRed (681 bp) | pSB1C3 | plasmid | plasmid bands (coiled, supercoiled, etc.) | |
| 4 | amilGFP (699 bp) | pSB1C3 | 2,0 kb | 0,7 kb | right part |
| 5 | mCFP (714 bp) | pSB1C3 | 2,0 kb | 0,7 kb | right part |
| 6 | RD29A promoter (1535 bp) | pSB1C3 | 2,0 kb | 1,5 kb | right part |
| 7 | amilCP (669 bp) | pSB1C3 | 2,0 kb | 0,7 kb | right part |
| 8 | CMV promoter (580 bp) | pSB1C3 | 2,0 kb | 0,6 kb | right part |
| 9 | middle linker (24 bp) | pMA-BBFR | 2,4 kb | short insert went through the gel | |
| 10 | GSAT (108 bp) | pMA-BBFR | 2,4 kb | 0,1 kb | right part |
| 11 | GFP(720 bp) | pSB1A2 | ? | ||
| 12 | long linker (36 bp) | pMA-BBFR | 2,4 kb | short insert went through the gel |
- Gel 2:
| lane | part | plasmid | band 1 | band 2 | result |
| 2 | SEG linker (108 bp) | pMA-BBFR | 2,4 kb | short insert went through the gel | |
| 3 | short linker (12 bp) | pMA-BBFR | 2,4 kb | short insert went through the gel | |
| 4 | FluA (522 bp) | pMA-BBFR | 2,4 kb | 0,5 kb | right part |
| 5 | mCherry (769 bp) | pSB2K3 | 4,4 kb | 0,75 kb | right part |
| 6 | Cerulean (778 bp) | pSB2K3 | ? | ||
| 7 | mRFP1 (706 bp) | pSB2K3 | 4,4 kb | 0,7 kb | right part |
| 8 | mOrange (769 bp) | pSB2K3 | 4,4 kb | 0,75 kb | right part |
| 9 | PIF3-20aa (366 bp) | pSB1C3 | 2,0 kb | 0,35 kb | right part |
| 10 | 20aa-PIF3 (366 bp) | pSB1C3 | 2,0 kb | 0,35 kb | two plasmids and two insert can be seen |
| 11 | LexA (ca. 6 kb) | pSB1C3 | > 8 kb | only one cut, huge plasmid |
Picking of Plasmid P23 (CaMV35S) and Ligation Product F8 + F15 (TEV)
Investigator: Jeff, Florian
Aim of the study: Picking of Plasmid P23 (CaMV35S) and Ligation Product F8 + F15 (TEV) for Miniprep.
Operational sequence:
- Picking and overnight culture after standard laboratory's protocol. (CamR LB-medium)
- 1 colony of P23 were picked.
- 7 colonies of F8 + F15 were picked.
Sequencing of P63 & P64
Investigators: Louise
Aim of the experiment: Sequencing of genes for which no prior sequencing results were available.
Procedure: The DNA was prepared for sequencing according to the Eurofins SmartSeq Kit protocol (15 µl of plasmid DNA (50 - 100 ng) and 2 µl sequencing primer).
The different genes we sequenced received the following barcodes:
| Label | Name | Barcode1 | Barcode2 |
|---|---|---|---|
| p63 | EreA | FR01002349 | FR01002350 |
| p64 | EreA | FR01002351 | FR01002352 |
Wednesday, May 15th
Preparative digestion of psB1C3-RFP-generator (P60) and another psB1C3 (P8) as well as P64 (EreA in defect psB1C3 (F8)) with XbaI & AgeI
Investigator: Louise,Johanna
Aim of the experiment:Preparative digestion of psB1C3-RFP-generator (P60) and another psB1C3 (P8) as well as P64 (EreA in defect psB1C3 (F8)) with XbaI & AgeI to recover EreA and prepare two new vectors for cloning.
Procedure:
- Batch for preparative digestion of PCR product and EreB (F2) with XbaI & AgeI.
| volume | reagent |
| 20 µl | Plasmid (P8,P60,P64) |
| 4 µl | NEBuffer 4 |
| 0.4 µl | BSA (100x) |
| 1 µl | AgeI-HF |
| 1 µl | XbaI |
| 13.6 µl | ddH2O |
| =40 µl | TOTAL |
- Incubation for 2.5 h at 37 °C
- The digested fragments were labelled F17(P8), F18(P60), F19(P64)
PCR, purification and analytical geleletrophoresis of and EreB (P16)
Investigator: Jeff, Louise
Aim of the experiment: PCR of EreB (P16).
Procedure:
Operational sequence:
- PCR reaction mixture
| volume | reagent |
| 10 µl | 5x OneTaq Standard Reaction Buffer |
| 1 µl | 10 mM dNTPs |
| 1 µl | 10 µM Forward Primer (O31 (EreB_for)) |
| 1 µl | 10 µM Reverse Primer (O32 (EreB_rev)) |
| 0.25 µL | OneTaq Hot Start DNA Polymerase (Finally: 1.25 units/50 µL) |
| 1 µl | Plasmid DNA (P15; P16) |
| 35.75 µL | ddH2O Water |
| =50 µL | TOTAL |
- Mix with pipette
- The PCR program TMKS was performed after following scheme:
| Initial denaturation | 94 °C | 30 s |
| 30 cycles | 94 °C | 30 s |
| 52 °C | 60 s | |
| 68 °C | 80 s | |
| Final extension | 68 °C | 5 min |
| Hold | 4 °C | infinite |
- After PCR, the product was purified with QIAquick PCR Purification Kit, Qiagen.
- 4 µl of the purified PCR products were mixed with 0.4 µl DNA loading buffer (10x) for analytical gelelectrophoresis
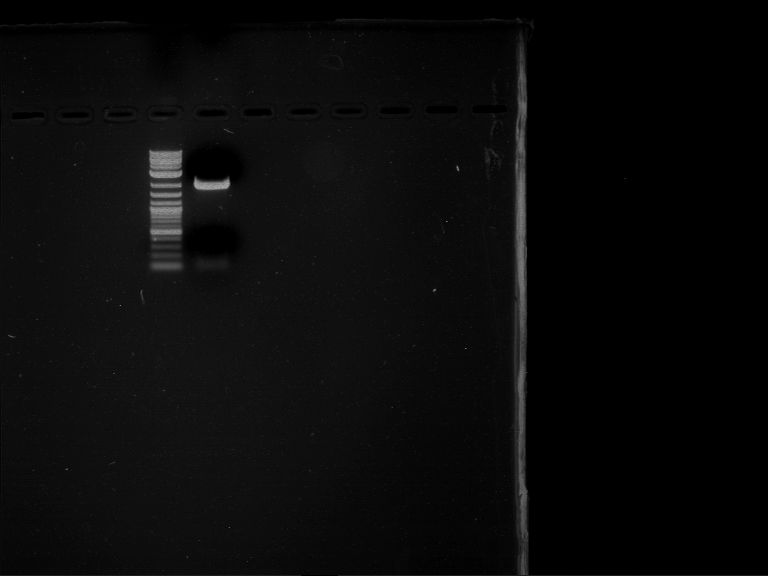
- Analytical gelelectrophoresis was performed at 90 V for 30 min in 1% agarose gel.
| 1 kbp ladder | F16 |
| Fragment-size like expected |
Purification and ligation of digested products F17, F18, F19
Investigator: Jeff, Louise
Aim of the experiment: Purification and ligation of digested products F17, F18, F19.
Procedure:
- To purify the digested Fragments the QIAquick PCR Purification Kit was used. The concentration of the purified product was found to be XX ng/µl via Nano drop.
Ligation of F19 + FXX(pSB1C3)
- Ligation batch for F19+FXX (positive control)
| volume | reagent |
| 1.35 µl | F8 (74.3 ng/µl, 2086 bp) |
| X µl | F14 (5 ng/µl, 1257 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- Negative control was also prepared. Instead of the insert, the same volume of ddH2O was added to the reaction batch.
- Cycled ligation has been performed after following protocol in a thermocycler:
| 99 cycles | 12 °C | 60 s |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| Hold | 16 °C | infinite |
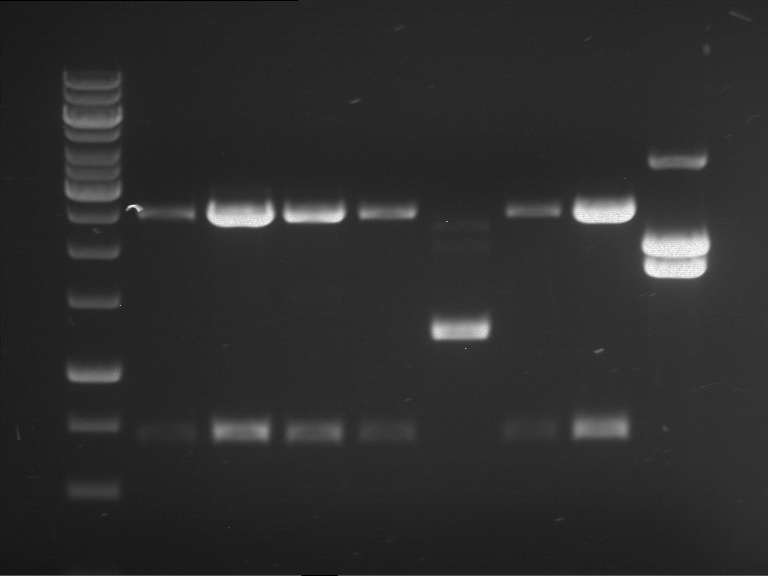
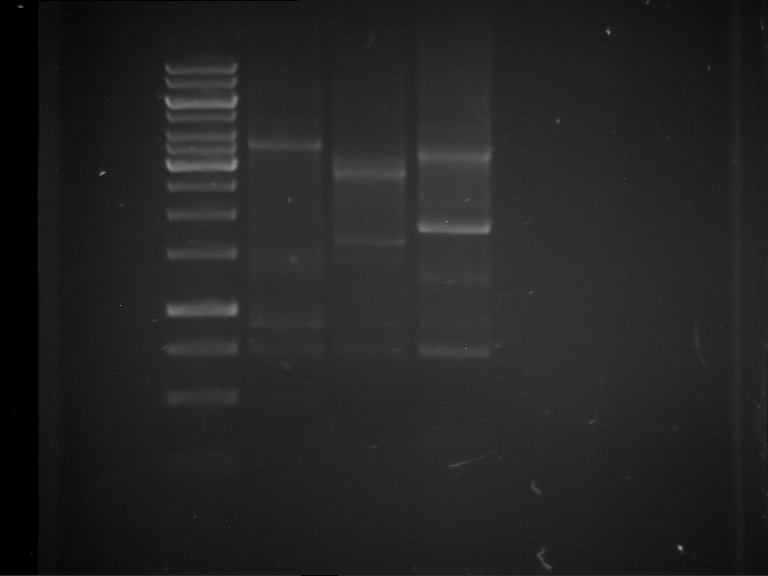
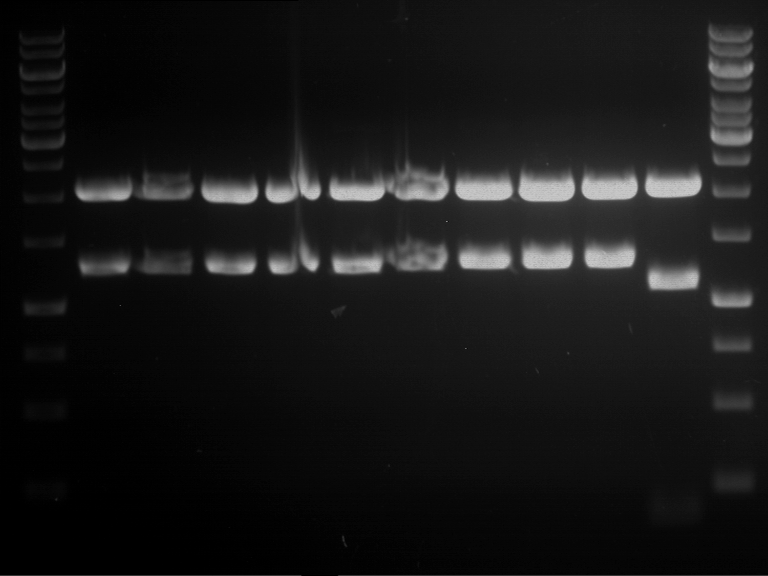
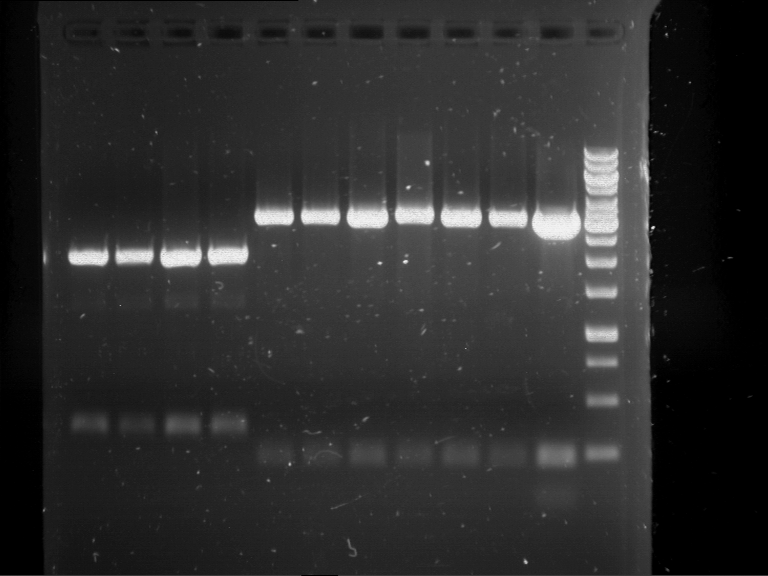
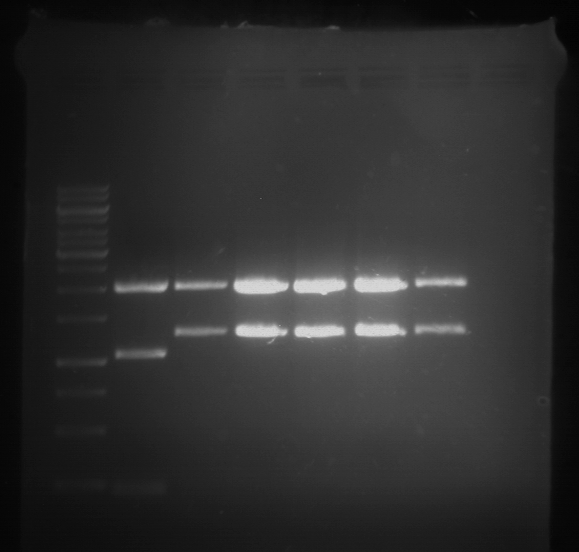
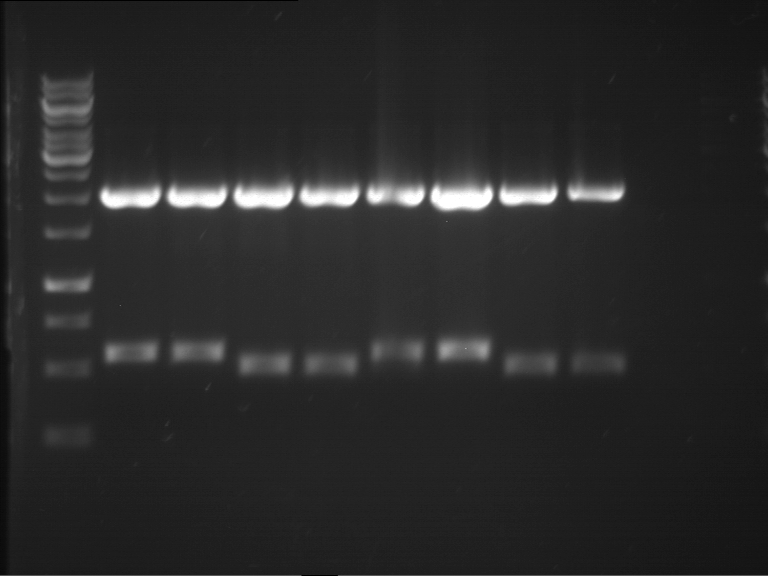
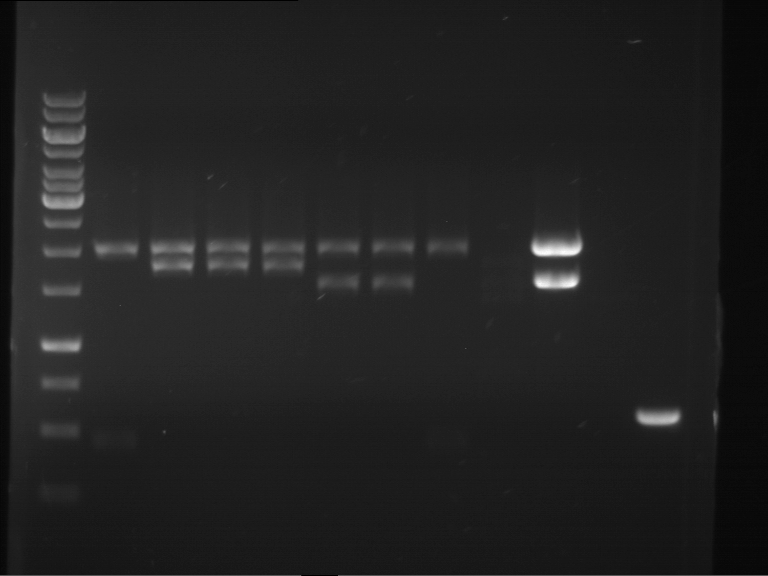
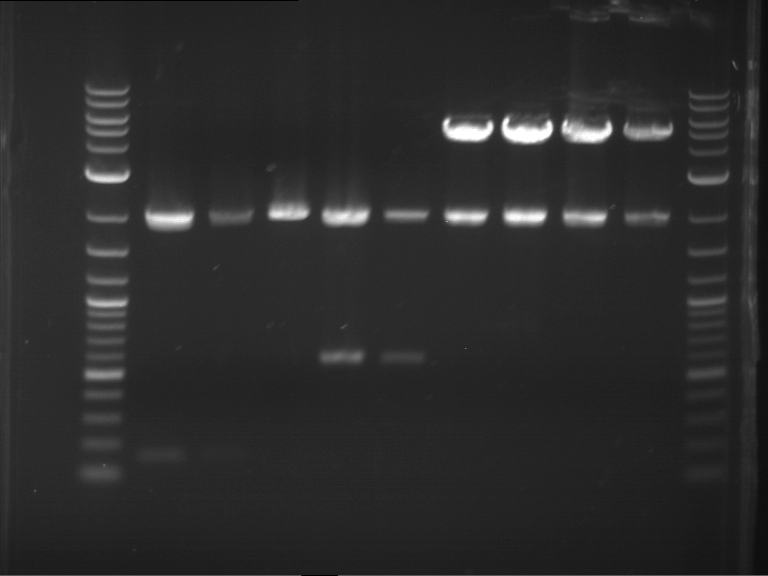
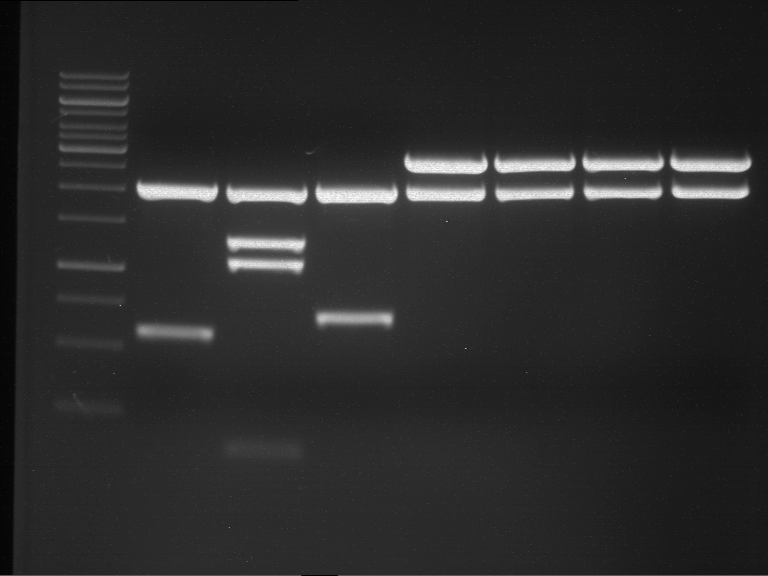
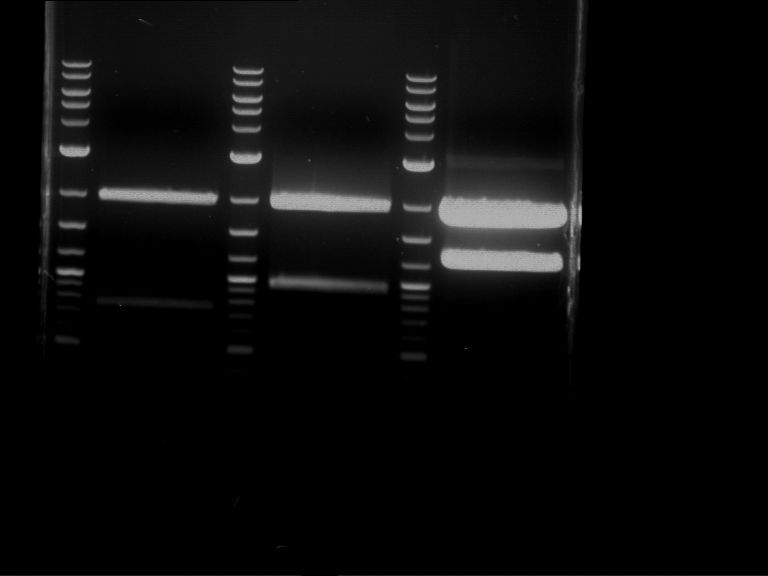
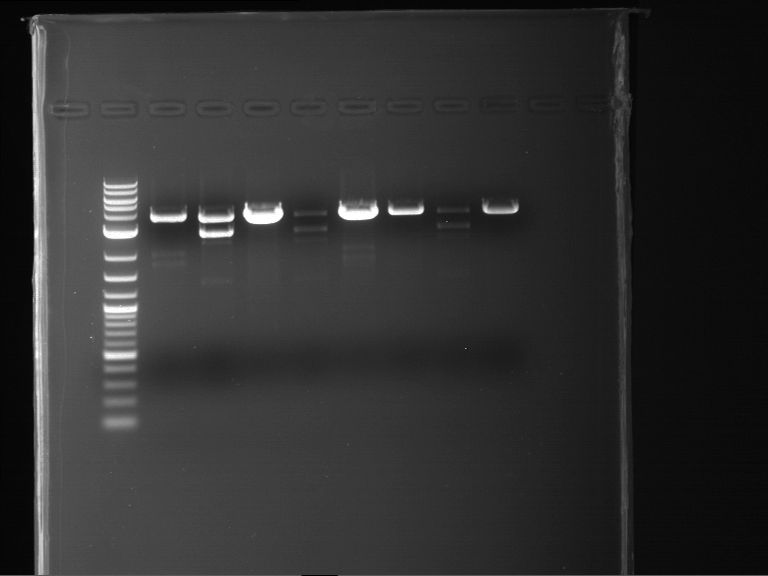
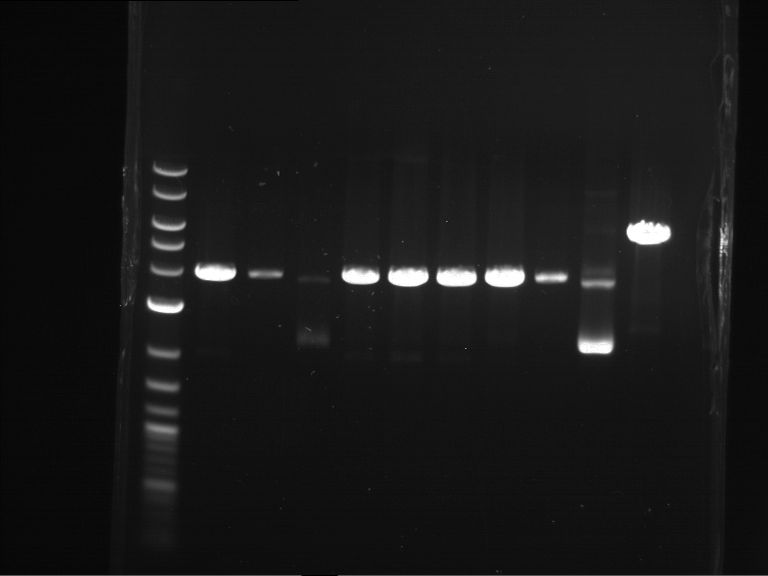
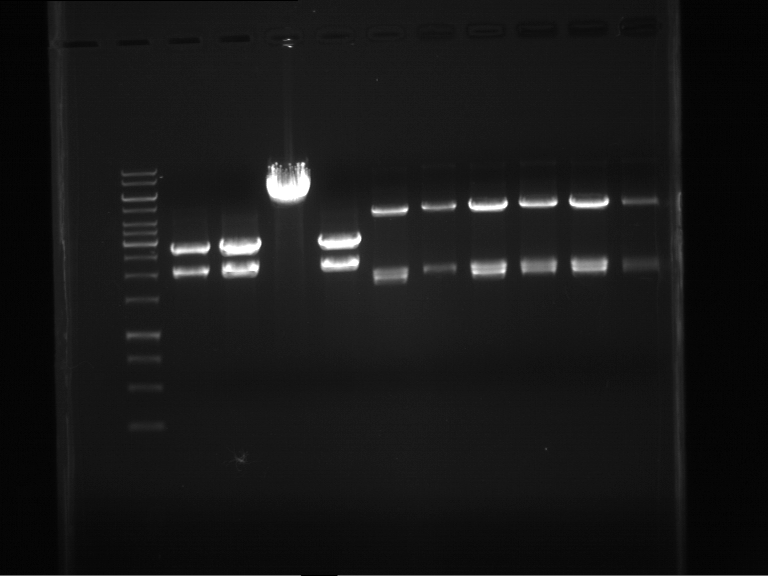
Analytical digestion and analytical gelelectrophoresis of P90-P97 (TEV protease, XbaI+AgeI) and P97 (CaMV p35S, XbaI+PstI)
Investigator: Jeff
Aim of the experiment: Analytical digestion and analytical gelelectrophoresis of P90-P97 (TEV protease, XbaI+AgeI) and P97 (CaMV p35S, XbaI+PstI).
Procedure:
- Mastermix for analytical digestion of P90-P96 with XbaI+AgeI-HF:
| volume | reagent |
| 16 µl | NEBuffer 4 (10x) |
| 1.6 µl | BSA (100x) |
| 2 µl | XbaI (20 U/µl) |
| 2 µl | AgeI-HF (20 U/µl) |
| 118.4 µl | ddH2O |
| =140 µl | TOTAL |
- Batch for digestion of P90-P96 with XbaI+AgeI-HF:
| volume | reagent |
| 2.5 µl | Plasmid DNA |
| 17.5 µl | Mastermix |
| =20 µl | TOTAL |
- Batch for digestion of P90-P96 with XbaI+AgeI-HF:
| volume | reagent |
| 2.5 µl | Plasmid DNA |
| 2 µl | NEBuffer 4 (10x) |
| 0.2 µl | BSA (100x) |
| 0.25 µl | XbaI (20 U/µl) |
| 0.25 µl | AgeI-HF (20 U/µl) |
| 14.8 µl | ddH2O |
| =20 µl | TOTAL |
- Incubation for 90 min at 37 °C.
- Analytical gelelectrophoresis was performed at 90 V for 60 min.
Results:
| 1 kbp ladder DNA ladder | Digestion of P90 | Digestion of P91 | Digestion of P92 | Digestion of P93 | Digestion of P94 | Digestion of P95 | Digestion of P96 | Digestion of P97 |
| Insert has expected length | Insert has expected length | Insert has expected length | Insert has expected length | Corrupt | Insert has expected length | Insert has expected length | Corrupt |
- The inserts for TEV semm to be right in the most cases, but the backbone has mutated and it should be cloned into a new pSB1C3 with new same restriction enzymes (XbaI+AgeI)!
- The forward sequencing of CaMV p35S is right but the analytical digestion is wrong. Maybe further parts are downstream of the promoter?! Reverse sequencing should be performed for further analysis.
Thursday, May 16th
Preparative digestion of pSH21 (P61), psB1A2(P41) and P96 (TEV S19V)
Investigator: Jeff, Katrin
Aim of the experiment: Preparative digestion of pSH21 (P61)and psB1A2(P41) with EcoRI and P96 (TEV S19V) with XbaI & AgeI for subsequent ligation.
Procedure:
- Batch for preparative digestion of pSH21 with EcoRI
| volume | reagent |
| 20 µl | Plasmid DNA P61 |
| 4 µl | Buffer 4(10x) |
| 1 µl | EcoRI (20 U/µl) |
| 15 µl | ddH2O |
| =40 µl | TOTAL |
- Batch for preparative digestion of psB1A2 with EcoRI
| volume | reagent |
| 20 µl | Plasmid DNA P41 |
| 4 µl | Buffer 4(10x) |
| 1 µl | EcoRI (20 U/µl) |
| 15 µl | ddH2O |
| =40 µl | TOTAL |
- Batch for preparative digestion of P96 (TEV S219V) with XbaI & AgeI.
| volume | reagent |
| 20 µl | Plasmid DNA P96 |
| 4 µl | Buffer 4(10x) |
| 1 µl | XbaI (20 U/µl) |
| 1 µl | AgeI-HF (20 U/µl) |
| 13.6 µl | ddH2O |
| =40 µl | TOTAL |
- Incubation for 3 h at 37 °C.
- The resulting fragments were named: F23 (P41) and F22 (P61)
- 4 µl of DNA loading buffer (10x) were added to the reaction batch containing pSH21 (P61) after digestion and were loaded on an 1% agarose gel for preparative gelelectrophoresis.
- Preparative gelelectrophoresis was performed at 70 V for 90 min.
| 1 kbp ladder | P61 digestion = F23 |
| Fragment-size like expected; band was cut out |
- The band had the expected length. They were cut out and purified using the QIAquick Gel Extraction Kit, QIAGEN
Preparative digestion of ereB (P64) with XbaI & AgeI.
Investigator: Jeff, Louise, Katrin
Aim of the experiment:Preparative digestion of EreB (P64) with XbaI & AgeI.
Procedure:
- Batch for preparative digestion of PCR product and EreB (F2) with XbaI & AgeI.
| volume | reagent |
| 20 µl | Plasmid |
| 4 µl | NEBuffer 4 |
| 0.4 µl | BSA (100x) |
| 1 µl | AgeI-HF |
| 1 µl | XbaI |
| 13.6 µl | ddH2O |
| =40 µl | TOTAL |
- Incubation for 2.5 h at 37 °C
- The digested fragments were labelled
Dephosphorylation of F23
Investigator: Jeff, Katrin
Aim of the experiment: Dephosphorylation of cut vector psB1A2 to prevent re-ligation
Procedure:
- the digestion product was purified with QIAquick PCR Purification Kit, Qiagen in order to change the buffer
- Batch for the dephosphorylation of F23
| volume | reagent |
| 30 µl | Plasmid DNA F23 (2 µg) |
| 4 µl | 10X Fast AP buffer |
| 2 µl | FastAP Thermosensitive Alkaline
Phosphatase |
| 4 µl | ddH2O |
| =40 µl | TOTAL |
- the reaction batch was spun briefly and incubated at 37 °C for 10 minutes
- the reaction was stopped by heating to 75 °C for 5 minutes
Ligation of pSH21 (F22) with psB1A2(F23), TEV (P96), EreA (P64), pSB1A2(P41)
Investigator: Jeff, Katrin
Aim of the experiment: ligation of pSH21 (F22) with pSB1A2(F23), TEV (F20) with pSB1C3 (F17), EreA (F19)with pSB1C3 (F17) and EreB (F21) with pSB1C3 (F17)
Procedure:
- Ligation batch for EreB (F21) with pSB1C3 (F17)
| volume | reagent |
| 1.6 µl | F17 (61,4 ng/µl) |
| 15,4 µl | F21 (8,3 ng/µl) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- Ligation batch for TEV (F20) with pSB1C3 (F17)
| volume | reagent |
| 1.6 µl | F17 (61,4 ng/µl) |
| 4,1 µl | F20 (24,7 ng/µl) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 11,3 µl | ddH20 |
| =20 µl | TOTAL |
- Ligation batch for pSH21 (F22) with pSB1A2(F23)
| volume | reagent |
| 0,85 µl | F23 (118,1 ng/µl) |
| 2,3 µl | F22 (119,0 ng/µl) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 13,85 µl | ddH20 |
| =20 µl | TOTAL |
- Ligation batch for EreA (F19) with pSB1C3 (F17)
| volume | reagent |
| 1.6 µl | F17 (61,4 ng/µl) |
| 6,4 µl | F19 (8,3 ng/µl) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 9 µl | ddH20 |
| =20 µl | TOTAL |
- Negative control was also prepared. Instead of the insert, the same volume of ddH2O was added to the reaction batch.
- the ligation was performed at 16 °C overnight
Friday, May 17th
Transformation of E. coli XL1 blue with F17+F19, F17+F20, F17+F21, F22+F23, F17 (negative control), F23 (negative control)
Investigator: Jeff, Katrin
Aim of the experiment: Transformation of ligation products F17+F21 (pSB1C3, EreB), F17+F20 (pSB1C3, TEV), F17+F19 (pSB1C3, EreA), F23+F22 (pSB1A2, pSH21) and negative controls
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 5 µl of DNA was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of the cell suspension was plated on chloramphenicol plates (F17+F19, F17+F20, F17+F21, F17 negative) or ampicillin plates (F23+F22, F23 negative control)
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was discarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on chloramphenicol plates (F17+F19, F17+F20, F17+F21) or ampicillin plates (F23+F22)
Quick Change mutagenesis to remove mutations found by sequencing in our gene constructs
Investigator: Jeff
Aim of the experiment: removal of forbidden restiction sites from laccase
Operational sequence
PCR general setup
| volume | reagent |
| 5 µl | 10x Pfu Ultra II buffer |
| 1 µl | DNA template (p28) |
| 0,5 µl each | 1:10 dilution of appropriate Oligos (1: O26 + O27, 2: O28 + O29, 3: O30 + O31, 4: O32 + O33 ) |
| 42 µl | ddH2O |
| 1 µl | dNTP mix |
| 1 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
PCR cycling parameters
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 30 sec |
| 2 | 12 | 95 °C | 30 s |
| 55 °C | 1 min | ||
| 68 °C | 4 min | ||
| 3 | 1 | 4 °C | hold |
Transformation of E. coli XL1 blue with the 4 Quickchangeproducts using 1: O26 + O27, 2: O28 + O29, 3: O30 + O31, 4: O32 + O33
Investigator: Jeff, Andi, Johanna
Aim of the experiment: Transformation of E. coli XL1 blue with the 4 Quickchangeproducts using 1: O26 + O27, 2: O28 + O29, 3: O30 + O31, 4: O32 + O33
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 5 µl of DNA was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of the cell suspension was plated on chloramphenicol plates 1: O26 + O27, 2: O28 + O29, 3: O30 + O31, 4: O32 + O33
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was discarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on chloramphenicol plates (1: O26 + O27, 2: O28 + O29, 3: O30 + O31, 4: O32 + O33)
Saturday, May 18th
Picking of transformed Plasmid E. coli XL1 blue with F17+F19, F17+F20, F17+F21, F22+F23
Investigator: Andi, Jeff
Aim of the study: Picking of transformed Plasmid F17+F19, F17+F20, F17+F21, F22+F23 .
Operational sequence:
- Picking and overnight culture after standard laboratory's protocol. (CamR/AmpR LB-medium)
- 2 colonies were picked for every Transformation product.
Sunday, May 19th
Miniprep of overnight culture of transformated E. coli XL1 blue with F17+F19, F17+F20, F17+F21, F22+F23
Investigator: Jeff, Andi
Aim of the experiment: Miniprep of overnight culture of transformated E. coli XL1 blue with F17+F19, F17+F20, F17+F21, F22+F23 of 2 clones picked the day before
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
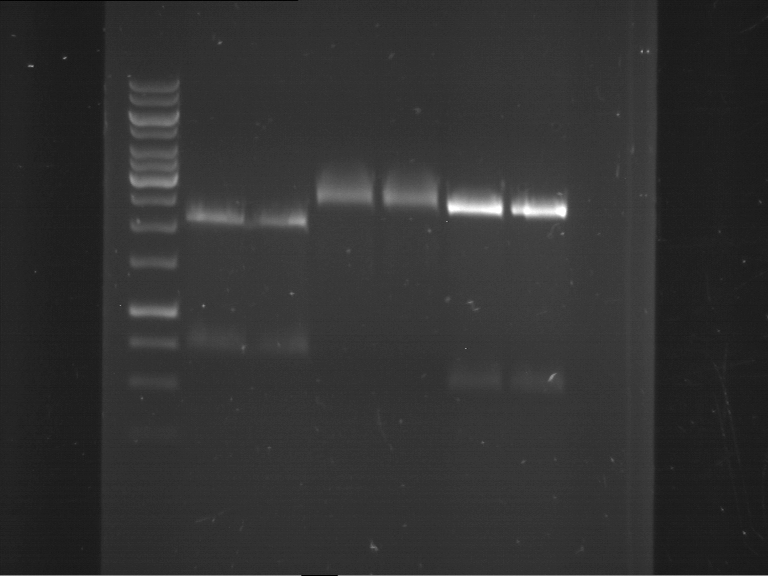
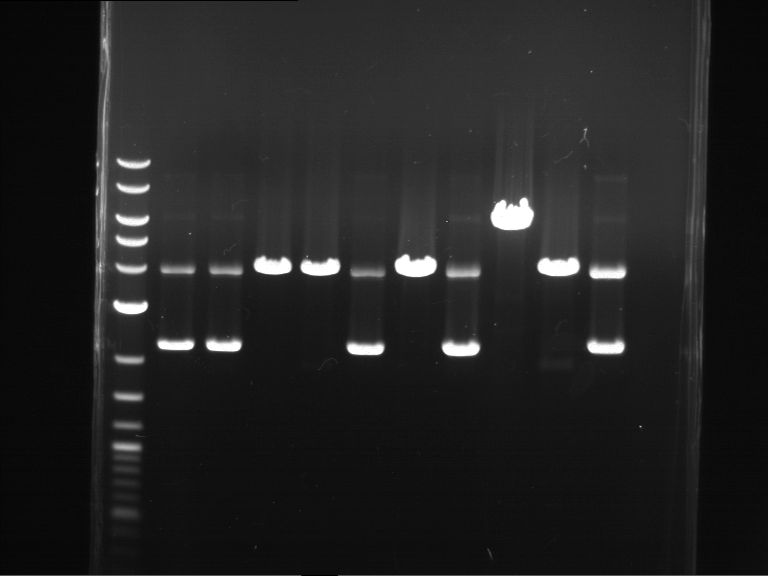
Analytical digestion and analytical gelelectrophoresis of F17+F19, F17+F20, F17+F21 and F22+F23
Investigator: Jeff, Andi
Aim of the experiment: Analytical digestion and analytical gelelectrophoresis of ligation products F17+F19, F17+F20, F17+F21, F22+F23.
Procedure:
- Mastermix for analytical digestion of F17+F19, F17+F20, F17+F21 with XbaI+PstI-HF:
| volume | reagent |
| 14 µl | NEBuffer 4 (10x) |
| 1.4 µl | BSA (100x) |
| 1.75 µl | XbaI (20 U/µl) |
| 1.75 µl | AgeI-HF (20 U/µl) |
| 103.6 µl | ddH2O |
| =122.5 µl | TOTAL |
- Batch for digestion of F22+F23 with EcoRI-HF:
| volume | reagent |
| 2.5 µl | Plasmid DNA |
| 2 µl | NEBuffer 4 (10x) |
| 15.25 µl | ddH2O |
| 0.25 µl | EcoRI-HF (20 U/µl) |
| =20 µl | TOTAL |
- Incubation for 90 min at 37 °C.
- Analytical gelelectrophoresis was performed two times for each ligation product at 90 V for 60 min.
Results:
| 1 kbp ladder DNA ladder | Digestion of F17+F19 | Digestion of F17+F19 | Digestion of F17+F20 | Digestion of F17+F20 | Digestion of F17+F21 | Digestion of F17+F21 | Digestion of F22+F23 | Digestion of F22+F23 |
| Insert has expected length | Insert has expected length, the first line represents the rest of the uncut Plasmid | Insert has expected length | Insert has expected length, the first line represents the rest of the uncut Plasmid | Insert has expected length | Insert has expected length | Insert has expected length, insert and cut Plasmid overlay each other, because of the same length | Corrupt |
Quick Change mutagenesis to remove forbidden restriction sites of P28 (Laccase), P100 (TEV S219V), P102 (EreB)
Investigator: Jeff, Andi
Aim of the experiment: Removal of forbidden restiction sites from P28 (Laccase), P100 (TEV S219V), P102 (EreB).
Operational sequence
PCR general setup
Procedure:
PCR
Reaction batch 1
| volume | reagent |
| 2.5 µl | 10x Pfu Ultra II buffer |
| 4 µl | Plasmid template (P28/P100/P102) |
| 1 µl | 1:10 dilution of O26 (for P28, 10 µM)/O16 (for P102, 10 µM)/O36 (for P100, 10 µM) |
| 16.5 µl | ddH2O |
| 0.5 µl | dNTP mix |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
Reaction batch 2
| volume | reagent |
| 2.5 µl | 10x Pfu Ultra II buffer |
| 4 µl | Plasmid P38 template |
| 1 µl | 1:10 dilution of O27 (for P28, 10 µM)/O17 (for P102, 10 µM)/O37 (for P100, 10 µM) |
| 16.5 µl | ddH2O |
| 0.5 µl | dNTP mix |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
PCR cycling parameters
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 30 s |
| 2 | 10 | 95 °C | 30 s |
| 55 °C | 1 min | ||
| 68 °C | 4 min |
- Having completed the PCR cycling parameters listed above both PCR reaction batches were mixed together and the cycling parameters listed above were one time more applied, but the amount of cycles have been increased to 20.
- Digestion of the parental DNA with DpnI: Addition of 1 µl DpnI to the reaction batch and incubate for 1 h at 37 °C.
Analytical gelelectrophoresis of QuickChange products of P100, P102 and P28 to check whether the QuickChange PCR and DpnI digestion were successful
Investigator:
Aim of the experiment:
Procedure:
Transformation of the Quickchange products with P28, P100 and P102 into E. coli XL1 blue
Investigator: Jeff, Andi
Aim of the experiment: Transformation of E. coli XL1 blue with P28, P102 and P100.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 2 µl of DNA was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of the cell suspension was plated on one chloramphenicol plate.
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on a new chlorampenicol plate.
Week 5
Tuesday, May 21st
Quick Change mutagenesis to remove forbidden restriction sites - SDMI - of P28 (Laccase), P100 (TEV S219V), P102 (EreB)
Investigator: Jeff, Andi, Johanna
Aim of the experiment: Removal of forbidden restiction sites from P28 (Laccase), P100 (TEV S219V), P102 (EreB).
Operational sequence
PCR general setup
Procedure:
PCR
Reaction batch 1
| volume | reagent |
| 2.5 µl | 10x Pfu Ultra II buffer |
| 4 µl | Plasmid template (P28/P100/P102) |
| 1 µl | 1:10 dilution of O26 (for P28, 10 µM)/O16 (for P102, 10 µM)/O36 (for P100, 10 µM) |
| 16.5 µl | ddH2O |
| 0.5 µl | dNTP mix |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
Reaction batch 2
| volume | reagent |
| 2.5 µl | 10x Pfu Ultra II buffer |
| 4 µl | Plasmid P38 template |
| 1 µl | 1:10 dilution of O27 (for P28, 10 µM)/O17 (for P102, 10 µM)/O37 (for P100, 10 µM) |
| 16.5 µl | ddH2O |
| 0.5 µl | dNTP mix |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
PCR cycling parameters
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 30 s |
| 2 | 10 | 95 °C | 30 s |
| 55 °C | 1 min | ||
| 68 °C | 4 min |
- Having completed the PCR cycling parameters listed above both PCR reaction batches were mixed together and the cycling parameters listed above were one time more applied, but the amount of cycles have been increased to 20.
- Digestion of the parental DNA with DpnI: Addition of 1 µl DpnI to the reaction batch and incubate for 1 h at 37 °C.
Analytical gelelectrophoresis of QuickChange products of P100, P102 and P28 to check whether the QuickChange PCR and DpnI digestion were successful
Investigator:
Aim of the experiment:
Procedure:
Transformation of the Quickchange products with P28, P100 and P102 into E. coli XL1 blue
Investigator: Jeff, Andi, Johanna
Aim of the experiment: Transformation of E. coli XL1 blue with P28, P102 and P100.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 2 µl of DNA was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of the cell suspension was plated on one chloramphenicol plate.
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on a new chlorampenicol plate.
Sequencing of P105, P28, P100, P102 & P98
Investigators: Jeff, Andi,Johanna
Aim of the experiment: Sequencing of genes for which no prior sequencing results were available.
Procedure: The DNA was prepared for sequencing according to the Eurofins SmartSeq Kit protocol (15 µl of plasmid DNA (50 - 100 ng) and 2 µl sequencing primer).
The different genes we sequenced received the following barcodes:
| Label | Name | Barcode1 | Barcode2 |
|---|---|---|---|
| p105 | IRES | FR01002288 | FR01002269 |
| p102 | EreB | FR01002270 | FR01002275 |
| p100 | TEV S219V | FR01002278 | FR01002279 |
| p98 | EreA | FR01002276 | FR01002277 |
| p28 | Laccase | FR01002280 |
Preparation of Knop-Medium for Moss
Investigator: Jeff, Andi, Johanna, Rosario
Aim of the experiment: Preparation of Knop-Medium for Moss
Procedure:
- All four prepared stocks have been: 25g/L KH2PO4; 25g/L KCl; 25g/L MgSO4 x 7 H2O; 100g/L Ca(NO3)2
- Volume of prepared Medium: 1L
- 10 ml of each stock were converted into a 1,8L Erlenmeyer flask and filled up to 1L with destilled water (ELGA); 12,5mg FeSO4 x 7 H2O were added; the pH was set to 5.8 with KOH
- The prepared Volume of 1L was shared equally to 500ml Erlenmeyer flasks, so each Erlenmeyer flask contained 200 ml of the Knop-Medium
- All flasks have been autoclaved
Picking of transformed Plasmid E. coli XL1 blue with QuickChange products of P100 (TEV, QC with O36/O37) and P102 (EreB, QC with O16/O17)
Investigator: Andi, Jeff, Johanna, Rosario
Aim of the study: Picking of transformed Plasmid E. coli XL1 blue with QuickChange products of P100 (TEV, QC with O36/O37) and P102 (EreB, QC with O16/O17) from Sunday, May 19th.
Operational sequence:
- Picking and overnight culture after standard laboratory's protocol. (CamR LB-medium)
- 3 colonies for P100QC (TEV, QC with O36/O37) and 5 colonies from P102QC (EreB, QC with O16/O17) were picked from the plates.
Wednesday, May 22nd
Miniprep of overnight culture of transformated E. coli XL1 blue with QuickChange products of P100 (TEV, QC with O36/O37) and P102 (EreB, QC with O16/O17)
Investigator: Jeff
Aim of the experiment: Miniprep of overnight culture of transformated E. coli XL1 blue with QuickChange products of P100 (TEV, QC with O36/O37) and P102 (EreB, QC with O16/O17).
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
Picking of transformed Plasmid E. coli XL1 blue with QuickChange products of P28 (Laccase, QC with O26/O27)
Investigator: Jeff
Aim of the study: Picking of transformed Plasmid E. coli XL1 blue with QuickChange products of P28 (Laccase, QC with O26/O27).
Operational sequence:
- Picking and overnight culture after standard laboratory's protocol. (CamR LB-medium)
- 8 colonies for P28QC (Laccase, QC with O26/O27) were picked from the plates.
Analytical digestion and analytical gelelectrophoresis of P106, P107, P108, P109, P110, P111, P112, P113
Investigator: Jeff, Rosario
Aim of the experiment: Analytical digestion and analytical gelelectrophoresis of Quickchanges to check whether the transformation of E. coli Xl1 Blue with QuickChanges products of P100 (TEV, QC with O36/O37: P111, P112, P113) and P102 (EreB, QC with O16/O17: P106, P107, P108, P109, P110) were successful
Procedure:
Mastermix for analytical digestion of P106, P107, P108, P109, P110, P111, P112, P113 with AgeI-HF and XbaI:
| volume | reagent |
| 20 µl | NEBuffer 4 (10x) |
| 2 µl | BSA (100x) |
| 2.5 µl | AgeI-HF (20 U/µl) |
| 2.5 µl | XbaI |
| 148 µl | ddH2O |
| = 175 µl | TOTAL |
- 17,5 µl Mastermix with 2,5 µl Plasmid
Mastermix for analytical digestion of P111, P112, P113 with XbaI and SpeI and P106, P107, P108, P109, P110 with XbaI and PstI-HF:
| volume | reagent |
| 20 µl | NEBuffer 4 (10x) |
| 2 µl | BSA (100x) |
| 2.5 µl | XbaI |
| 148 µl | ddH2O |
| = 172,5 µl | TOTAL |
- 17,25 µl Mastermix with 2,5 µl Plasmid and 0,25 µl SpeI or PstI respectively
Mastermix for analytical digestion of with P106, P107, P108, P109, P110 EcoRI and SpeI:
| volume | reagent |
| 12 µl | NEBuffer 4 (10x) |
| 1,2 µl | BSA (100x) |
| 1,5 µl | EcoRI-HF |
| 1,5 µl | SpeI-HF |
| 88.8 µl | ddH2O |
| = 105 µl | TOTAL |
- 17,5 µl Mastermix with 2,5 µl Plasmid
Mastermix for analytical digestion of with P106, P107, P108, P109, P110 XbaI and SpeI:
| volume | reagent |
| 12 µl | NEBuffer 4 (10x) |
| 1,2 µl | BSA (100x) |
| 1,5 µl | XbaI |
| 1,5 µl | SpeI-HF |
| 88.8 µl | ddH2O |
| = 105 µl | TOTAL |
- 17,5 µl Mastermix with 2,5 µl Plasmid
- Incubation for 90 min at 37 °C.
- Analytical gelelectrophoresis was performed at 90 V for 60 min.
Results:
| 1 kbp ladder DNA ladder | P111 XbaI + AgeI | P112 XbaI + AgeI | P113 XbaI + AgeI | P111 XbaI + SpeI | P112 XbaI + SpeI | P113 XbaI + SpeI |
| Ctrl | Ctrl | Ctrl | QC succesful | QC failed | QC succesful |
| 1 kbp ladder DNA ladder | P106 XbaI + AgeI | P107 XbaI + AgeI | P108 XbaI + AgeI | P109 XbaI + AgeI | P110 XbaI + AgeI | P106 XbaI + PstI | P107 XbaI + PstI | P108 XbaI + PstI | P109 XbaI + PstI | P110 XbaI + PstI |
| Ctrl | Ctrl | Ctrl | Ctrl | Ctrl | PstI inside as should be; useless experiment | PstI inside as should be; useless experiment | PstI inside as should be; useless experiment | PstI inside as should be; useless experiment | PstI inside as should be; useless experiment |
| 1 kbp ladder DNA ladder | P106 XbaI + SpeI | P107 XbaI + SpeI | P108 XbaI + SpeI | P109 XbaI + SpeI | P110 XbaI + SpeI | P106 EcoRI + SpeI | P107 EcoRI + SpeI | P108 EcoRI + SpeI | P109 EcoRI + SpeI | P110 EcoRI + SpeI |
| Ctrl | Ctrl | Ctrl | Ctrl | Ctrl | QC succesful | QC succesful | QC succesful | QC succesful | QC failed |
Thursday, May 23rd
Sequencing of P111, P113 and P109 (?)
Investigator: Jeff
Aim of the experiment: Sequencing of P111, P113 and P109 to check whether the QuickChange brought further unwanted mutations.
Procedure:
PCR of P111 (TEV S219V, SpeI-less) to generate Split-TEV
Investigator: Jeff
Aim of the experiment: PCR of P111 (TEV S219V, SpeI-less) to generate Split-TEV. To generate N-TEV, the primer O38 and O39 were used. To generate C-TEV, the primer O40 and O41 were used.
Procedure:
Operational sequence:
- PCR reaction mixture
| volume | reagent |
| 10 µl | 5x OneTaq Standard Reaction Buffer |
| 1 µl | 10 mM dNTPs |
| 1 µl | 10 µM Forward Primer (O38 (N_Split_TEV_fw) for generating N-TEV; O40 (C_Split_TEV_fw) for generating C-TEV) |
| 1 µl | 10 µM Reverse Primer (O39 (N_Split_TEV_rv) for generating N-TEV; O41 (C_Split_TEV_rv) for generating C-TEV) |
| 0.25 µL | OneTaq Hot Start DNA Polymerase (Finally: 1.25 units/50 µL) |
| 1 µl | Plasmid DNA (P111) |
| 35.75 µL | ddH2O Water |
| =50 µL | TOTAL |
- Mix with pipette
- The PCR program was performed after following scheme with following conditions:
| Initial denaturation | 94 °C | 30 s |
| 30 cycles | 94 °C | 30 s |
| 51 °C | 60 s | |
| 68 °C | 30 s | |
| Final extension | 68 °C | 5 min |
| Hold | 4 °C | infinite |
- After PCR, the product was purified with QIAquick PCR Purification Kit, Qiagen.
- Resulting product was labeled as F24 (N-TEV) and F25 (C-TEV).
Analytical gelelectrophoresis of F24 and F25 (N-TEV and C-TEV)
Investigator: Jeff, Rosario
Aim of the experiment: Analytical gelelectrophoresis of F24 and F25 (N-TEV and C-TEV) to check whether PCR has worked.
Procedure:
- 4 µl of F24 and F25 were mixed with seperately with 0.44 µl of DNA loading buffer (10x) and analytical gelelectrophoresis was performed at 1% agarose gel for 60 min at 90 V.
- PCR products has expected length, but there are unspecific products. So we have to be cautious and have to check if we ligate the right products.
Preparative digestion of PCR products (N-TEV and C-TEV) of P111 (TEV S219V, SpeI-less) with XbaI & AgeI
Investigator: Jeff, Rosario
Aim of the experiment: Preparative digestion of PCR products (N-TEV and C-TEV) of P111 (TEV S219V, SpeI-less) with XbaI & AgeI to clone it into pSB1C3.
Procedure:
- Batch for preparative digestion of PCR products (N-TEV and C-TEV) of P111 (TEV S219V, SpeI-less) with XbaI & AgeI.
| volume | reagent |
| 25 µl | PCR product F24/F25 |
| 5 µl | NEBuffer 4 |
| 0.5 µl | BSA (100x) |
| 1 µl | AgeI-HF |
| 1 µl | XbaI |
| 17.5 µl | ddH2O |
| =50 µl | TOTAL |
- Incubation for 150 min at 37 °C.
- Digested products were purified with QIAquick PCR Purification Kit, QIAGEN.
- The digested fragments were labelled F26 (digestion of F24 with XbaI&AgeI) and F27 (digestion of F25 with XbaI&AgeI)
Ligation of F17+F26 (N-TEV in pSB1C3) and F17+F27 (C-TEV in pSB1C3)
Investigator: Jeff, Rosario
Aim of the experiment: Ligation of F17+F26 (N-TEV in pSB1C3) and F17+F27 (C-TEV in pSB1C3).
Procedure:
- Ligation batch for F17+F26:
| volume | reagent |
| 1.63 µl | F17 (61.4 ng/µl, 2086 bp) |
| 3.20 µl | F26 (15.9 ng/µl, 354 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 12.17 µl | ddH2O |
| =20 µl | TOTAL |
- Ligation batch for F17+F27 (positive control)
| volume | reagent |
| 1.63 µl | F17 (61.4 ng/µl, 2086 bp) |
| 2.41 µl | F27 (21.1 ng/µl, 354 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 12.96 µl | ddH2O |
| =20 µl | TOTAL |
- Negative control was also prepared. Instead of the insert, the same volume of ddH2O was added to the reaction batch.
- Cycled ligation has been performed after following protocol in a thermocycler:
| 99 cycles | 12 °C | 60 s |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| Hold | 16 °C | infinite |
- Lid temperature = 37 °C
Miniprep of overnight culture of transformated E. coli XL1 blue with the first Quickchange (SDMI) Product of P28
Investigator: Jeff, Andi, Rosario, Johanna
Aim of the experiment: Miniprep of overnight culture of transformated E. coli XL1 blue with the first Quickchange Product of P28. 8 clones were picked the day before.
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
Analytical digestion and analytical gelelectrophoresis of P28, P117, P118, P119, P120, P121, P122, P123
Investigator: Rosario, Andi, Johanna, Jeff
Aim of the experiment: Analytical digestion and analytical gelelectrophoresis of P28 (promoter SV40) and quickchange products P117, P118, P119, P120, P121, P122, P123 (all QC - SDMI - of Bpul Laccase to remove forbidden AgeI).
Procedure:
- Mastermix for analytical digestion of P28, P117, P118, P119, P120, P121, P122, P123 AgeI-HF:
| volume | reagent |
| 20 µl | NEBuffer 4 (10x) |
| 2 µl | BSA (100x) |
| 2.5 µl | AgeI-HF (20 U/µl) |
| 150.5 µl | ddH2O |
| =175 µl | TOTAL |
- Incubation for 90 min at 37 °C.
- Analytical gelelectrophoresis was performed two times for each ligation product at 90 V for 60 min.
Results:
| 1 kbp ladder DNA ladder | Digestion of P28 | Digestion of P116 | Digestion of P117 | Digestion of P118 | Digestion of P119 | Digestion of P120 | Digestion of P121 | Digestion of P122 | Digestion of P123 |
| as expected | QC successful | QC successful | QC successful | QC successful | QC successful | QC successful | QC successful | QC successful |
Quick Change mutagenesis to remove forbidden restriction sites - SDMII - of P28 (Laccase), P106 (EreB)
Investigator: Jeff, Andi, Johanna, Rosario
Aim of the experiment: Removal of forbidden restiction sites from P28 (Laccase), P106 (EreB).
Procedure: Quickchange - PCR
Reaction batch - P106
| volume | reagent | Additional information |
| 5 µl | 10x Pfu Ultra II buffer | |
| 2 µl | Plasmid template (P106 - 1:10 dilution) | need to get to 50 ng/µl |
| 1.25 µl | 1:10 dilution of O18 (for P106, 10 µM) | |
| 1.25 µl | 1:10 dilution of O19 (for P106, 10 µM) | |
| 38.5 µl | ddH2O | |
| 1 µl | dNTP mix | |
| 1 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
PCR cycling parameters - P106
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 30 s |
| 2 | 18 | 95 °C | 30 s |
| 52 °C | 1 min | ||
| 68 °C | 4 min |
- Digestion of the parental DNA with DpnI: Addition of 1 µl DpnI to the reaction batch and incubate for 1 h at 37 °C.
Reaction batch - P116 (Laccase)
| volume | reagent | Additional information |
| 5 µl | 10x Pfu Ultra II buffer | |
| 2 µl | Plasmid template - 1:5 dilution | need to get to 50 ng/µl |
| 1.25 µl | 1:10 dilution of O28 for P116 | |
| 1.25 µl | 1:10 dilution of O29 for P116 | |
| 38.5 µl | ddH2O | |
| 1 µl | dNTP mix | |
| 1 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
PCR cycling parameters - P116
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 30 s |
| 2 | 18 | 95 °C | 30 s |
| 52 °C | 1 min | ||
| 68 °C | 4 min |
- Digestion of the parental DNA with DpnI: Addition of 1 µl DpnI to the reaction batch and incubate for 1 h at 37 °C.
Transformation of the Quickchange products with P28, P100 and P102 into E. coli XL1 blue
Investigator: Jeff, Andi, Johanna, Rosario
Aim of the experiment: Transformation of E. coli XL1 blue with Quickchangeproduct SDM II of P106 and P116.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 2 µl of DNA was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of the cell suspension was plated on one chloramphenicol plate.
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on a new chlorampenicol plate.
Friday, May 24th
Transformation of E. coli XL1 blue with ligation products F17+F26 (N-TEV S219V in pSB1C3) and F17+F27 (C-TEV S219V in pSB1C3)
Investigator: Jeff, Florian
Aim of the experiment: Transformation of E. coli XL1 blue with ligation products F17+F26 (N-TEV S219V in pSB1C3) and F17+F27 (C-TEV S219V in pSB1C3).
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 10 µl of DNA was added to 200 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of the cell suspension was plated on one chloramphenicol plate.
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellets were resuspended in 100 µl of LB-medium and these concentrated cell suspensions was plated again on a new chlorampenicol plates.
Quick Change mutagenesis to remove forbidden restriction sites - SDMII - of P116 (Laccase), P109 (EreB)
Investigator: Volker, Louise, Katrin, Flo
Aim of the experiment: Removal of forbidden restiction sites from P116 (Laccase), P109 (EreB).
Procedure: Quickchange - PCR
Reaction batch - P109
| volume | reagent | Additional information |
| 2.5 µl | 10x Pfu Ultra II buffer | |
| 1 µl | Plasmid template (P109 - 1:10 dilution) | need to get to 50 ng/µl |
| 1 µl | 1:10 dilution of O18 (for P109, 10 µM) | |
| 1 µl | 1:10 dilution of O19 (for P109, 10 µM) | |
| 18 µl | ddH2O | |
| 1 µl | dNTP mix 50mM | |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
PCR cycling parameters - P109
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 30 s |
| 2 | 21 | 95 °C | 30 s |
| 52 °C | 1 min | ||
| 68 °C | 4 min 50 s |
- Digestion of the parental DNA with DpnI: Addition of 1 µl DpnI to the reaction batch and incubate for 1 h at 37 °C.
Reaction batch - P116 (Laccase)
| volume | reagent | Additional information |
| 2.5 µl | 10x Pfu Ultra II buffer | |
| 1 µl | Plasmid template - 1:5 dilution | need to get to 50 ng/µl |
| 1 µl | 1:10 dilution of O28 for P116 | |
| 1 µl | 1:10 dilution of O29 for P116 | |
| 18.0 µl | ddH2O | |
| 1 µl | dNTP mix 50 mM | |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
PCR cycling parameters - P116
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 30 s |
| 2 | 21 | 95 °C | 30 s |
| 52 °C | 1 min | ||
| 68 °C | 4 min 50 s |
- Digestion of the parental DNA with DpnI: Addition of 1 µl DpnI to the reaction batch and incubate for 1 h at 37 °C.
Transformation of the Quickchange products into E. coli XL1 blue
Investigator: Volker
Aim of the experiment: Transformation of E. coli XL1 blue with Quickchangeproduct SDM II of P109 and P116.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 2 µl of DNA was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of the cell suspension was plated on one chloramphenicol plate.
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on a new chlorampenicol plate.
Saturday, May 25th
Analytical gelelectrophoresis of QuickChange products of P116 and P109 to check whether the QuickChange PCR and DpnI digestion were successful
Investigator: Volker, Louise, Katrin, Flo
Aim of the experiment: Analytical gelelectrophoresis of QuickChange products of P116 and P109 to check whether the QuickChange PCR and DpnI digestion were successful
Procedure:
- 5 µl of the PCR Quickchange products were either mixed with with 0.5 µl of DNA loading buffer (10x) and analytical gelelectrophoresis was performed at 1% agarose gel for 60 min at 90 V.
Picking of transformed Plasmid E. coli XL1 blue with QuickChange products of P116 (Laccase, QC with O28/O29) and P109 (EreB, QC with O18/O19)
Investigator: Volker
Aim of the study: Picking of transformed Plasmid E. coli XL1 blue with QuickChange products of P116 (Laccase, QC with O28/O29) and P109 (EreB, QC with O18/O19).
Procedure:
- Picking and overnight culture after standard laboratory's protocol. (CamR LB-medium)
- 5 colonies for P116QC (Laccase, QC with O28/O29) and 6 colonies for P109QC (EreB, QC with O18/O19) were picked from the plates.
Picking of E. coli XL1 blue with ligation products F17+F26 (N-TEV S219V in pSB1C3) and F17+F27 (C-TEV S219V in pSB1C3)
Investigator: Volker
Aim of the study: Picking of E. coli XL1 blue with ligation products F17+F26 (N-TEV S219V in pSB1C3) and F17+F27 (C-TEV S219V in pSB1C3).
Procedure:
- Picking and overnight culture after standard laboratory's protocol. (CamR LB-medium)
- 5 colonies were picked from plates for E. colis XL1 blue transformed with ligation products F17+F26 (N-TEV S219V in pSB1C3) and F17+F27 (C-TEV S219V in pSB1C3).
Sunday, May 26th
Miniprep of overnight cultures of transformated E. coli XL1 blue with QuickChange products of P116 (Laccase, QC with O28/O29) and P109 (EreB, QC with O18/O19)
Investigator: Katrin, Louise
Aim of the experiment: Miniprep of overnight cultures of transformated E. coli XL1 blue with QuickChange products of P116 (Laccase, QC with O28/O29) and P109 (EreB, QC with O18/O19).
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
Analytical digestion and gelelectrophoresis to check whether the transformation of E. coli Xl1 Blue with QuickChanges products of P116 (Laccase, QC with O28/O29) and P109 (EreB, QC with O18/O19) were successful
Investigator: Katrin, Louise
Aim of the experiment:Analytical digestion and gelelectrophoresis to check whether the transformation of E. coli Xl1 Blue with QuickChanges products of P116 (Laccase, QC with O28/O29) and P109 (EreB, QC with O18/O19)
Procedure:
- analytical digestion of Quickchange products of P109 (EreB, QC with O18/O19)
| volume | reagent |
| 2 µl | NEB CutSmart Buffer (10x) |
| 0,25 µl | PstI-HF (20 U/µl) |
| 2,5 µl | DNA from Miniprep |
| 15,25 µl | ddH2O |
| =20 µl | TOTAL |
- in order to check if the QC was successful, P109 (before QC) was also digested
- analytical digestion of Quickchange products of P116 (Laccase, QC with O28/O29)
| volume | reagent |
| 2 µl | NEB CutSmart Buffer (10x) |
| 0,25 µl | EcoRI-HF |
| 0,25 µl | NgoMIV |
| 2,5 µl | DNA from Miniprep |
| 15 µl | ddH2O |
| =20 µl | TOTAL |
- in order to check if the QC was successful, P116 (before QC) was also digested
- Incubation for 90 min at 37 °C.
- Analytical gelelectrophoresis was performed at 90 V for 60 min.
| 1 kbp ladder DNA ladder# | P116 (Laccase before QCII) digested with EcoRI-HF, NgoMIV | Laccase after QCII (P128) digested with EcoRI-HF, NgoMIV | Laccase after QCII (P129) digested with EcoRI-HF, NgoMIV | Laccase after QCII (P130) digested with EcoRI-HF, NgoMIV | Laccase after QCII (P131) digested with EcoRI-HF, NgoMIV | Laccase after QCII (P124) digested with EcoRI-HF, NgoMIV | belongs to other experiment | belongs to other experiment | belongs to other experiment | belongs to other experiment |
| as expected | as expected | as expected | as expected | as expected | as expected |
Laccase P124 was chosen for sequencing
| 1 kbp ladder DNA ladder | belongs to other experiment | belongs to other experiment | belongs to other experiment | belongs to other experiment | EreB after QCII (P125) digested with PstI | EreB after QCII (P132) digested with PstI | EreB after QCII (P133) digested with PstI | EreB after QCII (P134) digested with PstI | EreB after QCII (P135) digested with PstI | EreB after QCII (P136) digested with PstI | EreB before QCII (P109) digested with PstI |
| as expected | as expected | as expected | as expected | as expected | as expected | as expected |
EreB P125 was chosen for sequencing
Miniprep of ligation products of N-TEV (F17+F26) and C-TEV (F17+F25) with pSB1C3
Investigator: Katrin, Louise
Aim of the experiment: Miniprep of ligation products of N-TEV (F17+F26) and C-TEV (F17+F25) with pSB1C3
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
Analytical digestion and gelelectrophoresis of ligation products F17 + F26, F17 + F27
Investigator: Katrin, Louise
Aim of the experiment: Analytical digestion and gelelectrophoresis to check whether the ligation of N-TEV (F17+F26) and C-TEV (F17+F25) in pSB1C3 were successful
Procedure:
- analytical digestion of ligation products of N-TEV (F17+F26) and C-TEV (F17+F25)in pSB1C3
Mastermix:
| volume | reagent |
| 22 µl | NEB Buffer 4 (10x) |
| 2,2 µl | BSA (100X) |
| 2,75 µl | XbaI |
| 2,75 µl | AgeI-HF |
| 162,8 µl | ddH2O |
| =192,5 µl | TOTAL |
- 2,5 µl of miniprep products were added to 17,5 µl Mastermix
- Incubation for 90 min at 37 °C.
- Analytical gelelectrophoresis was performed at 90 V for 60 min.
| 1 kbp ladder DNA ladder# | belongs to other experiment | belongs to other experiment | belongs to other experiment | belongs to other experiment | belongs to other experiment | belongs to other experiment | N-TEV ligated into pSB1C3 (F17 + F26) (P126) digested with XbaI, AgeI-HF | N-TEV ligated into pSB1C3 (F17 + F26) (P137) digested with XbaI, AgeI-HF | N-TEV ligated into pSB1C3 (F17 + F26) (P138) digested with XbaI, AgeI-HF | N-TEV ligated into pSB1C3 (F17 + F26) (P139) digested with XbaI, AgeI-HF |
| as expected | as expected | as expected | as expected |
N-TEV ligated into pSB1C3 (F17 + F26) (P126) was chosen for sequencing
| 1 kbp ladder DNA ladder | C-TEV ligated into pSB1C3 (F17 + F27) (P127) digested with XbaI, AgeI-HF | C-TEV ligated into pSB1C3 (F17 + F27) (P140) digested with XbaI, AgeI-HF | C-TEV ligated into pSB1C3 (F17 + F27) (P141) digested with XbaI, AgeI-HF | C-TEV ligated into pSB1C3 (F17 + F27) (P142) digested with XbaI, AgeI-HF | belongs to other experiment | belongs to other experiment | belongs to other experiment | belongs to other experiment | belongs to other experiment | belongs to other experiment | belongs to other experiment |
| as expected | as expected | as expected | as expected |
C-TEV ligated into pSB1C3 (F17 + F27) (P127) was chosen for sequencing
Sequencing of P124-P127
Investigators: Louise, Katrin
Aim of the experiment: Sequencing of QCII products (EreB, Laccase) and ligation product of split-TEV
Procedure: The DNA was prepared for sequencing according to the Eurofins SmartSeq Kit protocol (15 µl of plasmid DNA (50 - 100 ng) and 2 µl sequencing primer).
The different genes we sequenced received the following barcodes:
| Label | Name | Barcode1 | Barcode2 |
|---|---|---|---|
| p124 | Miniprep of transformation of QC product of P116 (Laccase, QC with O28/O29)(QC - SDMII - of Bpul Laccase to remove forbidden NgoMIV) | FR01002337 (VR) | FR01002338 (VF2) |
| p125 | Miniprep of transformation of QC product of P109 (EreB, QC with O18/O19) (QC - SDMII - of EreB to remove forbidden PstI) | FR01002339 (VR) | FR01002340 (VF2) |
| p126 | Miniprep of ligation product F17 + F26 (N-TEV) | FR01002341 (VR) | FR01002342 (VF2) |
| p127 | Miniprep of ligation product F17 + F27 (C-TEV) | FR01002343 (VR) | FR01002344 (VF2) |
Week 6
Monday, May 27th
Oligohybridization of MiniMCS (O56,O57) and Spytag (O54,O55)
Investigator: Johanna
Aim of the experiment: Oligohybridization of MiniMCS (MiniMCSII_fw (O56) and MiniMCSII_rv (O57) and of Spytag (SpyTag_fw (O54) and SpyTag_rv (O55))
Operational sequence
- 25 µL of 100 pmol/µl of MiniMCS fw and 25 µL of 100 pmol/µl MiniMCS rev in one Eppi. 25 µL of 100 pmol/µl of Spytag fw and 25 µL of 100 pmol/µl Spytag rev in a second Eppi
- Heating up to 95 °C for 5 min
- Cooling at RT in a styropor box overnight
PCR of npt-Kasette (P115), PIF3 (P51), pActin (P114) and t35S (P114)
Investigator: Johanna
Aim of the experiment: PCR of npt-Kasette (P115), PIF3 (P51), pActin (P114) and t35S (P114).
Procedure:
Operational sequence:
- PCR reaction mixture
| volume | reagent |
| 10 µl | 5x OneTaq Reaction GC Buffer |
| 1 µl | 10 mM dNTPs |
| 1 µl | 10 µM Forward Primer (P114:O62 (pActin_for); P114:O60 (t35S_for); P115:O58 (npt-Kasette_for); P51:O66 (PIF3_for)) |
| 1 µl | 10 µM Reverse Primer (P114:O63 (pActin_rev); P114:O61 (t35S_rev); P115:O59 (npt-Kasette_rev); P51:O67 (PIF3_rev)) |
| 0.25 µL | OneTaq Hot Start DNA Polymerase (Finally: 1.25 units/50 µL) |
| 1 µl | Plasmid DNA (P114; P115; P51) |
| 35.75 µL | ddH2O Water |
| =50 µL | TOTAL |
- Mix with pipette
- The PCR program was performed after following scheme:
| Initial denaturation | 94 °C | 30 s |
| 30 cycles | 94 °C | 30 s |
| 49 °C | 60 s | |
| 68 °C | 100 s | |
| Final extension | 68 °C | 5 min |
| Hold | 4 °C | infinite |
- After PCR, the product was purified with QIAquick PCR Purification Kit, Qiagen.
Results:
- PCR did not work properly because primer concentration was wrong.
Preparative digestion and gelelectrophoresis of P45 (FluA-Psb1C3) with XbaI & AgeI, P39 with AgeI & PstI, P20 with NgoMIV & PstI
Investigator: Andi
Aim of the experiment:Preparative digestion of P45 (FluA-Psb1C3) with XbaI & AgeI, P39 with AgeI & PstI, P20 with NgoMIV & PstI to clone fragment of P45 into F17 (pSB1C3) and to fusion Middle linker of P39 with GFP of P20.
Procedure:
Batch for preparative digestion of P45 (FluA-Psb1C3) with XbaI & AgeIwith XbaI & AgeI.
| volume | reagent |
| 20 µl | P45 |
| 4 µl | NEBuffer 4 |
| 0.4 µl | BSA (100x) |
| 2 µl | AgeI-HF |
| 2 µl | XbaI |
| 11.6 µl | ddH2O |
| =40 µl | TOTAL |
Batch for preparative digestion of P39 (Middle-Linker) with PstI and AgeI.
| volume | reagent |
| 20 µl | P39 |
| 4 µl | NEBuffer 4 |
| 0.4 µl | BSA (100x) |
| 2 µl | AgeI-HF |
| 2 µl | PstI |
| 11.6 µl | ddH2O |
| =40 µl | TOTAL |
Batch for preparative digestion of P20 (GFP-Psb1C3) with NgoMIV and PstI.
| volume | reagent |
| 20 µl | P45 |
| 4 µl | NEBuffer 4 |
| 0.4 µl | BSA (100x) |
| 2 µl | NgoMIV |
| 2 µl | PstI |
| 11.6 µl | ddH2O |
| =40 µl | TOTAL |
- Incubation for 120 min at 37 °C.
- Preparative gelelectrophoresis was performed at 90 V for 1.5 h.
| 1 kbp ladder | P20 digested with NgoMIV&PstI (=F30) | P20 digested with NgoMIV&PstI (=F30) | P39 digested with AgeI&PstI (=F29) | P39 digested with AgeI&PstI (=F29) | P45 digested with XbaI&AgeI (=F28) | P45 digested with XbaI&AgeI (=F28) |
| lower band was cut out | lower band was cut out | band was cut out | band was cut out | lower band was cut out | lower band was cut out |
- Bands were extracted by QIAquick Gel Extraction Kit, QIAGEN.
Quick Change mutagenesis to remove forbidden restriction sites - SDMIII - of P124(Laccase), P125 (EreB)
Investigator: Andi
Aim of the experiment: Removal of forbidden restiction sites from P124 (Laccase), P125 (EreB).
Procedure: Quickchange - PCR
Reaction batch - P124
| volume | reagent | Additional information |
| 2.5 µl | 10x Pfu Ultra II buffer | |
| 1 µl | Plasmid template (P124 - 1:6 dilution) | need to get to 50 ng/µl |
| 1 µl | 1:10 dilution of O30 (for P125, 10 µM) | |
| 1 µl | 1:10 dilution of O31 (for P125, 10 µM) | |
| 18 µl | ddH2O | |
| 1 µl | dNTP mix 50mM | |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
Reaction batch - P125
| volume | reagent | Additional information |
| 2.5 µl | 10x Pfu Ultra II buffer | |
| 1 µl | Plasmid template (P125 - 1:6 dilution) | need to get to 50 ng/µl |
| 1 µl | 1:10 dilution of O20 (for P125, 10 µM) | |
| 1 µl | 1:10 dilution of O21 (for P125, 10 µM) | |
| 18 µl | ddH2O | |
| 1 µl | dNTP mix 50mM | |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
PCR cycling parameters for both batches
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 30 s |
| 2 | 21 | 95 °C | 30 s |
| 52 °C | 1 min | ||
| 68 °C | 4 min 50 s |
- Digestion of the parental DNA with DpnI: Addition of 1 µl DpnI to the reaction batch and incubate for 1 h at 37 °C.
Preparation of ordered primers
Investigators: Andi
Aim of the experiment: Dissolving of primer pallets to provide a concentration of 100 pmol/µl
Procedure: We added an amount of water which gave us a final primer concentration of 100 pmol/µl.
Primers were labeled as follows:
| Label | Oligonr. |
|---|---|
| O54 | Spytag_fw |
| O55 | Spytag_rev |
| O56 | MiniMCSII_fw |
| O57 | MiniMCSII_rev |
| O58 | Npt_for |
| O59 | Npt_rev |
| O60 | t35S_for |
| O61 | t35S_rev |
| O62 | pACT_for |
| O63 | pACT_rev |
| O64 | NucA_for |
| O65 | NucA_rev |
| O66 | Pif3_for |
| O67 | Pif3_rev |
Transformation of P45, P51, P100 and P102 into E. coli XL1 blue
Investigator: Andi, Johanna
Aim of the experiment: Transformation of E. coli XL1 blue with P45, P51, P102 and P100.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 2 µl of DNA was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of the cell suspension was plated on one chloramphenicol plate.
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on a new chlorampenicol plate.
Picking of transformed Plasmid E. coli XL1 blue with P114, P115
Investigator: Andi
Aim of the study: Picking of transformed Plasmid E. coli XL1 blue with P114, P115.
Procedure:
- Picking and overnight culture after standard laboratory's protocol. (AmpR LB-medium)
- 5 colonies of P114 and P115 were picked from the plates.
Ligation of F29+F30 (GFP in pSB1C3 after middle linker, ( Gly-Gly-Ser-Gly)x2) and F17+F27 (FluA in pSB1C3)
Investigator: Jeff
Aim of the experiment: Ligation of F29+F30 (GFP in pSB1C3 after middle linker, ( Gly-Gly-Ser-Gly)x2) and F17+F27 (FluA in pSB1C3).
Procedure:
- Ligation batch for F29+F30:
| volume | reagent |
| 4.22 µl | F29 (23.7 ng/µl, 2110 bp) |
| 10.48 µl | F30 (9.5 ng/µl, ~700 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 2.3 µl | ddH2O |
| =20 µl | TOTAL |
- Ligation batch for F17+F28:
| volume | reagent |
| 1.63 µl | F17 (61.4 ng/µl, 2086 bp) |
| 7.8 µl | F28 (9.6 ng/µl, 522 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 7.57 µl | ddH2O |
| =20 µl | TOTAL |
- Negative control was also prepared. Instead of the insert, the same volume of ddH2O was added to the reaction batch.
- Cycled ligation has been performed after following protocol in a thermocycler:
| 99 cycles | 12 °C | 60 s |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| Hold | 16 °C | infinite |
- Lid temperature = 37 °C
Tuesday, May 28th
Analytical gelelectrophoresis of PCR products of P114 (O62&O63), P114 (O60&O61), P115 (O58&O59), P51 (O66&O67)
Investigator: Jeff, Rosario
Aim of the experiment: Analytical gelelectrophoresis of PCR products of P114 (O62&O63), P114 (O60&O61), P115 (O58&O59), P51 (O66&O67).
Procedure:
- 4 µl of PCR products of P114 (O62&O63), P114 (O60&O61), P115 (O58&O59), P51 (O66&O67) were mixed with seperately with 0.44 µl of DNA loading buffer (10x) and analytical gelelectrophoresis was performed at 1% agarose gel for 40 min at 90 V.
| 1 kbp ladder | PCR products of P114 (O62&O63), P114 (O60&O61), P115 (O58&O59), P51 (O66&O67) | PCR products of P114 (O60&O61) | PCR products of P115 (O58&O59) | PCR products of P51 (O66&O67) |
| PCR did work, but strong primer dimer signal | PCR did work, but strong primer dimer signal | PCR did not work | PCR did work, but strong primer dimer signal |
- Wrong buffer (GC buffer, instead of standard reaction buffer, was used and 10x too much primers were also used)
Transformation of Quickchangeproduct SDMIII of P124 (Laccase) and P125 (EreB) + ligation product of F29+F30 + ligation product of F17+F19 into E. coli XL1 blue
Investigator: Andi, Johanna
Aim of the experiment: Transformation of Quickchangeproduct SDMIII of P124 (Laccase) and P125 (EreB) + ligation product of F29+F30 + ligation product of F17+F19 + ligation product of F17+NK + ligation product of F29+NK into E. coli XL1 blue.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 2 µl of DNA was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of the cell suspension was plated on one chloramphenicol plate.
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on a new chlorampenicol plate.
Sequencing of P133 and P134
Investigators: Louise, Johanna
Aim of the experiment: Sequencing of two more QCII products (EreB) Procedure: The DNA was prepared for sequencing according to the Eurofins SmartSeq Kit protocol (15 µl of plasmid DNA (50 - 100 ng) and 2 µl sequencing primer).
The different genes we sequenced received the following barcodes:
| Label | Name | Barcode1 | Barcode2 |
|---|---|---|---|
| p133 | Miniprep of transformation of QC product of P109 (EreB, QC with O18/O19) (QC - SDMII - of EreB to remove forbidden PstI) | FR01002287 (fw) | FR01002296 (rev) |
| p134 | Miniprep of transformation of QC product of P109 (EreB, QC with O18/O19) (QC - SDMII - of EreB to remove forbidden PstI) | FR01002295 (fw) | FR01002294 (rev) |
Miniprep of overnight cultures of transformated E. coli XL1 blue of P114 and P115
Investigator: Andi
Aim of the experiment: Miniprep of overnight cultures of transformated E. coli XL1 blue of P114 and P115.
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
Picking of of E. coli XL1 blue transformed with P45, P51, P100, P102
Investigator: Jeff, Rosario
Aim of the experiment: Picking of of E. coli XL1 blue transformed with P45, P51, P100, P102.
Procedure:
- Picked pipette tips was transferred into cell-culture tubes with air-permeable, sterile cover. Each tube contain 4 mL of LB-medium + 4 µL chloramphenicol(1000x) (P51, P100, P102)/4 µL Ampicillin (1000x) (P45).
- 1 colony for each plasmid were picked.
- These tubes were transferred in a cell culture shaker at 37 °C and were incubated overnight.
PCR of npt-Kasette (P144), PIF3 (P51), pActin (P143) and t35S (P143)
Investigator: Rosario, Jeff
Aim of the experiment: PCR of npt-Kasette (P144), PIF3 (P51), pActin (P143) and t35S (P143).
Procedure:
Operational sequence:
- PCR reaction mixture
| volume | reagent |
| 10 µl | 5x OneTaq Reaction Standard Reaction Buffer |
| 1 µl | 10 mM dNTPs |
| 1 µl | 10 µM Forward Primer (P143:O62 (pActin_for); P143:O60 (t35S_for); P144:O58 (npt-Kasette_for); P51:O66 (PIF3_for)) |
| 1 µl | 10 µM Reverse Primer (P143:O63 (pActin_rev); P143:O61 (t35S_rev); P144:O59 (npt-Kasette_rev); P51:O67 (PIF3_rev)) |
| 0.25 µL | OneTaq Hot Start DNA Polymerase (Finally: 1.25 units/50 µL) |
| 1 µl | Plasmid DNA (P114; P115; P51) |
| 35.75 µL | ddH2O Water |
| =50 µL | TOTAL |
- Mix with pipette
- The PCR program was performed after following scheme:
| Initial denaturation | 94 °C | 30 s |
| 30 cycles | 94 °C | 30 s |
| 47 °C | 60 s | |
| 68 °C | 120 s | |
| Final extension | 68 °C | 5 min |
| Hold | 4 °C | infinite |
- After PCR, the product was purified with QIAquick PCR Purification Kit, Qiagen.
Analytical gelelectrophoresis of F33, F34, F35, F36
Investigator: Jeff, Rosario
Aim of the experiment: Analytical gelelectrophoresis of F33, F34, F35, F36.
Procedure:
- 4 µl of F33, F34, F35 and F36 were mixed with seperately with 0.44 µl of DNA loading buffer (10x) and analytical gelelectrophoresis was performed at 1% agarose gel for 40 min at 90 V.
| 1 kbp ladder | F33 | F33 | F33 | F33 |
| PCR did not work, maybe high GC content? | PCR products has expected length, some byproducts | PCR products has expected length, some byproducts | PCR products has expected length |
Wednesday, May 29th
Transformation of E. coli XL1 blue with SDMIII product of P124, ligation product of F29+F30 and ligation product of F17+F18
Investigator: Andi
Aim of the experiment: Transformation of E. coli XL1 blue with SDMIII product of P124, ligation product of F29+F30 and ligation product of F17+F18.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 5 µl of DNA was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of the cell suspension was plated on one chloramphenicol plate.
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on a new chlorampenicol plate.
Miniprep of overnight cultures of transformated E. coli XL1 blue of P100, P102, P51, P45
Investigator: Rosario
Aim of the experiment: Miniprep of overnight cultures of transformated E. coli XL1 blue of P100, P102, P51, P45 .
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
Picking of of E. coli XL1 blue transformed with ligation product of F17+F28
Investigator: Andi
Aim of the experiment: Picking of of E. coli XL1 blue transformed with ligation product of F17+F28.
Procedure:
- Picked pipette tips was transferred into cell-culture tubes with air-permeable, sterile cover. Each tube contain 4 mL of LB-medium + 4 µL chloramphenicol(1000x) (
- 3 colonies were picked.
- These tubes were transferred in a cell culture shaker at 37 °C and were incubated overnight.
Quick Change mutagenesis to remove forbidden restriction sites - SDMIII - of P133 (EreB)
Investigator: Louise
Aim of the experiment: Removal of forbidden restiction site from P133 (EreB).
Procedure: Quickchange - PCR
Reaction batch
| volume | reagent | Additional information |
| 2.5 µl | 10x Pfu Ultra II buffer | |
| 1 µl | Plasmid template (P133 - 1:10 dilution) | need to get to 50 ng/µl |
| 1 µl | 1:10 dilution of O20 (for P133, 10 µM) | |
| 1 µl | 1:10 dilution of O21 (for P133, 10 µM) | |
| 18 µl | ddH2O | |
| 1 µl | dNTP mix 50mM | |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
PCR cycling parameters - P133
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 30 s |
| 2 | 19 | 95 °C | 30 s |
| 52 °C | 1 min | ||
| 68 °C | 3 min 30 s |
- Digestion of the parental DNA with DpnI: Addition of 1 µl DpnI to the reaction batch and incubate for 1 h at 37 °C.
Transformation of the Quickchange product into E. coli XL1 blue
Investigator: Louise
Aim of the experiment: Transformation of E. coli XL1 blue with Quickchangeproduct SDM III of P133.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 10 µl of DNA was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of the cell suspension was plated on one chloramphenicol plate.
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on a new chlorampenicol plate.
Preparative digestion of t35s(F34) with XbaI & PstI, npt(F25) with EcoRI & SpeI, PIF3(F36) with XbaI & AgeI-HF and P10 with XbaI & PstI as well as with EcoRI & SpeI
Investigator: Louise
Aim of the experiment: Preparative digestion of t35s(F34) with XbaI & PstI, npt(F25) with EcoRI & SpeI, PIF3(F36) with XbaI & AgeI-HF and P10 with XbaI & PstI as well as with EcoRI & SpeI.
Procedure:
- Batch for preparative digestion of t35s(F34) with XbaI and PstI
| volume | reagent |
| 25 µl | Plasmid DNA P61 |
| 5 µl | CutSmart Buffer |
| 1 µl | XbaI (20 U/µl) |
| 1 µl | PstI (20 U/µl) |
| 18 µl | ddH2O |
| =50 µl | TOTAL |
- Batch for preparative digestion of npt(F35) with EcoRI and SpeI
| volume | reagent |
| 25 µl | Plasmid DNA P61 |
| 5 µl | CutSmart Buffer |
| 1 µl | EcoRI (20 U/µl) |
| 1 µl | SpeI (20 U/µl) |
| 18 µl | ddH2O |
| =50 µl | TOTAL |
- Batch for preparative digestion of PIF3(F36) with XbaI and AgeI-HF
| volume | reagent |
| 25 µl | Plasmid DNA P61 |
| 5 µl | CutSmart Buffer |
| 1 µl | XbaI (20 U/µl) |
| 1 µl | AgeI-HF (20 U/µl) |
| 18 µl | ddH2O |
| =50 µl | TOTAL |
- Batch for preparative digestion of P10 with XbaI & PstI for backbone isolation.
| volume | reagent |
| 20 µl | Plasmid DNA P96 |
| 4 µl | CutSmart Buffer |
| 1 µl | XbaI (20 U/µl) |
| 1 µl | PstI (20 U/µl) |
| 14 µl | ddH2O |
| =40 µl | TOTAL |
- Batch for preparative digestion of P10 with EcoRI and SpeI for backbone isolation.
| volume | reagent |
| 20 µl | Plasmid DNA P96 |
| 4 µl | CutSmart Buffer |
| 1 µl | EcoRI (20 U/µl) |
| 1 µl | SpeI (20 U/µl) |
| 14 µl | ddH2O |
| =40 µl | TOTAL |
- Incubation for 2.5 h at 37 °C.
- The resulting fragments were named: ?????
- 5 µl of DNA loading buffer (10x) were added to the 50 µl reaction batches and 4 µl of DNA loading buffer (10x) were added to the 40 µl reaction batches after digestion and were loaded on a 1% agarose gel for preparative gelelectrophoresis.
- Preparative gelelectrophoresis was performed at 90 V for 90 min.
| 1 kbp ladder DNA ladder | Digestion of PCR product t35s (F34) with XbaI & PstI = F38 | Digestion of PCR product npt casette (F25) with EcoRI & SpeI = F39 | Digestion of PCR product PIF3 (F36) with XbaI & AgeI-HF = F40 |
| as expected, lower band was cut out | as expected, bright band was cut out | as expected, bright band was cut out |
| 1 kbp ladder DNA ladder | Digestion of P10 with EcoRI & SpeI = F41 | Digestion of P10 with XbaI & PstI = F42 |
| as expected, lower band was cut out | as expected, lower band was cut out |
- Bands were extracted by QIAquick Gel Extraction Kit, QIAGEN.
Transformation of E. coli XL1 blue with pSB1C3 containing a NLS from SV40 (BBa_K801030)
Investigator: Florian, Jeff
Aim of the experiment: Transformation of E. coli XL1 blue with pSB1C3 containing a NLS from SV40 (BBa_K801030).
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 10 µl of DNA was added to 200 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of the cell suspension was plated on one chloramphenicol plate.
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on a new chlorampenicol plate.
PCR of pActin (P143)
Investigator: Florian, Jeff
Aim of the experiment: PCR of pActin (P143).
Procedure:
Operational sequence:
- PCR reaction mixture
| volume | reagent |
| 10 µl | 5x OneTaq Reaction Buffer |
| 1 µl | 10 mM dNTPs |
| 1 µl | 10 µM Forward Primer (P143:O62 (pActin_for)) |
| 1 µl | 10 µM Reverse Primer (P143:O63 (pActin_rev)) |
| 0.25 µL | OneTaq Hot Start DNA Polymerase (Finally: 1.25 units/50 µL) |
| 1 µl | Plasmid DNA (P143) |
| 35.75 µL | ddH2O Water |
| =50 µL | TOTAL |
- Mix with pipette
- The PCR program was performed after following scheme:
| Initial denaturation | 94 °C | 30 s |
| 30 cycles | 94 °C | 30 s |
| 51 °C | 60 s | |
| 68 °C | 80 s | |
| Final extension | 68 °C | 5 min |
| Hold | 4 °C | infinite |
- After PCR, the product was purified with QIAquick PCR Purification Kit, Qiagen.
Results:
- After analytical gelelectrophoresis there was no product visible. PCR did't work!
Ligation of F17+F40 (PIF3 in pSB1C3(RFC25)), F42+F38 (t35S in pSB1C3(RFC10)) and F41+F39(npt-casette in pSB1C3(RFC10))
Investigator: Jeff
Aim of the experiment: Ligation of F17+F40 (PIF3 in pSB1C3(RFC25)), F42+F38 (t35S in pSB1C3(RFC10)) and F41+F39(npt-casette in pSB1C3(RFC10)).
Procedure:
- Ligation batch for F17+F40:
| volume | reagent |
| 1.63 µl | F17 (61.4 ng/µl, 2086 bp) |
| 1.88 µl | F40 (22.8 ng/µl, ~297 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 13.49 µl | ddH2O |
| =20 µl | TOTAL |
- Ligation batch for F42+F38:
| volume | reagent |
| 1.36 µl | F42 (73.5 ng/µl, 2070 bp) |
| 3.42 µl | F38 (9.2 ng/µl, 217 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 12.22 µl | ddH2O |
| =20 µl | TOTAL |
- Ligation batch for F41+F39:
| volume | reagent |
| 1.11 µl | F41 (89.9 ng/µl, 2070 bp) |
| 13.74 µl | F39 (16.0 ng/µl, 1517 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 2.15 µl | ddH2O |
| =20 µl | TOTAL |
- Negative control was also prepared. Instead of the insert, the same volume of ddH2O was added to the reaction batch.
- Cycled ligation has been performed after following protocol in a thermocycler:
| 99 cycles | 12 °C | 60 s |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| Hold | 16 °C | infinite |
- Lid temperature = 37 °C
Thursday, May 30th
Gradient PCR of pActin (P143)
Investigator: Ingmar
Aim of the experiment: PCR of pActin
Procedure:
Operational sequence:
- PCR reaction mixture
| volume | reagent |
| 10 µl | 5x OneTaq Reaction Standard Reaction Buffer |
| 1 µl | 10 mM dNTPs |
| 1 µl | 10 µM Forward Primer O62 (pActin_for) |
| 1 µl | 10 µM Reverse Primer O63 (pActin_rev) |
| 0.25 µL | OneTaq Hot Start DNA Polymerase (Finally: 1.25 units/50 µL) |
| 1 µl | Plasmid DNA P143 |
| 35.75 µL | ddH2O Water |
| =50 µL | TOTAL |
- PCR reaction mixture
| volume | reagent |
| 10 µl | 5x OneTaq GC Reaction Buffer |
| 1 µl | 10 mM dNTPs |
| 1 µl | 10 µM Forward Primer O62 (pActin_for) |
| 1 µl | 10 µM Reverse Primer O63 (pActin_rev) |
| 0.25 µL | OneTaq Hot Start DNA Polymerase (Finally: 1.25 units/50 µL) |
| 1 µl | Plasmid DNA P143 |
| 35.75 µL | ddH2O Water |
| =50 µL | TOTAL |
- Mix with pipette
- The PCR program was performed after following scheme:
| Initial denaturation | 94 °C | 30 s |
| 30 cycles | 94 °C | 30 s |
| 47 °C | 60 s | |
| 68 °C | 120 s | |
| Final extension | 68 °C | 5 min |
| Hold | 4 °C | infinite |
A temperature gradient was applied beginning at 47°C and ranging to 51°C. For each integer (47, 48, 49, 50 and 51°C) a single PCR batch was used.
- After PCR, the product was purified with QIAquick PCR Purification Kit, Qiagen.
Analytical gelelectrophoresis
Procedure:
- 4 µl of each PCR product were mixed seperately with 0.44 µl of DNA loading buffer (10x) and analytical gelelectrophoresis was performed at 1% agarose gel for 60 min at 90 V.
| 1 kbp ladder | F45 GC buffer | F46 GC buffer | F47 GC buffer | F47 GC buffer | F49 GC buffer | normal buffer | normal buffer | normal buffer | normal buffer | normal buffer | 1 kbp ladder |
| PCR products has expected length | PCR products has expected length | PCR products has expected length | PCR products has expected length | PCR products has expected length | PCR products has expected length, some byproducts | PCR products has expected length, some byproducts | PCR products has expected length, some byproducts | PCR products has expected length, some byproducts | PCR products has expected length, some byproducts |
Miniprep of overnight cultures of transformated E. coli XL1 blue of F17+F28 (FluA in pSB1C3 RFC25)
Investigator: Jeff, Flo
Aim of the experiment: Miniprep of overnight cultures of transformated E. coli XL1 blue of F17+F28 (FluA in pSB1C3 RFC25).
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
Analytical digestion and gelelectrophoresis of ligation products F17+F28 (FluA in pSB1C3 RFC25)
Investigator: Flo, Jeff
Aim of the experiment: Analytical digestion and gelelectrophoresis of ligation products F17+F28 (FluA in pSB1C3 RFC25).
Procedure:
- Analytical digestion of ligation products of F17+F28 (FluA in pSB1C3 RFC25) with XbaI & AgeI.
| volume | reagent |
| 2.5 µl P150/P151/P152/P153 | |
| 2 µl | CutSmart Buffer (10x) |
| 0.25 µl | XbaI |
| 0.25 µl | AgeI-HF |
| 15 µl | ddH2O |
| =20 µl | TOTAL |
- Incubation for 90 min at 37 °C.
- Analytical gelelectrophoresis was performed at 90 V for 60 min.
| 1 kbp ladder DNA ladder# | Digestion of P150 with XbaI&AgeI-HF | Digestion of P151 with XbaI&AgeI-HF | Digestion of P152 with XbaI&AgeI-HF | Digestion of P153 with XbaI&AgeI-HF |
| as expected | as expected | as expected | as expected |
Preparative gelelectrophoresis of oligohybridization products F31 & F32
Investigator: Flo, Jeff
Aim of the experiment: Preparative gelelectrophoresis of oligohybridization products F31 & F32.
Procedure:
- 50 µl of the 1:10 dilution of F31 and F32 were mixed with 5 µl of DNA loading buffer (10x).
- Preparative gelelectrophoresis was performed 1.5 h on a 2% agarose gel at 90 V.
| 1 kbp ladder | F31 | F32 |
- Bands were extracted by QIAquick Gel Extraction Kit, QIAGEN.
- Gelpurified F31 and F32 were labelled as F43 and F44.
Transformation of E. coli XL1 blue with F17+F40 (PIF3 in pSB1C3(RFC25)), F42+F38 (t35S in pSB1C3(RFC10)) and F41+F39(npt-casette in pSB1C3(RFC10))
Investigator: Florian, Jeff
Aim of the experiment: Transformation of E. coli XL1 blue with F17+F40 (PIF3 in pSB1C3(RFC25)), F42+F38 (t35S in pSB1C3(RFC10)) and F41+F39(npt-casette in pSB1C3(RFC10)).
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 10 µl of DNA were added to 200 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of the cell suspension was plated on one chloramphenicol plate.
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellets were resuspended in 100 µl of LB-medium and these concentrated cell suspensions were plated again on a new chlorampenicol plates.
Ligation of F42+F43 and F17+F32
Investigator: Rosario
Aim of the experiment: Ligation of F42+F43 and F17+F32.
Procedure:
- Ligation batch for F42+F43:
| volume | reagent |
| 1.36 µl | F42 (73.5 ng/µl, 2070 bp) |
| 1.51 µl | F43 (5.4 ng/µl, 34 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 14.13 µl | ddH2O |
| =20 µl | TOTAL |
- Ligation batch for F17+F32:
| volume | reagent |
| 1.63 µl | F17 (61.4 ng/µl, 2086 bp) |
| 0.25 µl | F32 (2510.8 ng/µl, 49 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 15.12 µl | ddH2O |
| =20 µl | TOTAL |
- Negative control was also prepared. Instead of the insert, the same volume of ddH2O was added to the reaction batch.
- Cycled ligation has been performed after following protocol in a thermocycler:
| 99 cycles | 12 °C | 60 s |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| Hold | 16 °C | infinite |
- Lid temperature&
Picking of of E. coli XL1 blue transformed with EreB SDMIII (P149) and BBa_K801030 (SV40 NLS)
Investigator: Louise
Aim of the experiment: Picking of of E. coli XL1 blue transformed with EreB SDMIII (P149) and BBa_K801030 (SV40 NLS).
Procedure:
- Picked pipette tips was transferred into cell-culture tubes with air-permeable, sterile cover. Each tube contain 4 mL of LB-medium + 4 µL chloramphenicol(1000x).
- 5 colonies of EreB SDMIII (P149) and 2 colonies of BBa_K801030 (SV40 NLS) were picked.
- These tubes were transferred in a cell culture shaker at 37 °C and were incubated overnight.
Friday, May 31st
Miniprep of overnight cultures of transformated E. coli XL1 blue with P149 (QCIII of EreB) and BBa_K801030 (SV40 NLS)
Investigator: Louise, Jeff, Katrin
Aim of the experiment: Miniprep of overnight cultures of transformated E. coli XL1 blue with P149 (QCIII of EreB) and BBa_K801030 (SV40 NLS)
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
Analytical digestion and gelelectrophoresis of P149 (QCIII of EreB) with EcoRI-HF and PstI-HF
Investigator: Louise, Jeff, Katrin
Aim of the experiment: Analytical digestion and gelelectrophoresis of P154-P158 (QCIII of EreB) with EcoRI-HF and PstI-HF to check if QCII was successful
Procedure:
- Analytical digestion of P154-P158 (QCIII of EreB) with EcoRI-HF and PstI-HF
| volume | reagent |
| 2.5 µl | P149-P158/P133 (control before QCIII) |
| 2 µl | CutSmart Buffer (10x) |
| 0.25 µl | EcoRI-HF |
| 0.25 µl | PstI-HF |
| 15 µl | ddH2O |
| =20 µl | TOTAL |
- Incubation for 90 min at 37 °C.
- Analytical gelelectrophoresis was performed at 90 V for 60 min.
| 1 kbp ladder DNA ladder# | Digestion of P133 with EcoRI-HF and PstI-HF | Digestion of P154 with EcoRI-HF and PstI-HF | Digestion of P155 with EcoRI-HF and PstI-HF | Digestion of P156 with EcoRI-HF and PstI-HF | Digestion of P157 with EcoRI-HF and PstI-HF | Digestion of P158 with EcoRI-HF and PstI-HF |
| as expected | as expected | as expected | as expected | as expected | as expected |
Sequencing of P157 and P160
Investigators: Louise, Jeff, Katrin
Aim of the experiment: Sequencing of QCIII product (EreB) and BBa_K801030 (SV40 NLS)
Procedure: The DNA was prepared for sequencing according to the Eurofins SmartSeq Kit protocol (15 µl of plasmid DNA (50 - 100 ng) and 2 µl sequencing primer).
The different genes we sequenced received the following barcodes:
| Label | Name | Barcode1 | Barcode2 |
|---|---|---|---|
| P157 | Miniprep of transformation of QCIII product of P149 (QC - SDMIII - of EreB to remove forbidden PstI) | FR01002289 (VR) | FR01002290 (VF2) |
| P160 | Miniprep of transformation of BBa_K801030 (SV40 NLS) | FR01002291 (VR) | FR01002292 (VF2) |
Ligation of F29+F30
Investigator: Katrin
Aim of the experiment: Ligation of F29+F30
Procedure:
- Ligation batch
| volume | reagent |
| 4.22 µl | F29 (23.7 ng/µl, 2110 bp)) |
| 10.48 µl | F30 (9,5 ng/µl, about 700 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 2.3 µl | ddH2O |
| =20 µl | TOTAL |
No negative control was prepared because there was not enough vector backbone left
Ligation was performed at 37 °C overnight
Quick Change mutagenesis to remove forbidden restriction sites - SDMIII - of P124 (Laccase)
Investigator: Louise, Katrin
Aim of the experiment: Removal of forbidden restiction site from P124 (Laccase).
Procedure: Quickchange - PCR
Reaction batch
| volume | reagent | Additional information |
| 2.5 µl | 10x Pfu Ultra II buffer | |
| 1 µl | Plasmid template (P124 - 1:10 dilution) | need to get between 5 and 50 ng/µl |
| 1 µl | 1:10 dilution of O30 (for P124, 10 µM) | |
| 1 µl | 1:10 dilution of O31 (for P124, 10 µM) | |
| 18 µl | ddH2O | |
| 1 µl | dNTP mix 50mM | |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
PCR cycling parameters - P124
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 30 s |
| 2 | 19 | 95 °C | 30 s |
| 52 °C | 1 min | ||
| 68 °C | 4 min | ||
| 4 °C | hold |
After the PCR; 1 µl of Dpn1 was added and the reaction mixture was incubated at 37°C for one hour
Preparative digestion of pActin(F46) with XbaI & PstI
Investigator: Ingmar
Aim of the experiment: Preparative digestion of pActin(F46) in order to ligat this fragment in pSB1C3 later on.
Procedure:
- Batch for preparative digestion of pActint(F46) with XbaI and PstI
| volume | reagent |
| 25 µl | F46 PCR product of pActin |
| 5 µl | CutSmart Buffer |
| 1 µl | XbaI (20 U/µl) |
| 1 µl | PstI (20 U/µl) |
| 18 µl | ddH2O |
| =50 µl | TOTAL |
- Incubation for 3 h at 37 °C.
- 5 µl of DNA loading buffer (10x) were added to the 50 µl reaction batchesand loaded on a 1% agarose gel for preparative gelelectrophoresis.
- Preparative gelelectrophoresis was performed at 90 V for 90 min.
- The band was extracted by QIAquick Gel Extraction Kit, QIAGEN. The resulting fragment was named: F50
Ligation of F42+F50 (pActin in pSB1C3)
Investigator: Ingmar
Aim of the experiment: Ligation of F42+F50 (pActin in pSB1C3(RFC25).
Procedure:
- Ligation batch
| volume | reagent |
| 1.37 µl | F42 |
| 13.13 µl | F50 |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 2.5 µl | ddH2O |
| =20 µl | TOTAL |
- Negative control was also prepared. Instead of the insert, the same volume of ddH2O was added to the reaction batch.
- Cycled ligation has been performed after following protocol in a thermocycler:
| 99 cycles | 12 °C | 60 s |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| Hold | 16 °C | infinite |
- Lid temperature = 37 °C
Picking of of E. coli XL1 blue transformed with F42+F38 (t35S), F17+F40 (Pif 3) and F41+F39 (npt)
Investigator: Ingmar
Aim of the experiment: Picking of of E. coli XL1 blue transformed with F42+F38 (t35S), F17+F40 (Pif 3) and F41+F39 (npt).
Procedure:
- The picked colonies were transferred into cell-culture tubes with air-permeable, sterile cover containing 5 mL of LB-medium + 5 µL chloramphenicol(1000x).
- 3 colonies of each agar plate were picked.
- These tubes were transferred in a cell culture shaker at 37 °C and were incubated overnight.
Transformation of E. coli XL1 blue with ligation products F42+F43 and F17+F32
Investigator: Louise, Jeff
Aim of the experiment: Transformation of E. coli XL1 blue with ligation products F42+F43 and F17+F32.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 10 µl of DNA was added to 200 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of the cell suspension was plated on one chloramphenicol plate.
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellets were resuspended in 100 µl of LB-medium and these concentrated cell suspensions were plated again on new chlorampenicol plates.
Saturday, June 1st
Transformation of E. coli XL1 blue with ligation products F29+F30 (GFP in pSB1C3), F42+F50 (actin promoter in pSB1C3), F42+F50 (negative control)
Investigator: Katrin
Aim of the experiment: Transformation of E. coli XL1 blue with ligation products F29+F30 (GFP in pSB1C3), F42+F50 (actin promoter in pSB1C3), F42+F50 (negative control)
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 5 µl of DNA were added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of the cell suspension was plated on one chloramphenicol plate.
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellets were resuspended in 100 µl of LB-medium and these concentrated cell suspensions were plated again on a new chlorampenicol plates.
Transformation of E. coli XL1 blue with QCIII product of Laccase (SDMIII of P124)
Investigator: Katrin
Aim of the experiment: Transformation of E. coli XL1 blue with QCIII product of Laccase
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 10 µl of DNA were added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of the cell suspension was plated on one chloramphenicol plate.
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellets were resuspended in 100 µl of LB-medium and these concentrated cell suspensions were plated again on a new chlorampenicol plates.
Miniprep of overnight cultures of transformated E. coli XL1 blue with F39 + F41 (npt-casette in pSC1C3),F42+F38 (t35s in pSB1C3), F17+F40 (PIF3 in pSB1C3)
Investigator: Katrin
Aim of the experiment: Miniprep of overnight cultures of transformated E. coli XL1 blue with F39 + F41 (npt-casette in pSC1C3),F42+F38 (t35s in pSB1C3), F17+F40 (PIF3 in pSB1C3)
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
Analytical digestion and gelelectrophoresis of P161-163 (F39 + F41, npt-casette in pSB1C3), P164-166 (F42+F38, t35s in pSB1C3), P167-169 (F17+F40, PIF3 in pSB1C3)
Investigator: Katrin
Aim of the experiment: Analytical digestion and gelelectrophoresis of P161-163 (F39 + F41, npt-casette in pSC1C3) with EcoRI-HF and SpeI-HF, P164-166 (F42+F38, t35s in pSB1C3) with EcoRI-HF and SpeI-HF and P167-169 (F17+F40, PIF3 in pSB1C3) with AgeI-HF and XbaI
Procedure:
- Analytical digestion of P161-163 (F39 + F41, npt-casette in pSC1C3), P164-166 (F42+F38, t35s in pSB1C3) with EcoRI-HF and SpeI-HF
| volume | reagent |
| 2.5 µl | P161-P166 |
| 2 µl | CutSmart Buffer (10x) |
| 0.25 µl | EcoRI-HF |
| 0.25 µl | SpeI-HF |
| 15 µl | ddH2O |
| =20 µl | TOTAL |
- Incubation for 90 min at 37 °C.
- Analytical gelelectrophoresis was performed at 90 V for 60 min.
- Analytical digestion of P167-169 (F17+F40, PIF3 in pSB1C3) with AgeI-HF and XbaI
| volume | reagent |
| 2.5 µl | P167-P169 |
| 2 µl | CutSmart Buffer (10x) |
| 0.25 µl | AgeI-HF |
| 0.25 µl | XbaI |
| 15 µl | ddH2O |
| =20 µl | TOTAL |
- Incubation for 90 min at 37 °C.
- Analytical gelelectrophoresis was performed at 90 V for 60 min.
| 1 kbp ladder DNA ladder# | Digestion of P161 with EcoRI-HF and SpeI-HF | Digestion of P162 with EcoRI-HF and SpeI-HF | Digestion of P163 with EcoRI-HF and SpeI-HF | Digestion of P164 with EcoRI-HF and SpeI-HF | Digestion of P165 with EcoRI-HF and SpeI-HF | Digestion of P166 with EcoRI-HF and SpeI-HF | Digestion of P167 with AgeI-HF and XbaI | Digestion of P168 with AgeI-HF and XbaI | Digestion of P169 with AgeI-HF and XbaI |
| as expected | as expected | as expected | as expected | as expected | as expected | as expected | as expected | as expected |
Sunday, June 2nd
Picking of of E. coli XL1 blue transformed with QCIII product of P124 and the ligation products of F17+F44 and F42+F32
Investigator: Andi
Aim of the experiment: Picking of of E. coli XL1 blue transformed with QCIII product of P124 and the ligation products of F17+F32 and F42+F32.
Procedure:
- The picked colonies were transferred into cell-culture tubes with air-permeable, sterile cover containing 4 mL of LB-medium + 4 µL chloramphenicol(1000x).
- 4 colonies of each agar plate were picked.
- These tubes were transferred in a cell culture shaker at 37 °C and were incubated overnight.
Week 7
Monday, June 3rd
Analytical digestion and gelelectrophoresis of P170-P173 (F17+F32, SpyTag in pSB1C3 RFC25), P174-P177 (F42+F43, miniMCSII in pSB1C3 RFC10), P178-181 (QCIII von P142, Laccase in pSB1C3 RFC10)
Investigator: Andi, Johanna, Jeff
Aim of the experiment: Analytical digestion and gelelectrophoresis of P170-P173 (F17+F32, SpyTag in pSB1C3 RFC25), P174-P177 (F42+F43, miniMCSII in pSB1C3 RFC10), P178-181 (QCIII von P142, Laccase in pSB1C3 RFC10).
Procedure:
- Mastermix for analytical digestion of P170-P173 (F17+F32, SpyTag in pSB1C3 RFC25), P174-P177 (F42+F43, miniMCSII in pSB1C3 RFC10) with XbaI+PstI.
| volume | reagent |
| 18 µl | CutSmart Buffer (10x) |
| 2.25 µl | EcoRI-HF |
| 2.25 µl | SpeI-HF |
| 135 µl | ddH2O |
| =157.5 µl | TOTAL |
- 17.5 µl of the mastermix was added to 2.5 µl of plasmid DNA (P170-P177)
- Incubation for 90 min at 37 °C.
- Analytical gelelectrophoresis was performed at 90 V for 60 min.
- Analytical digestion of P178-181 (QCIII von P142, Laccase in pSB1C3 RFC10):
| volume | reagent |
| 2.5 µl | P178-P181 |
| 2 µl | CutSmart Buffer (10x) |
| 0.25 µl | NgoMIV |
| 0.25 µl | PstI-HF |
| 15 µl | ddH2O |
| =20 µl | TOTAL |
- Incubation for 90 min at 37 °C.
- Analytical gelelectrophoresis was performed at 90 V for 60 min.
| 1 kbp ladder DNA ladder | Digestion of P170 with EcoRI-HF and PstI-HF | Digestion of P162 with EcoRI-HF and PstI-HF | Digestion of P163 with EcoRI-HF and PstI-HF | Digestion of P164 with EcoRI-HF and PstI-HF | Digestion of P164 with EcoRI-HF and PstI-HF | Digestion of P164 with EcoRI-HF and PstI-HF | Digestion of P164 with EcoRI-HF and PstI-HF | Digestion of P164 with EcoRI-HF and PstI-HF | Digestion of P164 with EcoRI-HF and PstI-HF |
| as expected, insert too small to be visible | as expected, insert too small to be visible | as expected, insert too small to be visible | as expected, insert too small to be visible | as expected, insert too small to be visible | as expected, insert too small to be visible | as expected, insert too small to be visible | as expected, insert too small to be visible | as expected, insert too small to be visible |
| 1 kbp ladder DNA ladder | Digestion of P142 with NgoMIV and PstI-HF | Digestion of P178 with NgoMIV and PstI-HF | Digestion of P179 with NgoMIV and PstI-HF | Digestion of P180 with NgoMIV and PstI-HF | Digestion of P181 with NgoMIV and PstI-HF |
| Ctrl (before QuickChange III) | as expected, plasmid is only linearized, all NgoMIV sites are removed | as expected, plasmid is only linearized, all NgoMIV sites are removed | as expected, plasmid is only linearized, all NgoMIV sites are removed | as expected, plasmid is only linearized, all NgoMIV sites are removed |
Tuesday, June 4th
Sequencing of P162(npt-cassete), P165(t35s), P168(PIF3), P179(LaccQCIII), P177(MiniMCS), P170(Spytag)
Investigators: Louise, Johanna
Aim of the experiment: Sequencing of P162(npt-cassete), P165(t35s), P168(PIF3), P179(LaccQCIII), P177(MiniMCS), P170(Spytag).
Procedure: The DNA was prepared for sequencing according to the Eurofins SmartSeq Kit protocol (15 µl of plasmid DNA (50 - 100 ng) and 2 µl sequencing primer).
The different genes we sequenced received the following barcodes:
| Label | Name | Barcode1 | Barcode2 |
|---|---|---|---|
| P162 | Miniprep of ligation product F39 + F41 (npt-casette in pSB1C3) | FR01002331 (VR) | FR01002330 (VF2) |
| P165 | Miniprep of ligation product F42+F38 (t35s in pSB1C3) | FR01002332 (VF2) | |
| P168 | Miniprep of ligation product F17+F40 (PIF3 in pSB1C3) | FR01002333 (VF2) | |
| P179 | Miniprep of ligation product Laccase QCIII (P142) | FR01002335 (VR) | FR01002334 (VF2) |
| P177 | Miniprep of ligation product MiniMCSII F42+F43 | FR01002336 (VF2) | |
| P170 | Miniprep of ligation product F17+F32 (SpyTag) | FR01002328 (VF2) |
Preparative digestion and gelelectrophoresis of P39(middle-linker) with PstI and AgeI-HF, P20(GFP-Psb1C3) with NgoMIV and PstI, F45(pActin) with XbaI and PstI
Investigator: Louise, Johanna
Aim of the experiment: Preparative digestion of P39(middle-linker) with PstI and AgeI-HF, P20(GFP-Psb1C3) with NgoMIV and PstI, F45(pActin) with XbaI and PstI.
Procedure:
- Batch for preparative digestion of P39(middle-linker) with PstI and AgeI-HF
| volume | reagent |
| 15 µl | Plasmid DNA P39 |
| 4 µl | CutSmart Buffer |
| 1 µl | AgeI-HF(20 U/µl) |
| 1 µl | PstI (20 U/µl) |
| 19 µl | ddH2O |
| =40 µl | TOTAL |
- Batch for preparative digestion of P20(GFP-Psb1C3) with NgoMIV and PstI
| volume | reagent |
| 20 µl | Plasmid DNA P20 |
| 4 µl | CutSmart Buffer |
| 1 µl | NgoMIV (20 U/µl) |
| 1 µl | PstI (20 U/µl) |
| 14 µl | ddH2O |
| =40 µl | TOTAL |
- Batch for preparative digestion of F45(pActin) with XbaI and PstI
| volume | reagent |
| 25 µl | Fragment DNA F45 |
| 5 µl | CutSmart Buffer |
| 1 µl | XbaI (20 U/µl) |
| 1 µl | PstI (20 U/µl) |
| 18 µl | ddH2O |
| =50 µl | TOTAL |
- Incubation for 3 h at 37 °C.
- 5 µl of DNA loading buffer (10x) were added to the 50 µl reaction batches and 4 µl of DNA loading buffer (10x) were added to the 40 µl reaction batches after digestion and were loaded on a 1% agarose gel for preparative gelelectrophoresis.
- Preparative gelelectrophoresis was performed at 90 V for 90 min.
| 1 kbp DNA ladder | Digestion of P20 with NgoMIV & PstI = F53 | Digestion of P39 with AgeI & PstI = F52 | Digestion of F45 with XbaI & PstI = F51 |
| as expected, lower band was cut out | as expected, band was cut out | as expected, lower band was cut out |
- Bands were extracted by QIAquick Gel Extraction Kit, QIAGEN.
Quick Change mutagenesis to remove forbidden restriction sites - SDMIV - of P179 (Laccase)
Investigator: Andi, Jeff
Aim of the experiment: Removal of forbidden restiction site SDM IV from P179 (Laccase).
Procedure: Quickchange - PCR
Reaction batch
| volume | reagent | Additional information |
| 2.5 µl | 10x Pfu Ultra II buffer | |
| 1 µl | Plasmid template (P179 - 1:6 dilution) | need to get between 5 and 50 ng/µl |
| 1 µl | 1:10 dilution of O32 (for P179, 10 µM) | |
| 1 µl | 1:10 dilution of O33 (for P179, 10 µM) | |
| 18 µl | ddH2O | |
| 1 µl | dNTP mix 50mM | |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
PCR cycling parameters - P179
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 30 s |
| 2 | 19 | 95 °C | 30 s |
| 52 °C | 1 min | ||
| 68 °C | 4 min | ||
| 4 °C | hold |
After the PCR; 1 µl of Dpn1 was added and the reaction mixture was incubated at 37°C for one hour
Preparative digestion and gelelectrophoresis of P182 (Gensynthesis 1) with XbaI and AgeI-HF, P183 (Gensynthesis 2) with XbaI and AgeI-HF
Investigator: Andi, Jeff
Aim of the experiment: Preparative digestion and gelelectrophoresis of P182 (Gensynthese 1) with XbaI and AgeI-HF, P183 (Gensynthese 2) with XbaI and AgeI-HF
Procedure:
- Batch for preparative digestion of P182 with XbaI and AgeI-HF
| volume | reagent |
| 15 µl | Plasmid DNA P182 |
| 4 µl | CutSmart Buffer |
| 1 µl | AgeI-HF(20 U/µl) |
| 1 µl | XbaI (20 U/µl) |
| 19 µl | ddH2O |
| =40 µl | TOTAL |
- Batch for preparative digestion of P183 with XbaI and AgeI- HF
| volume | reagent |
| 15 µl | Plasmid DNA P183 |
| 4 µl | CutSmart Buffer |
| 1 µl | AgeI-HF (20 U/µl) |
| 1 µl | XbaI (20 U/µl) |
| 19 µl | ddH2O |
| =40 µl | TOTAL |
- Incubation for 2.5 h at 37 °C.
- 4 µl of DNA loading buffer (10x) were added to the 40 µl reaction batches after digestion and were loaded on a 1.5% agarose gel for preparative gelelectrophoresis.
- Preparative gelelectrophoresis was performed at 70 V for 60 min.
| 1 kbp DNA ladder | Digestion of Gensynthesis 1 with XbaI & AgeI | 100 bp DNA ladder | Digestion of Gensynthesis 2 with XbaI & AgeI | 1 kbp DNA ladder |
| as expected, fragments (78 bp and 530 bp) were cut out | as expected, fragments (111 bp, 206 bp and 359 bp) were cut out |
- Bands were extracted by QIAquick Gel Extraction Kit, QIAGEN.
Preparative digestion and gelelectrophoresis of P9 with XbaI and AgeI-HF
Investigator: Andi, Jeff
Aim of the experiment: Preparative digestion and gelelectrophoresis of P9 with XbaI and AgeI-HF
Procedure:
- Batch for preparative digestion of P9 with XbaI and AgeI-HF
| volume | reagent |
| 15 µl | Plasmid DNA P9 |
| 4 µl | CutSmart Buffer |
| 1 µl | AgeI-HF(20 U/µl) |
| 1 µl | XbaI (20 U/µl) |
| 19 µl | ddH2O |
| =40 µl | TOTAL |
- Incubation for 2.5 h at 37 °C.
- The resulting fragments were named: ?????
- 4 µl of DNA loading buffer (10x) were added to the 40 µl reaction batches after digestion and were loaded on a 1% agarose gel for preparative gelelectrophoresis.
- Preparative gelelectrophoresis was performed at 70 V for 60 min.
- Gelfragmentation was like expected, both bands were cut out (upper band = PhyB, lower band = pSB1C3 RFC25)
- Bands were extracted by QIAquick Gel Extraction Kit, QIAGEN.
Transformation of E. coli XL1 blue with Quickchange product SDM IV of P179 (Laccase)
Investigator: Andi, Jeff
Aim of the experiment: Transformation of E. coli XL1 blue with Quickchange product SDM IV of P179 (Laccase).
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 2x 10 µl of DNA was added seperately into 2x 200 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of the cell suspension was plated on one chloramphenicol plate.
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellets were resuspended in 100 µl of LB-medium and these concentrated cell suspensions were plated again on new chlorampenicol plates.
Transformation of E. coli XL1 blue with P39
Investigator: Louise, Johanna
Aim of the experiment: Transformation of E. coli XL1 blue with P39
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 10 µl of DNA was added seperately into 200 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of the cell suspension was plated on one chloramphenicol plate.
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellets were resuspended in 100 µl of LB-medium and these concentrated cell suspensions were plated again on new chlorampenicol plates.
Preparative digestion and gelelectrophoresis of P39 with AgeI & PstI, P20 with NgoMIV & PstI
Investigator: Johanna, Louise
Aim of the experiment:Preparative digestion of P39 with AgeI & PstI, P20 with NgoMIV & PstI to fusion Middle linker of P39 with GFP of P20.
Procedure:
Batch for preparative digestion of P39 (Middle-Linker) with PstI and AgeI.
| volume | reagent |
| 20 µl | P39 |
| 4 µl | NEBuffer 4 |
| 0.4 µl | BSA (100x) |
| 2 µl | AgeI-HF |
| 2 µl | PstI |
| 11.6 µl | ddH2O |
| =40 µl | TOTAL |
Batch for preparative digestion of P20 (GFP-Psb1C3) with NgoMIV and PstI.
| volume | reagent |
| 20 µl | P45 |
| 4 µl | NEBuffer 4 |
| 0.4 µl | BSA (100x) |
| 2 µl | NgoMIV |
| 2 µl | PstI |
| 11.6 µl | ddH2O |
| =40 µl | TOTAL |
- Incubation for 150 min at 37 °C.
- Preparative gelelectrophoresis was performed at 90 V for 1 h.
| 1 kbp ladder | P20 digested with NgoMIV&PstI (=F30) | P39 digested with NgoMIV&PstI (=F30) |
| lower band was cut out | band was cut out |
- Bands were extracted by QIAquick Gel Extraction Kit, QIAGEN.
Ligation of F58+F59, F55+F59, F54+F59, F56+F59, F57+F59, F52+F53, F42+F51
Investigator: Jeff, Andi, Johanna, Louise
Aim of the experiment: Ligation of F53+F59, F58+F59, F55+F59, F54+F59, F56+F59, F57+F59, F52+F53, F42+F51.
Procedure:
- Ligation batch for F54+F59 :
| volume | reagent |
| 2.2 µl | F54 (5.9 ng/µl, 90 bp) |
| 1.5 µl | F59 (66.7 ng/µl, 2100 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 13.3 µl | ddH2O |
| =20 µl | TOTAL |
- Ligation batch for F55+F59:
| volume | reagent |
| 11.6 µl | F55 (6.4 ng/µl, 520 bp) |
| 1.5 µl | F59 (66.7 ng/µl, 2100 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 3.9 µl | ddH2O |
| =20 µl | TOTAL |
- Ligation batch for F56+F59:
| volume | reagent |
| 3.3 µl | F56 (4.7 ng/µl, 110 bp) |
| 1.5 µl | F59 (66.7 ng/µl, 2100 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 12.2 µl | ddH2O |
| =20 µl | TOTAL |
- Ligation batch for F57+F59:
| volume | reagent |
| 3.1 µl | F57 (9.1 ng/µl, 200 bp) |
| 1.5 µl | F59 (66.7 ng/µl, 2100 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 12.4 µl | ddH2O |
| =20 µl | TOTAL |
- Ligation batch for F58+F59:
| volume | reagent |
| 4.4 µl | F58 (12.9 ng/µl, 400 bp) |
| 1.5 µl | F59 (66.7 ng/µl, 2100 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 11.1 µl | ddH2O |
| =20 µl | TOTAL |
- Negative Control Ligation batch for F59:
| volume | reagent |
| 1.5 µl | F59 (66.7 ng/µl, 2100 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 15.5 µl | ddH2O |
| =20 µl | TOTAL |
- Ligation batch for F52+F53:
| volume | reagent |
| 10 µl | F52 (9.9 ng/µl, XXX bp) |
| 7 µl | F53 (12.2 ng/µl, XXX bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- Ligation batch for F42+F51:
| volume | reagent |
| 1.36 µl | F42 (XXX ng/µl, XXX bp) |
| 15.64 µl | F51 (8.2 ng/µl, 1237 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- Negative control was also prepared. Instead of the insert, the same volume of ddH2O was added to the reaction batch.
- Cycled ligation has been performed after following protocol in a thermocycler:
| 99 cycles | 12 °C | 60 s |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| Hold | 16 °C | infinite |
- Lid temperature = 37 °C
Plating of received E. coli containing biobricks
Investigator: Jeff, Andreas
Aim of the experiment: The received biobricks were already transformed in E. coli and were in an agar stabs. These E. coli cells were transferred with an inoculation loop on antibiotic selection plates and were incubated over night.
Operational sequence:
- Bacterias containing plasmids with biobricks were transferred with a sterile inoculation loop on antibiotic plates and were incubated at 37 °C overnight.
- The biobricks were:
| Name | Function |
|---|---|
| BBa_J61032 | Alkaline Phosphatase |
| BBa_K426020 | Self lysis device |
| BBa_K729004 | Nuclease from S. Aureus |
| BBa_K365001 | PIF6 |
| BBa_K648011 | RBS B0034 with cI repressor C0051 under the control of constitutive promoter J23113 |
Wednesday, June 5th
Transformation of E. coli XL1 blue transformed with F59+F54, F59+F55, F59+F56, F59+F57, F59+F58, F42+F51 and F52+F53
Investigator: Florian, Rosario, Louise, Jeff
Aim of the experiment: Transformation of E. coli XL1 blue transformed with F59+F54, F59+F55, F59+F56, F59+F57, F59+F58, F42+F51 and F52+F53.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 10 µl of DNA was added to 200 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- The cell suspensions were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellets were resuspended in 100 µl of LB-medium and these concentrated cell suspensions were plated again on plates containing antibiotics.
Picking of of E. coli XL1 blue transformed with BBa_K365001 (PIF6), BBa_K729004 (Nuclease from Staphylococcus aureus), BBa_K648011 (XylE RFC25), BBa_J61032 (Alkaline phosphotase), BBa_K426020 (BBa_K426020)
Investigator: Rosario, Florian, Louise, Jeff
Aim of the experiment: Picking of of E. coli XL1 blue transformed with BBa_K365001 (PIF6), BBa_K729004 (Nuclease from Staphylococcus aureus), BBa_K648011 (XylE RFC25), BBa_J61032 (Alkaline phosphotase), BBa_K426020 (BBa_K426020).
Procedure:
- Picked pipette tips was transferred into cell-culture tubes with air-permeable, sterile cover. Each tube contain 4 mL of LB-medium + 4 µL chloramphenicol(1000x) or ampicillin(1000x).
- 2 colonies of each plate were picked.
- These tubes were transferred in a cell culture shaker at 37 °C and were incubated overnight.
Thursday, June 6th
Miniprep of overnight cultures of transformated E. coli XL1 blue with P39, P20, genesynthesis 1&2 and BBa_J61032, BBa_K426020, BBa_K729004, BBa_K365001, BBa_K648011
Investigator: Ingmar
Aim of the experiment:Miniprep of overnight cultures of transformated E. coli XL1 blue with P39, P20, genesynthesis 1&2 and BBa_J61032, BBa_K426020, BBa_K729004, BBa_K365001, BBa_K648011
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
- The resulting DNA were eluted in tubes labeled as follows:
| Content | New Tube |
| P39 | P184 |
| P39 | P185 |
| P20 | P186 |
| P20 | P187 |
| BBa_J61032 | P188 |
| BBa_J61032 | P189 |
| BBa_K426020 | P190 |
| BBa_K426020 | P191 |
| BBa_K729004 | P192 |
| BBa_K729004 | P193 |
| BBa_K365001 | P194 |
| BBa_K365001 | P195 |
| BBa_K648011 | P196 |
| BBa_K648011 | P197 |
| Genesynthesis 2 | P198 |
| Genesynthesis 2 | P199 |
| Genesynthesis 1 | P200 |
| Genesynthesis 1 | P201 |
Analytical digestion and gelelectrophoresis of P188-P197 (latest received biobricks: BBa_J61032, BBa_K426020, BBa_K729004, BBa_K365001, BBa_K648011)
Investigator: Jeff
Aim of the experiment: Analytical digestion and gelelectrophoresis of P188-P197 (latest received biobricks: BBa_J61032, BBa_K426020, BBa_K729004, BBa_K365001, BBa_K648011).
Procedure:
| volume | reagent |
| 2 µl | CutSmart Buffer (10x) |
| 0.25 µl | EcoRI-HF |
| 0.25 µl | SpeI-HF |
| 15 µl | ddH2O |
| 2.5 µl | Plasmid |
| =20 µl | TOTAL |
Mastermix (12x)
| volume | reagent |
| 24 µl | CutSmart Buffer (10x) |
| 3 µl | EcoRI-HF |
| 03 µl | SpeI-HF |
| 180 µl | ddH2O |
| =210 µl | TOTAL |
- 17.5 µl Mastermix + 2.5 µl Plasmid each
- Incubation for 90 min at 37 °C.
- Analytical gelelectrophoresis was performed at 90 V
| 1 kb Marker | P188 Alk.Phos. | P189 Alk.Phos. | P190 self lysis device | P191 self lysis device | P192 Nuclease | P193 Nuclease | P194 PIF6 | P195 PIF6 | P196 RBS | P197 RBS |
| successful | successful | successful | successful | successful | successful | successful | successful | successful | successful |
Picking of of E. coli XL1 blue transformed with the ligation products of F52+F53, F59+F58, F59+F54, F59+F55, F59+F57 and F59+F56
Investigator: Ingmar
Aim of the experiment: Picking of of E. coli XL1 blue transformed with the ligation products of F52+F53, F59+F58, F59+F54, F59+F55, F59+F57 and F59+F56
Procedure:
- The picked colonies were transferred into cell-culture tubes with air-permeable, sterile cover containing 4 mL of LB-medium + 4 µL chloramphenicol(1000x) or ampecillin (1000x).
- 2 colonies of each agar plate were picked.
- These tubes were transferred in a cell culture shaker at 37 °C and were incubated overday.
- The ligation of F42 and F51 was not successful and has to be repeated. One reason for the failure could be secondary structures of the promoter. Therefore the new ligation batch should be heated to 95°C for a while before adding the T4 ligase.
PCR of P192 (Thermonuclease, O64 & O65) and P194 (PIF6, O42 & O43)
Investigator: Jeff
Aim of the experiment: PCR of P192 (Thermonuclease, O64 & O65) and P194 (PIF6, O42 & O43).
Procedure:
Operational sequence:
- PCR reaction mixture
| volume | reagent |
| 10 µl | 5x OneTaq Standard Reaction Buffer |
| 1 µl | 10 mM dNTPs |
| 1 µl | 10 µM Forward Primer (P192: O64 (NucA_fw); P194: O42 (PIF6_fw)) |
| 1 µl | 10 µM Reverse Primer (P192: O65 (NucA_rv); P194: O43 (PIF6_rv)) |
| 0.25 µL | OneTaq Hot Start DNA Polymerase (Finally: 1.25 units/50 µL) |
| 1 µl | 1:1000 dilution of plasmid DNA (P192; P194) |
| 35.75 µL | ddH2O Water |
| =50 µL | TOTAL |
- Mix with pipette
- The gradient PCR program was performed after following scheme with following conditions:
| Initial denaturation | 94 °C | 30 s |
| 30 cycles | 94 °C | 30 s |
| 55 °C | 60 s | |
| 68 °C | 30 s | |
| Final extension | 68 °C | 5 min |
| Hold | 4 °C | infinite |
- After PCR, the product was purified with QIAquick PCR Purification Kit, Qiagen.
- PCR product of P192 = F61; PCR product of 194 = F62
QuikChange mutagenesis to remove forbidden restriction sites - SDMIV - of P179 (Laccase)
Investigator: Jeff
Aim of the experiment: Removal of forbidden restiction site from P179 (Laccase).
Procedure: Quickchange - PCR
Reaction batch
| volume | reagent | Additional information |
| 2.5 µl | 10x Pfu Ultra II buffer | |
| 0.5 µl | Plasmid template (P179 - 1:10 dilution) | need to get between 2.5 ng and 25 ng for a reaction batch of 25 µl |
| 1 µl | 1:10 dilution of O32 (10 µM) | |
| 1 µl | 1:10 dilution of O33 (10 µM) | |
| 18.5 µl | ddH2O | |
| 1 µl | dNTP mix 50 mM | |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U/µl) |
PCR cycling parameters - P179
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 30 s |
| 2 | 19 | 95 °C | 30 s |
| 52 °C | 1 min | ||
| 68 °C | 4 min | ||
| 4 °C | hold |
After the PCR; 1 µl of Dpn1 was added and the reaction mixture was incubated at 37°C for one hour
Analytical gelelectrophoresis of F61 (PCR product of P192, Thermonuclease), F62 (PCR product of P194, PIF6) and the DpnI digestion of the SDMIV product of P179
Investigator: Rosario, Andi, Jeff
Aim of the experiment: Analytical gelelectrophoresis of F61 (PCR product of P192, Thermonuclease), F62 (PCR product of P194, PIF6) and the DpnI digestion of the SDMIV product of P179.
Procedure:
- 4 µl of F61 or F62 or the DpnI digestion of the SDMIV product of P179 were mixed with 0.444 µl of DNA loading buffer (10x)
- Analytical gelelectrophoresis was performed at 90 V for 1 h on a 1% agarose gel.
| 1 kbp DNA ladder | PCR product F61 | PCR product F62 | DpnI digestion of QCIV of P179 |
| successful | successful | successful |
Ligation of F42+F51
Investigator: Ingmar
Aim of the experiment: Ligation of F42+F51.
Procedure:
- Ligation batch for F42+F51:
| volume | reagent |
| 0.68 µl | F42 (73.5 ng/µl, 2070 bp) |
| 15.64 µl | F51 (10.93 ng/µl, 1237 bp) |
| 5.39 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- Negative control was also prepared. Instead of the insert, the same volume of ddH2O was added to the reaction batch.
- The reaction batch was heated up to 95°C for 5 min before the enzyme and buffer were added.
- The ligation was performed for two hours at room temperature.
- Lid temperature = 37 °C
Transformation of E. coli XL1 blue with F42+F51 (pActin in pSB1C3 RFC10) and DpnI digestion of the SDMIV product of P179 (Laccase)
Investigator: Katrin
Aim of the experiment: Transformation of E. coli XL1 blue with F42+F51 (pActin in pSB1C3 RFC10 )and DpnI digestion of the SDMIV product of P179 (Laccase).
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 10 µl of DNA was added to 200 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- The cell suspensions were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellets were resuspended in 100 µl of LB-medium and these concentrated cell suspensions were plated again on plates containing antibiotics (CamR).
Sequencing of Biobricks P188/189 (alk.Phos., 1475 bp), P190/191 (self lysis device, 2910 bp), P192/193 (Nuclease, 561 bp), P194/195 (PIF6, 300 bp), P196/197 (RBS, 918 bp)
Investigators: Johanna
Aim of the experiment: Sequencing of the Biobrick MiniPreps
Procedure: Peparation of DNA for sequencing according to the Eurofins SmartSeq Kit protocol (15 µl of plasmid DNA (50 - 100 ng/µl) and 2 µl sequencing primer 10 µM):
| Label | Konz. ng/µl | Vol. BB µl | Vol H2O µl |
|---|---|---|---|
| P188 | 160.6 | 5 | 10 |
| P189 | 123.6 | 7.5 | 7.5 |
| P190 | 319.2 | 3 | 12 |
| P191 | 127.0 | 10 | 5 |
| P192 | 336.9 | 3 | 12 |
| P193 | 230.6 | 4 | 11 |
| P194 | 156.6 | 5 | 10 |
| P195 | 197.4 | 4 | 11 |
| P196 | 90.3 | 10 | 5 |
| P197 | 230.2 | 4 | 11 |
The different genes received the following barcodes:
| Label | Name | Barcode1 | Barcode2 |
|---|---|---|---|
| P188 | alk.Phos. | FR01002327 (O3) | FR01002326 (O4) |
| P189 | alk.Phos. | FR01002325 (O3) | FR01002320 (O4) |
| P190 | self lysis device | FR01002319 (O3) | FR01002318 (O4) |
| P191 | self lysis device | FR01002317 (O3) | FR01002316 (O4) |
| P192 | Nuclease | FR01002315 (O3) | - (O4) |
| P193 | Nuclease | FR01002314 (O3) | - (O4) |
| P194 | PIF6 | FR01002313 (O3) | - (O4) |
| P195 | PIF6 | FR01002312 (O3) | - (O4) |
| P196 | RBS | FR01002324 (O3) | FR01002323 (O4) |
| P197 | RBS | FR01002322 (O3) | FR01002321 (O4) |
Preparative digestion and gelelectrophoresis of F61 (Thermonuclease), F62 (PIF6) , P165 (t35S), F31 (miniMCSII)
Investigator: Rosario, Andi, Florian, Johanna, Jeff
Aim of the experiment: Preparative digestion and gelelectrophoresis of F61 (Thermonuclease), F62 (PIF6) , P165 (t35S), F31 (miniMCSII.
Procedure:
- Batch for preparative digestion of F61 (Thermonuclease) with XbaI & AgeI:
| volume | reagent |
| 25 µl | PCR product F61 |
| 5 µl | CutSmart Buffer |
| 1 µl | XbaI(20 U/µl) |
| 1 µl | AgeI-HF (20 U/µl) |
| 18 µl | ddH2O |
| =50 µl | TOTAL |
- Batch for preparative digestion of F62 (PIF6) with XbaI & AgeI:
| volume | reagent |
| 25 µl | PCR product F62 |
| 5 µl | CutSmart Buffer |
| 1 µl | XbaI(20 U/µl) |
| 1 µl | AgeI-HF (20 U/µl) |
| 18 µl | ddH2O |
| =50 µl | TOTAL |
- Batch for preparative digestion of P165 (t35S) with XbaI:
| volume | reagent |
| 20 µl | Plasmid DNA P165 |
| 4 µl | CutSmart Buffer |
| 1 µl | XbaI (20 U/µl) |
| 15 µl | ddH2O |
| =40 µl | TOTAL |
- Batch for preparative digestion of F31 (miniMCSII):
| volume | reagent |
| 10 µl | Oligohybridization F31 |
| 5 µl | CutSmart Buffer |
| 1 µl | SpeI-HF (20 U/µl) |
| 34 µl | ddH2O |
| =40 µl | TOTAL |
- Incubation for 3 h at 37 °C.
- 5 µl of DNA loading buffer (10x) were added to the 50 µl reaction batches and 4 µl of DNA loading buffer (10x) were added to the 40 µl reaction batches after digestion and were loaded on an 2% agarose gel for preparative gelelectrophoresis (1% agarose gel for digestion of P165).
- Preparative gelelectrophoresis was performed at 90 V for 60 min.
| 1 kbp DNA ladder | Digestion of F31 with SpeI-HF | Digestion of F61 with XbaI & AgeI-HF | Digestion of F62 with XbaI & AgeI-HF |
| Fragment length as expected; band was cut out | Fragment length as expected; band was cut out | Fragment length as expected; band was cut out |
| 1 kbp DNA ladder | Digestion of P165 with XbaI |
| Fragment length as expected; band was cut out |
- Bands were extracted by QIAquick Gel Extraction Kit, QIAGEN.
Dephosphorylation of preparative digestion of P165 with FastAP
Investigator: Rosario, Jeff
Aim of the experiment: Dephosphorylation of preparative digestion of P165 with FastAP to prevent re-ligation.
Procedure:
- After preparative digestion, gelelectrophoresis and gelextraction of P165, the digestion product was purified with QIAquick PCR Purification Kit, Qiagen.
- Batch for the dephosphorylation of digestion product of P165
| volume | reagent |
| 30 µl | Digestion product of P165 |
| 4 µl | 10X Fast AP buffer |
| 2 µl | FastAP Thermosensitive Alkaline Phosphatase |
| 4 µl | ddH2O |
| =40 µl | TOTAL |
- The reaction batch was spun briefly and incubated at 37 °C for 10 min.
- The reaction was stopped by heating to 75 °C for 5 min.
- The dephosphorylized digestion product of P165 product was purified with QIAquick PCR Purification Kit, Qiagen.
- The resulting product was labelled as F63
Ligation of F58+F59, F55+F59, F54+F59, F56+F59, F57+F59, F52+F53, F42+F51
Investigator: Jeff, Rosario
Aim of the experiment: Ligation of F53+F59, F58+F59, F55+F59, F54+F59, F56+F59, F57+F59, F52+F53, F42+F51.
Procedure:
- Ligation batch for F59+F65 (Thermonuclease in pSB1C3 RFC25):
| volume | reagent |
| 1.5 µl | F59 (66.7 ng/µl, 2086 bp) |
| 4.56 µl | F65 (14.1 ng/µl, 447 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 10.94 µl | ddH2O |
| =20 µl | TOTAL |
- Ligation batch for F59+F66:
| volume | reagent |
| 1.5 µl | F59 (66.7 ng/µl, 2086 bp) |
| 3.78 µl | F66 (11.3 ng/µl, 297 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 11.72 µl | ddH2O |
| =20 µl | TOTAL |
- Ligation batch for F63+F64:
| volume | reagent |
| 12.05 µl | F63 (8.3 ng/µl, 2287 bp) |
| 4.95 µl | F64 (4.3 ng/µl, 22 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- Negative control was also prepared. Instead of the insert, the same volume of ddH2O was added to the reaction batch.
- Ligation was performed at 16 °C overnight.
Friday, June 7th
Picking of QC IV Laccase
Investigator: Johanna
Aim of the study: Picking of QC IV Laccase (concentrated) and QC IV Laccase (diluted)
Operational sequence:
- Picking and overnight culture after standard laboratory's protocol. (4 µl CamR and 4 ml LB-medium)
- 3 colonies were picked for every Transformation product.
Preparation of Knop-Medium for Moss
Investigator: Andi
Aim of the experiment: Preparation of Knop-Medium for Moss
Procedure:
- All four prepared stocks have been: 25g/L KH2PO4; 25g/L KCl; 25g/L MgSO4 x 7 H2O; 100g/L Ca(NO3)2
- Volume of prepared Medium: 2.6L
- 10 ml of each stock were converted into a 5L Erlenmeyer flask and filled up to 2.6L with destilled water (ELGA); 32.5mg FeSO4 x 7 H2O were added; the pH was set to 5.8 with KOH/NaOH
- 200ml of the prepared medium was filled into three 500ml Erlenmeyer flasks, so each Erlenmeyer flask contained 200 ml of the Knop-Medium; the rest (2L) was left in the 5L EM flask
- All flasks have been autoclaved
Miniprep of overnight cultures of transformated E. coli XL1 blue with F52+F53, F59+F54, F59+F55, F59+F56, F59+F57, F59+F58
Investigator: Rosario
Aim of the experiment:Miniprep of overnight cultures of transformated E. coli XL1 blue with F52+F53, F59+F54, F59+F55, F59+F56, F59+F57, F59+F58
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
- The resulting DNA were eluted in tubes labeled as follows:
| Content | New Tube |
| F52+F53 I | P202 |
| F52+F53 II | 203 |
| F59+F54 I | P204 |
| F59+F54 II | P205 |
| F59+F55 I | P206 |
| F59+F55 II | P207 |
| F59+F56 I | P208 |
| F59+F56 II | P209 |
| F59+F57 I | P210 |
| F59+F57 II | P211 |
| F59+F58 I | P212 |
| F59+F58 II | P213 |
Transformation of E. coli XL1 blue with F59 + F65 (Thermonuclease in pSB1C3), F59 + F66 (PIF6 in pSB1C3) and F63 + F64 (MiniMCS + t35S in pSB1C3)
Investigator: Florian
Aim of the experiment: Transformation of E. coli XL1 blue with F59 + F65 (Thermonuclease in pSB1C3), F59 + F66 (PIF6 in pSB1C3) and F63 + F64 (MiniMCS + t35S in pSB1C3).
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 10 µl of ligation product were added to 200 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of each tube were plated on agarose plates containing antibiotics (CamR).
- The cell suspensions were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellets were resuspended in 100 µl of LB-medium and these concentrated cell suspensions were plated again on plates containing antibiotics (CamR).
Sequencing of P204 (Ligation Product F59+F54), P206 (Ligation Product F59+F55), P208 (Ligation Product F59+F56), P210 (Ligation Product F59+F57), P212 (Ligation Product F59+F58)
Investigators: Louise
Aim of the experiment: Sequencing of Ig-Kappa (P204), Nanoluciferase (P206), SigP (P208), Transmembrand. (P210) and SpyCatcher (P212) Ligations in pSB1C3 RFC25
Procedure: The DNA was prepared for sequencing according to the Eurofins SmartSeq Kit protocol (15 µl of plasmid DNA (50 - 100 ng) and 2 µl sequencing primer).
The different genes we sequenced received the following barcodes:
| Label | Name | Barcode (O3) |
|---|---|---|
| P204 | Ig-Kappa-SigP + pSB1C3 RFC25 | FR01002304 |
| P206 | Nanoluciferase + pSB1C3 RFC25 | FR01002303 |
| P208 | SERK-SigP + pSB1C3 RFC25 | FR01002302 |
| P210 | Transmembrand + pSB1C3 RFC25 | FR01002301 |
| P212 | SpyCatcher + pSB1C3 RFC25 | FR01002300 |
Analytical digestion and gelelectrophoresis of P202-P213
Investigator: Katrin
Aim of the experiment: Analytical digestion and gelelectrophoresis of P202-P213
Procedure:
- Mastermix for analytical digestion of P202-P213 with XbaI+AgeI
| volume | reagent |
| 30 µl | CutSmart Buffer (10x) |
| 3.75 µl | XbaI |
| 3.75 µl | SpeI-HF |
| 225 µl | ddH2O |
| =262.5 µl | TOTAL |
- 17.5 µl of the mastermix was added to 2.5 µl of plasmid DNA (P202-P203)
- Incubation for 90 min at 37 °C.
- Inaddition, P203 was digested for 4 and for 8 seconds in the microwave, a negative control was incubated at room temperature
- Analytical gelelectrophoresis was performed at 90 V for 60 min.
| 1 kbp ladder DNA ladder | Digestion of P202 with XbaI und AgeI (GFP + linker) | Digestion of P203 with XbaI und AgeI (GFP + linker) | Digestion of P204 with XbaI und AgeI | Digestion of P205 with XbaI und AgeI | Digestion of P206 with XbaI und AgeI | Digestion of P207 with XbaI und AgeI | Digestion of P208 with XbaI und AgeI | Digestion of P209 with XbaI und AgeI | Digestion of P210 with XbaI und AgeI |
| as expected | as expected | not as expected | not as expected | not as expected | not as expected | not as expected | not as expected | as expected (SERK-TMD) |
| 1 kbp ladder DNA ladder | Digestion of P211 with XbaI und AgeI | Digestion of P212 with XbaI und AgeI | Digestion of P213 with XbaI und AgeI | Digestion of P203 with XbaI und AgeI (microwave 4 seconds) | Digestion of P203 with XbaI und AgeI (microwave 8 seconds) | Digestion of P203 with XbaI und AgeI (control) |
| as expected (SERK-TMD) | not as expected | as expected | interesting | interesting | interesting |
Saturday, June 8th
Miniprep of overnight cultures of transformated E. coli XL1 blue QC IV Laccase
Investigator: Rosario
Aim of the experiment:Miniprep of overnight cultures of transformated E. coli XL1 blue QC IV Laccase
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
- The resulting DNA were eluted in tubes labeled as follows:
| Content | New Tube |
| QC IV Laccase clone 1 | P214 |
| QC IV Laccase clone 2 | P215 |
| QC IV Laccase clone 3 | P216 |
| QC IV Laccase clone 4 | P217 |
| QC IV Laccase clone 5 | P218 |
| QC IV Laccase clone 6 | P219 |
Picking of F59 + F65 (Thermonuclease in pSB1C3), F59 + F66 (PIF6 in pSB1C3) and F63 + F64 (MiniMCS + t35S in pSB1C3)
Investigator: Rosario
Aim of the study: Picking of F59 + F65 (Thermonuclease in pSB1C3), F59 + F66 (PIF6 in pSB1C3) and F63 + F64 (MiniMCS + t35S in pSB1C3) Operational sequence:
- Picking and overnight culture after standard laboratory's protocol. (4 µl CamR and 4 ml LB-medium)
- 2 colonies were picked for every Transformation product.
Preparative digestion and gelelectrophoresis of F47 (pActin, fragment named F68) with XbaI/SpeI, P202 (GFP+linker, fragment named F69) with NgoMIV/SpeI, P210 (SERK-TMD, fragment named F67)
Investigator: Jeff, Katrin
Aim of the experiment: Preparative digestion and gelelectrophoresis of F47 (pActin, fragment named F68) with XbaI/SpeI, P202 (GFP+linker, fragment named F69) with NgoMIV/SpeI, P210 (SERK-TMD, fragment named F67) with AgeI/SpeI
Procedure:
- Batch for preparative digestion of F47 (pActin, fragment named F68) with XbaI/SpeI
| volume | reagent |
| 25 µl | PCR product F47 |
| 5 µl | CutSmart Buffer |
| 1 µl | XbaI(20 U/µl) |
| 1 µl | SpeI-HF (20 U/µl) |
| 18 µl | ddH2O |
| =50 µl | TOTAL |
- Batch for preparative digestion of P202 (GFP+linker, fragment named F69) with NgoMIV/SpeI
| volume | reagent |
| 20 µl | plasmid P202 |
| 5 µl | CutSmart Buffer |
| 1 µl | SpeI-HF(20 U/µl) |
| 1 µl | NgoMIV (20 U/µl) |
| 23 µl | ddH2O |
| =50 µl | TOTAL |
- Batch for preparative digestion of P210 (SERK-TMD, fragment named F67) with AgeI/SpeI
| volume | reagent |
| 20 µl | Plasmid DNA P210 |
| 5 µl | CutSmart Buffer |
| 1 µl | AgeI-HF (20 U/µl) |
| 1 µl | SpeI-HF (20 U/µl) |
| 23 µl | ddH2O |
| =50 µl | TOTAL |
- Incubation for 3 h at 37 °C.
- 5 µl of DNA loading buffer (10x) were added to the 50 µl reaction batches after digestion and were loaded on a 1% agarose gel for preparative gelelectrophoresis
- Preparative gelelectrophoresis was performed at 90 V for 60 min.
| 1 kbp DNA ladder | digestion of F47 (pActin, fragment named F68) with XbaI/SpeI |
| Fragment length as expected; band was cut out |
| 1 kbp DNA ladder | digestion of P202 (GFP+linker, fragment named F69) with NgoMIV/SpeI |
| Fragment length as expected; band was cut out |
| 1 kbp DNA ladder | digestion of P210 (SERK-TMD, fragment named F67) with AgeI/SpeI |
| Fragment length as expected; band was cut out |
- Bands were extracted by QIAquick Gel Extraction Kit, QIAGEN.
- Concentration determined with Nanodrop:
| F68 | pActin | 10,4 ng/µl |
| F69 | GFP+linker | 13,0 ng/µl |
| F67 | SERK-TMD | 14,3 ng/µl |
Preparative digestion and gelelectrophoresis of P7 (PhyB in pSB1C3, fragment named F75) with XbaI/SpeI and P8 (PhyB in pSC1C3, fragment named F76) with NgoMIV/SpeI
Investigator: Jeff, Katrin
Aim of the experiment: Preparative digestion and gelelectrophoresis of P7 (PhyB in pSB1C3, fragment named F75) with XbaI/SpeI and P8 (PhyB in pSC1C3, fragment named F76) with NgoMIV/SpeI (backbones for ligation)
Procedure:
- Batch for preparative digestion of P7 (PhyB in pSB1C3, fragment named F75) with XbaI/SpeI
| volume | reagent |
| 20 µl | Plasmid P7 |
| 5 µl | CutSmart Buffer |
| 1 µl | XbaI(20 U/µl) |
| 1 µl | SpeI-HF (20 U/µl) |
| 23 µl | ddH2O |
| =50 µl | TOTAL |
- Batch for preparative digestion of P8 (PhyB in pSC1C3, fragment named F76) with NgoMIV/SpeI
| volume | reagent |
| 20 µl | plasmid P8 |
| 5 µl | CutSmart Buffer |
| 1 µl | SpeI-HF(20 U/µl) |
| 1 µl | NgoMIV (20 U/µl) |
| 23 µl | ddH2O |
| =50 µl | TOTAL |
- Incubation for 3 h at 37 °C.
- before gelelectrophoresis, P7 was dephosphorylated
- 5 µl of DNA loading buffer (10x) were added to the 50 µl reaction batches after digestion and were loaded on a 0.5% agarose gel for preparative gelelectrophoresis
- Preparative gelelectrophoresis was performed at 90 V for 60 min.
| 1 kbp DNA ladder | digestion of P7 (PhyB in pSB1C3, fragment named F75) with XbaI/SpeI | digestion of P8 (PhyB in pSC1C3, fragment named F76) with NgoMIV/SpeI |
| Fragment lengths as expected; lower band was cut out | Fragment lengths as expected; lower band was cut out |
- Bands were extracted by QIAquick Gel Extraction Kit, QIAGEN.
- Concentration determined with Nanodrop:
| F75 | dephosphorylated Backbone (P7) | 39,4 ng/µl |
| F76 | dephosphorylated Backbone (P8) | 52,0 ng/µl |
Dephosphorylation of F75
Investigator: Jeff, Katrin
Aim of the experiment: Dephosphorylation of cut vector pSB1C3 prevent re-ligation
Procedure:
- the digestion product was purified with QIAquick PCR Purification Kit, Qiagen in order to change the buffer
- Batch for the dephosphorylation of F75
| volume | reagent |
| 30 µl | Plasmid DNA F75 from digestion |
| 3 µl | 10X Fast AP buffer |
| 1.3 µl | FastAP Thermosensitive Alkaline
Phosphatase |
| =34.3 µl | TOTAL |
- the reaction batch was spun briefly and incubated at 37 °C for 10 minutes
- the reaction was stopped by heating to 75 °C for 5 minutes
Preparative digestion and gelelectrophoresis of P200 (gene synthesis 1, fragments named F70+F71) with XbaI/AgeI and P198 (gene synthesis 2, fragments named F72-F74) with XbaI/AgeI
Investigator: Jeff, Katrin
Aim of the experiment: Preparative digestion and gelelectrophoresis of P200 (gene synthesis 1, fragments named F70+F71) with XbaI/AgeI and P198 (gene synthesis 2, fragments named F72-F74) with XbaI/AgeI
Procedure:
- Batch for preparative digestion of P200 (gene synthesis 1, fragments named F70+F71) with XbaI/AgeI
| volume | reagent |
| 20 µl | Plasmid P7 |
| 5 µl | CutSmart Buffer |
| 1 µl | XbaI(20 U/µl) |
| 1 µl | AgeI-HF (20 U/µl) |
| 23 µl | ddH2O |
| =50 µl | TOTAL |
- Batch for preparative digestion of P198 (gene synthesis 2, fragments named F72-F74) with XbaI/AgeII
| volume | reagent |
| 20 µl | Plasmid P7 |
| 5 µl | CutSmart Buffer |
| 1 µl | XbaI(20 U/µl) |
| 1 µl | AgeI-HF (20 U/µl) |
| 23 µl | ddH2O |
| =50 µl | TOTAL |
- Incubation for 3 h at 37 °C.
- 5 µl of DNA loading buffer (10x) were added to the 50 µl reaction batches after digestion and were loaded on a 2% agarose gel for preparative gelelectrophoresis
- Preparative gelelectrophoresis was performed at 90 V for 60 min.
| 1 kbp DNA ladder | digestion of P200 (gene synthesis 1, fragments named F70+F71) with XbaI/AgeI | digestion of P198 (gene synthesis 2, fragments named F72-F74) with XbaI/AgeII |
| Fragment lengths as expected; the two lower bands were cut out (upper:F70, lower: F71) | Fragment lengths as expected; all bands were cut out (upper: F72, middle: F73, lower: F74) |
- Bands were extracted by QIAquick Gel Extraction Kit, QIAGEN.
- Concentrations measured with Nano-Drop
| Fragment | Name | Length | Concentration |
| F70 | Nanoluciferase | 510 bp | 9.2 ng/µl |
| F71 | IgKappa-SigP | 67 bp | 3.1 ng/µl |
| F72 | SpyCatcher | 339 bp | 10.8 ng/µl |
| F73 | SERK-TMD | 186 bp | 8.6 ng/µl |
| F74 | SERK-SigP | 100 bp | 4.8 ng/µl |
Analytical digestion and gelelectrophoresis of P214-219 (Miniprep of Laccase QC IV), P179 (control before QC IV)
Investigator: Jeff, Katrin
Aim of the experiment: Analytical digestion and gelelectrophoresis of P214-219 (Miniprep of Laccase QC IV), P179 (control before QC IV)
Procedure:
- Mastermix for analytical digestion of P214-219 (Miniprep of Laccase QC IV), P179 (control before QC IV)
- 17.5 µl of the mastermix was added to 2.5 µl of plasmid DNA (P214-219, P179)
- Incubation for 90 min at 37 °C.
- digestion for 4 x 10 seconds in the microwave (600 watt), with two minute breaks in between the microwaving
- Analytical gelelectrophoresis was performed at 90 V for 60 min.
| 1 kbp DNA ladder | digestion of P214 (Miniprep of Laccase QC IV clone 1) with NgoMVI/AgeI | digestion of P215 (Miniprep of Laccase QC IV clone 2) with NgoMVI/AgeI | digestion of P216 (Miniprep of Laccase QC IV clone 3) with NgoMVI/AgeI | digestion of P217 (Miniprep of Laccase QC IV clone 4) with NgoMVI/AgeI | digestion of P218 (Miniprep of Laccase QC IV clone 5) with NgoMVI/AgeI | digestion of P219 (Miniprep of Laccase QC IV clone 6) with NgoMVI/AgeI | digestion of P179 (Miniprep of ligation product Laccase QCIII clone 2) with NgoMVI/AgeI |
| as expected | as expected | as expected | as expected | as expected | as expected | as expected |
Ligation of F75+F69 (Linker+GFP into pSB1C3) and F67+F69 (Linker+GFP into TMD-pSB1C3-Suffix)
Investigator: Jeff
Aim of the experiment: Ligation of F75+F69 (Linker+GFP into pSB1C3) and F67+F69 (Linker+GFP into TMD-pSB1C3-Suffix).
Procedure:
| volume | reagent |
| 2.54 µl | F75 (39.4 ng/µl, ? bp) |
| 8.30 µl | F69 (13.0 ng/µl, ? bp) |
| 6.16 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
| volume | reagent |
| 7.00 µl | F67 (14.3 ng/µl, ? bp) |
| 7.62 µl | F69 (13.0 ng/µl, ? bp) |
| 2.38 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- Incubation: 1 h 22°C
Transformation of E. coli XL1 blue with F75+F69 (Linker+GFP into pSB1C3) and F67+F69 (Linker+GFP into pSB1C3-TMD-Suffix)
Investigator: Jeff
Aim of the experiment: Transformation of E. coli XL1 blue with F75+F69(Linker+GFP into pSB1C3) and F67+F69 (Linker+GFP into pSB1C3-TMD-Suffix)
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 10 µl of DNA was added to 200 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- The cell suspensions were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellets were resuspended in 100 µl of LB-medium and these concentrated cell suspensions were plated again on plates containing antibiotics (CamR).
Sunday, June 9th
Miniprep of overnight cultures of transformated E. coli XL1 blue of F59+F65 (Nuclease), F59+F66 (PIF6), F59+F54 (Ig-K), F59+F55 (Luciferase), F59+F56 (SERK-SigP), F59+F57 (SERK-TMD), F59+F58 (SpyCatcher), F42+F51 (pActin), F63+F64
Investigator: Katrin
Aim of the experiment:Miniprep of overnight cultures of transformated E. coli XL1 blue of F59+F65 (Nuclease), F59+F66 (PIF6), F59+F54 (Ig-K), F59+F55 (Luciferase), F59+F56 (SERK-SigP), F59+F57 (SERK-TMD), F59+F58 (SpyCatcher), F42+F51 (pActin), F63+F64
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
- The resulting DNA were eluted in tubes labeled as follows:
| Content | New Tube |
| F59+F65 clone 1 | P220 |
| F59+F65 clone 2 | P221 |
| F59+F66 clone 1 | P222 |
| F59+F66 clone 2 | P223 |
| F59+F56 clone 1 | P224 |
| F59+F56 clone 2 | P225 |
| F59+F55 clone 1 | P226 |
| F59+F55 clone 2 | P227 |
| F59+F57 clone 1 | P228 |
| F59+F57 clone 2 | P229 |
| F59+F58 clone 1 | P230 |
| F59+F58 clone 2 | P231 |
| F63+F64 | P232 |
| F42+F51 | P233 |
| F59+F54 clone 1 | P234 |
| F59+F54 clone 2 | P235 |
Analytical digestion and gelelectrophoresis of P220-P235 ((P220-231, P233-P235 with XbaI/SpeI, P232 with MfeI/Pst1)
Investigator: Katrin
Aim of the experiment: Analytical digestion and gelelectrophoresis of P220-P235 (Minipreps)
Procedure:
- Mastermix for analytical digestion of P220-231, P233-P235 with XbaI, SpeI
| volume | reagent |
| 34 µl | CutSmart Buffer |
| 4.25 µl | XbaI(20 U/µl) |
| 4.25 µl | SpeI-HF (20 U/µl) |
| 255 µl | ddH2O |
| =266.5 µl | TOTAL |
- 17.5 µl of the mastermix was added to 2.5 µl of plasmid DNA
- reaction batch for analytical digestion of P232 with MfeI/PstI
| volume | reagent |
| 2.5 µl | P232 |
| 2 µl | CutSmart Buffer |
| 0.25 µl | MfeI(20 U/µl) |
| 0.25 µl | PstI-HF(20 U/µl) |
| 15 µl | ddH2O |
| =20 µl | TOTAL |
- Incubation for 60 min at 37 °C.
- Analytical gelelectrophoresis was performed at 90 V for 60 min
- for P224, P225, P228, P229, P230, P231, P234, P235,a 2% agarose gel was used, gelelectrophoresis was performed at 90 V for 45 min
| 1 kbp DNA ladder | 100 bp DNA ladder | digestion of P224 (SERK-SigP, clone 1) with XbaI, SpeI | digestion of P225 (SERK-SigP, clone 2) with XbaI, SpeI | digestion of P228 (SERK TMD, clone 1) with XbaI, SpeI | digestion of P229 (SERK TMD, clone 2) with XbaI, SpeI | digestion of P230 (SpyCatcher, clone 1) with XbaI, SpeI | digestion of P231 (SpyCatcher, clone 2) with XbaI, SpeI | digestion of P234 (Ig-K-SigP, clone 1) with XbaI, SpeI | digestion of P235 (Ig-K-SigP, clone 2) with XbaI, SpeI |
| not as expected | not as expected | as expected | as expected | as expected | as expected | fragment not visible due to small size | not as expected |
| 1 kbp DNA ladder | digestion of P220 (Thermonuclease, clone 1) with XbaI, SpeI | digestion of P221 (Thermonuclease, clone 2) with XbaI, SpeI | digestion of P222 (PIF6, clone 1) with XbaI, SpeI | digestion of P223 (PIF6, clone 2) with XbaI, SpeI | digestion of P226 (Luciferase, clone 1) with XbaI, SpeI | digestion of P227 (Luciferase, clone 2) with XbaI, SpeI | digestion of P232 (miniMCS) with MfeI/PstI | digestion of P233 (pActin) with XbaI, SpeI |
| not as expected | not as expected | as expected | as expected | as expected | as expected | not as expected | as expected |
Picking of F67+F69 (SERK-TMD+linker GFP)
Investigator: Katrin
Aim of the study: Picking of F67+F69 (SERK-TMD+linker GFP)
Operational sequence:
- Picking and overnight culture after standard laboratory's protocol. (5 µl CamR and 5 ml LB-medium)
- 3 colonies were picked for every Transformation product.
- There were no colonies on the plates for ligations F68+F75 (pActin, pSB1C3),
(probably due to dephosphorylation)
Preparative digestion and gelelectrophoresis of P220 (thermonuclease, fragment corrupted) with NgoMIV/SpeI, P160 (NLS, fragment named F77) with AgeI/SpeI, P40 (GSAT linker, fragment named F79) with NgoMIV/SpeI, P7 (PhyB, fragment named F78) with AgeI/SpeI
Investigator: Katrin
Aim of the experiment: Preparative digestion and gelelectrophoresis of P220 (thermonuclease, fragment corrupted) with NgoMIV/SpeI, P160 (NLS, fragment named F77) with AgeI/SpeI, P40 (GSAT linker, fragment named F79) with NgoMIV/SpeI, P7 (PhyB, fragment named F78) with AgeI/SpeI
Procedure:
- Batch for preparative digestion of P220 (thermonuclease, fragment corrupted) with NgoMIV/SpeI
| volume | reagent |
| 20 µl | P220 |
| 5 µl | CutSmart Buffer |
| 1 µl | NgoMIV(20 U/µl) |
| 1 µl | SpeI-HF (20 U/µl) |
| 14 µl | ddH2O |
| =40 µl | TOTAL |
- Batch for preparative digestion of P160 (NLS, fragment named F77) with AgeI/SpeI
| volume | reagent |
| 20 µl | plasmid P160 |
| 5 µl | CutSmart Buffer |
| 1 µl | SpeI-HF(20 U/µl) |
| 1 µl | AgeI-HF (20 U/µl) |
| 14 µl | ddH2O |
| =40 µl | TOTAL |
- Batch for preparative digestion of P40 (GSAT linker, fragment named F79) with NgoMIV/SpeI
| volume | reagent |
| 20 µl | plasmid P40 |
| 5 µl | CutSmart Buffer |
| 1 µl | SpeI-HF(20 U/µl) |
| 1 µl | NgoMIV (20 U/µl) |
| 14 µl | ddH2O |
| =40 µl | TOTAL |
- Batch for preparative digestion of P7 (PhyB, fragment named F78) with AgeI/SpeI
| volume | reagent |
| 20 µl | plasmid P40 |
| 5 µl | CutSmart Buffer |
| 1 µl | SpeI-HF(20 U/µl) |
| 1 µl | AgeI-HF (20 U/µl) |
| 14 µl | ddH2O |
| =40 µl | TOTAL |
- Incubation for 2 h at 37 °C.
- 4 µl of DNA loading buffer (10x) were added to the 40 µl reaction batches containing P220 and P40 after digestion and were loaded on a 1% agarose gel for preparative gelelectrophoresis
- the vector backbones where only a few base pairs were cut out were purified via PCR-purification
- Preparative gelelectrophoresis was performed at 90 V for 60 min.
| digestion of P220 (thermonuclease, fragment corrupted) with NgoMIV/SpeI | 1 kbp DNA ladder | digestion of P40 (GSAT linker, fragment named F79) with NgoMIV/SpeI |
| not as expected, band was discarded | as expected, lower band (108 bp) was cut out |
- the band was extracted with QIAquick Gel Extraction Kit, QIAGEN.
PCR Purification of vector backbones P7 (PhyB in pSB1C3, fragment F78), P160 (NLS in pSB1C3, fragment F77)
Investigator: Katrin
Aim of the study: PCR Purification of vector backbones P7 (PhyB in pSB1C3, fragment F78), P160 (NLS in pSB1C3, fragment F77)
- in order to get rid of the few base pairs that were cut out during digestion, the digestion batches were purified
with QIAquick PCR Purification Kit, Qiagen
- the yiedls were 56.2 ng/µl for F77 and 277,6 ng/µl for F78
Ligation of F78+F79(PhyB+linker) and F59+F71 (pSB1C3+Ig-K)
Investigator: Katrin Aim of the experiment: Ligation of F78+F79(PhyB+linker) and F59+F71 (pSB1C3+Ig-K)
Procedure:
- Ligation batch for F78+F79(PhyB+linker)
| volume | reagent |
| 0.36 µl | F78 (277.6 ng/µl, 4807 bp) |
| 1.77 µl | P79 (3.8 ng/µl, 108 bp) |
| 14.87 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- Ligation batch for F59+F71 (pSB1C3+Ig-K)
| volume | reagent |
| 1.5 µl | F59 (66.7 ng/µl, 2086 bp) |
| 3.12 µl | P71 (3.1 ng/µl, 67 bp) |
| 12.38 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- Negative control was also prepared. Instead of the insert, the same volume of ddH2O was added to the reaction batch.
- The ligation was performed for 1 hour at room temperature.
Transformation of E. coli XL1 blue with F78+F79(PhyB+linker) and F59+F71 (pSB1C3+Ig-K)
Investigator: Katrin
Aim of the experiment: Transformation of E. coli XL1 blue with F78+F79(PhyB+linker) and F59+F71 (pSB1C3+Ig-K) and negative controls F78 and F59
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 10 µl of DNA was added to 200 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- The cell suspensions were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellets were resuspended in 100 µl of LB-medium and these concentrated cell suspensions were plated again on plates containing antibiotics (CamR).
PCR of P192 (Thermonuclease, O64 & O65)
Investigator: Katrin
Aim of the experiment: PCR of P192 (Thermonuclease, O64 & O65)
Procedure:
Operational sequence:
- PCR reaction mixture
| volume | reagent |
| 10 µl | 5x OneTaq Standard Reaction Buffer |
| 1 µl | 10 mM dNTPs |
| 1 µl | 10 µM Forward Primer O64 (NucA_fw) |
| 1 µl | 10 µM Reverse Primer O65 (NucA_rv) |
| 0.25 µL | OneTaq Hot Start DNA Polymerase (Finally: 1.25 units/50 µL) |
| 1 µl | 1:1000 dilution of plasmid DNA (P192) |
| 35.75 µL | ddH2O Water |
| =50 µL | TOTAL |
- Mix with pipette
- The gradient PCR program was performed after following scheme with following conditions:
| Initial denaturation | 94 °C | 30 s |
| 30 cycles | 94 °C | 30 s |
| 55 °C | 60 s | |
| 68 °C | 30 s | |
| Final extension | 68 °C | 5 min |
| Hold | 4 °C | infinite |
Week 8
Monday, June 10rd
Analytical Gel of PCR of Thermonuclease (P192)
Investigator: Florian
Aim of the study: Analysis of PCR of Thermonuclease (P192); supposed letgth: 447 bp
| Lane: | DNA ladder 1kb | P192 PCR product | DNA ladder 1kb |
| Result: | - | as expected | - |
PCR Purification of Thermonuclease (P192)
Investigator: Andi
Aim of the study: PCR Purification of Thermonuclease (P192)
- After the PCR the batch was purified with QIAquick PCR Purification Kit, Qiagen
- Eluation in 35 µl EB buffer -> F80
Miniprep of F67+F69 Ligation (Linker+GFP in Suffix of TMD)
Investigator: Johanna
Aim of the study: Preparation of DNA of the F67+F69 Ligation from 09.06.13 (Linker+GFP in Suffix of TMD)
Procedure: according to MiniPrep Kit QIAGEN; measurement of concentration (NanoDrop)
- Eluation in 35 µl EB buffer each
| Plamsid | Concentration |
| P236 | 108.0 ng/µl |
| P237 | 116.0 ng/µl |
| P238 | 165.4 ng/µl |
Picking of F78 + F79 (PhyB + Linker) and F59 + F71 (pSB1C3 + lgkappa)
Investigator: Johanna
Aim of the study: Picking of F78 + F79 (PhyB + Linker) and F59 + F71 (pSB1C3 + lgkappa)
Operational sequence:
- Picking and overnight culture after standard laboratory's protocol. (4 µl CamR and 4 ml LB-medium)
- 3 colonies were picked for every Transformation product.
Sequencing of P150 (FluA), P233 (pActin), P213 (Spycatcher) and F67+F69 Ligation (Linker+GFP + TMD)
Investigator: Johanna
Aim of the study: Sequencing of P150 (FluA), P233 (pActin), P213 (Spycatcher) and F67+F69 Ligation (Linker+GFP + TMD)
Procedure:Eurofins SmartSeq Kit protocol (15 µl plasmid DNA (50 - 100 ng), 2 µl sequencing primer).
| # | P150 | P233 | P213 | P236 | P237 |
| Name | FluA | pActin | Spycatcher | Linker+GFP+TMD clone1 | Linker+GFP+TMD clone2 |
| O3 (forward) | FR01002308 | FR01002307 | FR01002305 | FR01002299 | FR01002298 |
| O4 (reverse) | n.a. | FR01002306 | n.a. | FR01002297 | FR01002311 |
PCR of BPul Laccase P219 + Analytical Gelelectrophoresis
Investigator: Andi, Johanna
Aim of the experiment: PCR of Bpul Laccase P219.
Procedure:
Operational sequence:
- PCR reaction mixture
| volume | reagent |
| 10 µl | 5x OneTaq Reaction Standard Reaction Buffer |
| 1 µl | 10 mM dNTPs |
| 1 µl | 10 µM Forward Primer (P219:O24 (Bpul_for)) |
| 1 µl | 10 µM Reverse Primer (P143:O25 (Bpul_rev)) |
| 0.25 µL | OneTaq Hot Start DNA Polymerase (Finally: 1.25 units/50 µL) |
| 1 µl | Plasmid DNA (P219)- 1:1000 dilution |
| 35.75 µL | ddH2O Water |
| =50 µL | TOTAL |
- Mix with pipette
- The PCR program was performed after following scheme:
| Initial denaturation | 94 °C | 30 s |
| 30 cycles | 94 °C | 30 s |
| 55 °C | 60 s | |
| 68 °C | 90 s | |
| Final extension | 68 °C | 5 min |
| Hold | 4 °C | infinite |
- After PCR, the product was purified with QIAquick PCR Purification Kit, Qiagen.
- The resulting fragment was called F82
- Further on an analytif Gelelectrophoresis was performed to be sure that the expected fragment was amplified.
| DNA ladder 1kb | P219 PCR product |
- As we can see, the fragment has the expected length of about 1600 BP.
Preparative digestion of F80 (PCR purification of Thermonuclease) with XbaI/AgeI
Investigator: Florian, Johanna
Aim of the experiment: Preparative digestion of F80 (PCR purification of Thermonuclease) with XbaI/AgeI.
Procedure:
- Batch for preparative digestion of F80 (PCR purification of Thermonuclease) with XbaI/AgeI.
| volume | reagent |
| 25 µl | F80 |
| 5 µl | CutSmart Buffer |
| 1 µl | AgeI (20 U/µl) |
| 1 µl | XbaI (20 U/µl) |
| 18 µl | ddH2O |
| =50 µl | TOTAL |
- Incubation for 2.5 h at 37 °C.
- The digested fragment was purified via PCR-purification (Qiagen-kit)
- A yield of 26.2 ng/µl could be measured for F81
Ligation of F59 + F81 (Thermonuclease in pSB1C3)
Investigator: Florian, Johanna
Aim of the experiment: Ligation of F59 + F81 (Thermonuclease in pSB1C3).
Procedure:
- Ligation batch for F59 + F81 (Thermonuclease in pSB1C3)
| volume | reagent |
| 1.5 µl | F59 (66.7 ng/µl, 2086 bp) |
| 2.45 µl | P81 (26.2 ng/µl, 447 bp) |
| 13.55 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 0.5 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- Negative control was also prepared. Instead of the insert, the same volume of ddH2O was added to the reaction batch.
- The ligation was performed for 1 hour at room temperature.
Transformation of E. coli XL1 blue with F59+F81(Thermonuclease in pSB1C3)
Investigator: Katrin
Aim of the experiment: Transformation of E. coli XL1 blue with F59+F81(Thermonuclease in pSB1C3) and the negative control F59.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 10 µl of DNA was added to 200 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Addition of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of the cell suspensions were plated on plates containing antibiotics (CamR).
- The cell suspensions were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellets were resuspended in 100 µl of LB-medium and these concentrated cell suspensions were plated again on plates containing antibiotics (CamR).
Preparative digestion and purification of F82 (PCR product of Bpul Laccase) with XbaI/AgeI-HF and PCR purification kit of Qiagen
Investigator: Andi, Johanna
Aim of the experiment: Preparative digestion and purification of F82 (Bpul Laccase) with XbaI+AgeI and PCR purification kit of Qiagen
Procedure:
- Batch for preparative digestion of F82 with XbaI/AgeI
| volume | reagent |
| 25 µl | F82 |
| 5 µl | CutSmart Buffer |
| 1 µl | AgeI-HF(20 U/µl) |
| 1 µl | XbaI (20 U/µl) |
| 18 µl | ddH2O |
| =50 µl | TOTAL |
- Incubation for 2.5 h at 37 °C.
- the digested fragment was purified via PCR-purification
- the resulting fragment was called F83 (F82 digested with XbaI+AgeI)
Ligation of F83(PCR product of Bpul Laccase digested with AgeI-HF + XbaI)and F59 (psB1C3 digested with AgeI-HF + XbaI)
Investigator: Andi, Johanna
Aim of the experiment: Ligation of F83(PCR product of Bpul Laccase digested with AgeI-HF + XbaI)and F59 (psB1C3 digested with AgeI-HF + XbaI)
Procedure:
- Ligation batch for F83+F59 (psB1C3+Bpul Laccase)
| volume | reagent |
| 15.5 µl | F83 (9.2 ng/µl, 1600 bp) |
| 1.5 µl | F59 (66.7 ng/µl, 2100 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- Negative control was also prepared. Instead of the insert, the same volume of ddH2O was added to the reaction batch.
- The ligation was performed over night at 16°C.
Tuesday, June 11th
Transformation of E. coli XL1 blue with ligation products F59+F81 and F59+F83
Investigator: Louise, Andi
Aim of the experiment: Transformation of E. coli XL1 blue with ligation products F59+F81 and F59+F83.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 10 µl of DNA was added to 200 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of the cell suspension was plated on one chloramphenicol plate.
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellets were resuspended in 100 µl of LB-medium and these concentrated cell suspensions were plated again on new chlorampenicol plates.
Miniprep of overnight cultures of transformated E. coli XL1 blue with F78+F79 (PhytochromB +Linker) and F59+F71 (IgKappa)
Investigator: Louise, Johanna
Aim of the experiment:Miniprep of overnight cultures of transformated E. coli XL1 blue with F78+F79 (PhytochromB +Linker) and F59+F71 (IgKappa).
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
- The resulting DNA were eluted in tubes labeled as follows:
| Content | New Tube |
| F78+F79 (PhytochromB +Linker) clone 1 | P239 |
| F78+F79 (PhytochromB +Linker) clone 2 | P240 |
| F78+F79 (PhytochromB +Linker) clone 3 | P241 |
| F59+F71 (IgKappa) clone 4 | P242 |
| F59+F71 (IgKappa) clone 5 | P243 |
| F59+F71 (IgKappa) clone 6 | P244 |
Analytical digestion and gelelectrophoresis of P239-P241 (PhytochromB+Linker) and P242-P244 (IgKappa) with XbaI and AgeI
Investigator: Louise, Rosario, Florian
Aim of the experiment: Analytical digestion and gelelectrophoresis of P239-P241 (PhytochromB+Linker) and P242-P244 (IgKappa) with XbaI and AgeI
Procedure:
- Batch
| volume | reagent |
| 2 µl | CutSmart Buffer (10x) |
| 0.25 µl | AgeI |
| 0.25 µl | XbaI |
| 15 µl | ddH2O |
| 2.5 µl | Plasmid DNA (P293-P244) |
| =20 µl | TOTAL |
- Incubation for 60 min at 37 °C.
- Analytical gelelectrophoresis was performed at 90 V for 60 min
- For P242-P244 a 2% agarose gel was used and for P293-P241 a 0.5% agarose gel was used.
| Lane: | Digestion of P242 (IgK) with XbaI & AgeI, clone 1 | Digestion of P243 (IgK) with XbaI & AgeI, clone 2 | Digestion of P244 (IgK) with XbaI & AgeI, clone 3 | DNA ladder 100bp |
| Result: | as expected | as expected | as expected | - |
| Lane: | Digestion of P239 (PhyB+Linker) with XbaI & AgeI, clone 1 | Digestion of P240 (PhyB+Linker) with XbaI & AgeI, clone 2 | Digestion of P241 (PhyB+Linker) with XbaI & AgeI, clone 3 | DNA ladder 1kb |
| Result: | failed | as expected | as expected | - |
Preparative digestion and gelelectrophoresis of P204 (IgKappa-SigP, fragment named F90) with AgeI/SpeI, P157 (EreB, fragment named F86) with NgoMIV/SpeI, P208 (NanoLuc, fragment named F87) with NgoMIV/SpeI, P213 (Spycatcher, fragment named F88) with AgeI/SpeI and P170 (Spytag, fragment named F89) with NgoMIV/SpeI
Investigator: Johanna, Rosario, Flo
Aim of the experiment: Preparative digestion and gelelectrophoresis of P204 (IgKappa-SigP, fragment named F90) with AgeI/SpeI, P157 (EreB, fragment named F86) with NgoMIV/SpeI, P208 (NanoLuc, fragment named F87) with NgoMIV/SpeI, P213 (Spycatcher, fragment named F88) with AgeI/SpeI and P170 (Spytag, fragment named F89) with NgoMIV/SpeI
Procedure:
- Batch for preparative digestion of P204 (IgKappa-SigP, fragment named F90) with AgeI/SpeI
| volume | reagent |
| 20 µl | P204 |
| 5 µl | CutSmart Buffer |
| 1 µl | AgeI(20 U/µl) |
| 1 µl | SpeI-HF (20 U/µl) |
| 23 µl | ddH2O |
| =50 µl | TOTAL |
- Batch for preparative digestion of P157 (EreB, fragment named F86) with NgoMIV/SpeI
| volume | reagent |
| 20 µl | P157 |
| 5 µl | CutSmart Buffer |
| 1 µl | NgoMIV(20 U/µl) |
| 1 µl | SpeI-HF (20 U/µl) |
| 23 µl | ddH2O |
| =50 µl | TOTAL |
- Batch for preparative digestion of P208 (NanoLuc, fragment named F87) with NgoMIV/SpeI
| volume | reagent |
| 20 µl | P208 |
| 5 µl | CutSmart Buffer |
| 1 µl | NgoMIV(20 U/µl) |
| 1 µl | SpeI-HF (20 U/µl) |
| 23 µl | ddH2O |
| =50 µl | TOTAL |
- Batch for preparative digestion of P213 (Spycatcher, fragment named F88) with NgoMIV/SpeI
| volume | reagent |
| 20 µl | P213 |
| 5 µl | CutSmart Buffer |
| 1 µl | NgoMIV(20 U/µl) |
| 1 µl | SpeI-HF (20 U/µl) |
| 23 µl | ddH2O |
| =50 µl | TOTAL |
- Batch for preparative digestion of P170 (Spytag, frament named F89) with NgoMIV/SpeI
| volume | reagent |
| 20 µl | P170 |
| 5 µl | CutSmart Buffer |
| 1 µl | NgoMIV(20 U/µl) |
| 1 µl | SpeI-HF (20 U/µl) |
| 23 µl | ddH2O |
| =50 µl | TOTAL |
- Incubation for 2.5 h at 37 °C.
- 5 µl of DNA loading buffer (10x) were added to the 50 µl reaction batches after digestion and were loaded on a 1% agarose gel (P204, P157, P208, P213) and 2 % agarose gel (P170) for preparative gelelectrophoresis
- Preparative gelelectrophoresis was performed at 90 V for 60 min.
| 1 kbp DNA ladder | digestion of P204 (IgKappa-SigP) with AgeI/SpeI | 100 bp DNA ladder (Generuler) | digestion of P157 (EreB) with NgoMIV/SpeI |
| as expected, lower band was cut out | as expected, lower band was cut out |
| 1 kbp DNA ladder | digestion of P208 (Nanoluc) with NgoMIV/SpeI | 100 bp DNA ladder (Generuler) | digestion of P213 (Spycatcher) with NgoMIV/SpeI |
| as expected, lower band was cut out | as expected, lower band was cut out |
| 100 bp DNA ladder (Generuler) | digestion of P170 (Spytag) with NgoMIV/SpeI |
| as expected, lower band was cut out |
- the band was extracted with QIAquick Gel Extraction Kit, QIAGEN.
Ligation of F90+F86 (IgKappa-SigP + EreB), F90+F87 (IgKappa-SigP + Nanoluciferase), F90+F88 (IgKappa-SigP + SpyCatcher), F90+F89 (IgKappa-SigP + SpyTag) and negativ control of F90 (IgKappa-SigP)
Investigator: Johanna
Aim of the experiment: Ligation of F90+F86 (IgKappa-SigP + EreB), F90+F87 (IgKappa-SigP + Nanoluciferase), F90+F88 (IgKappa-SigP + SpyCatcher), F90+F89 (IgKappa-SigP + SpyTag) and negativ control of F90 (IgKappa-SigP)
Procedure:
- Ligation batch for F90+F86 (IgKappa-SigP + EreB)
| volume | reagent |
| 5.7 µl | F86 (31.0 ng/µl, 1254 bp) |
| 6.5 µl | F90 (15.3 ng/µl, 2144 bp) |
| 4.8 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- Ligation batch for F90+F87 (IgKappa-SigP + Nanoluciferase)
| volume | reagent |
| 7.0 µl | F87 (10.2 ng/µl, 510 bp) |
| 6.5 µl | F90 (15.3 ng/µl, 2144 bp) |
| 3.5 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- Ligation batch for F90+F88 (IgKappa-SigP + SpyCatcher)
| volume | reagent |
| 6.3 µl | F88 (8.7 ng/µl, 339 bp) |
| 6.5 µl | F90 (15.3 ng/µl, 2144 bp) |
| 4.2 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- Ligation batch for F90+F89 (IgKappa-SigP + SpyTag)
| volume | reagent |
| 10.5 µl | F89 (8.7 ng/µl, 39 bp) |
| 6.5 µl | F90 (15.3 ng/µl, 2144 bp) |
| 0.0 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- Negative control of F 90 (IgKappa-SigP) was also prepared. Instead of the insert, the same volume of ddH2O was added to the reaction batch.
- The ligation was performed for 1 hour at room temperature.
PCR of pActin (P143) and Thermonuclease (P192) to find the best conditions for PCR + Measurement via Nanodrop + Analytical Gelelectrophoresis to get to know about it!
Investigator: Andi
Aim of the experiment: PCR of pActin (P143) and Thermonuclease (P192) to find the best conditions for PCR + Measurement via Nanodrop + Analytical Gelelectrophoresis to get to know about it!
Procedure:
Operational sequence:
- The PCR for P143 (pActin) was performed three times under different conditions
- First batch - PCR reaction mixture
| volume | reagent |
| 10 µl | 5x OneTaq Reaction Standard Reaction Buffer |
| 1 µl | 10 mM dNTPs |
| 1 µl | 10 µM Forward Primer (P143:O62 (pAct_for)) |
| 1 µl | 10 µM Reverse Primer (P143:O63 (pAct_rev)) |
| 0.25 µL | OneTaq Hot Start DNA Polymerase (Finally: 1.25 units/50 µL) |
| 1 µl | Plasmid DNA (P143)- 1:1000 dilution |
| 35.75 µL | ddH2O Water |
| =50 µL | TOTAL |
- Second batch - PCR reaction mixture
| volume | reagent |
| 10 µl | 5x OneTaq GC Reaction Buffer |
| 1 µl | 10 mM dNTPs |
| 1 µl | 10 µM Forward Primer (P143:O62 (pAct_for)) |
| 1 µl | 10 µM Reverse Primer (P143:O63 (pAct_rev)) |
| 0.25 µL | OneTaq Hot Start DNA Polymerase (Finally: 1.25 units/50 µL) |
| 1 µl | Plasmid DNA (P143)- 1:1000 dilution |
| 35.75 µL | ddH2O Water |
| =50 µL | TOTAL |
- Third batch - PCR reaction mixture
| volume | reagent |
| 10 µl | 5x Herculase Fusion Polymerase reaction buffer |
| 2.5 µl | 10 mM dNTPs |
| 1.25 µl | 10 µM Forward Primer (P143:O62 (Bpul_for)) |
| 1.25 µl | 10 µM Reverse Primer (P143:O63 (Bpul_rev)) |
| 1 µL | Herculase II Fusion Polymerase |
| 2 µl | Plasmid DNA (P143)- 1:1000 dilution |
| 0.5 µl | DMSO |
| 35.75 µL | ddH2O Water |
| =50 µL | TOTAL |
- The content of all batches has been mixed with a pipette
- The PCR program was performed after following scheme:
| Initial denaturation | 94 °C | 30 s |
| 30 cycles | 94 °C | 30 s |
| 51 °C | 60 s | |
| 68 °C | 90 s | |
| Final extension | 68 °C | 5 min |
| Hold | 4 °C | infinite |
- The PCR for P192(Thermonuclease) was performed three times under different conditions
- First batch - PCR reaction mixture
| volume | reagent |
| 10 µl | 5x OneTaq Reaction Standard Reaction Buffer |
| 1 µl | 10 mM dNTPs |
| 1 µl | 10 µM Forward Primer (P192:O64 (NucA_fw)) |
| 1 µl | 10 µM Reverse Primer (P192:O65(NucA_rev)) |
| 0.25 µL | OneTaq Hot Start DNA Polymerase (Finally: 1.25 units/50 µL) |
| 1 µl | Plasmid DNA (P192)- 1:1000 dilution |
| 35.75 µL | ddH2O Water |
| =50 µL | TOTAL |
- Second batch - PCR reaction mixture
| volume | reagent |
| 10 µl | 5x OneTaq GC Reaction Buffer |
| 1 µl | 10 mM dNTPs |
| 1 µl | 10 µM Forward Primer (P192:O64 (NucA_fw)) |
| 1 µl | 10 µM Reverse Primer (P192:O65(NucA_rev)) |
| 0.25 µL | OneTaq Hot Start DNA Polymerase (Finally: 1.25 units/50 µL) |
| 1 µl | Plasmid DNA (P192)- 1:1000 dilution |
| 35.75 µL | ddH2O Water |
| =50 µL | TOTAL |
- Third batch - PCR reaction mixture
| volume | reagent |
| 10 µl | 5x Herculase II DNA Fusion Polymerase reaction Buffer |
| 2.5 µl | 10 mM dNTPs |
| 1.25 µl | 10 µM Forward Primer (P192:O64 (NucA_fw)) |
| 1.25 µl | 10 µM Reverse Primer (P192:O65(NucA_rev)) |
| 1 µL | Herculase II Fusion DNA Polymerase |
| 3 µl | Plasmid DNA (P192)- 1:1000 dilution |
| 0.5 µL | DMSO |
| 30.5 µL | ddH2O Water |
| =50 µL | TOTAL |
- The PCR program of the Thermonuclease (P192) was performed after following scheme:
| Initial denaturation | 94 °C | 30 s |
| 30 cycles | 94 °C | 30 s |
| 54 °C | 60 s | |
| 68 °C | 40 s | |
| Final extension | 68 °C | 5 min |
| Hold | 4 °C | infinite |
- Mix the content of the batches with pipette
- After PCR, the product was purified with QIAquick PCR Purification Kit, Qiagen.
- The concentrations of all PCR products have been measured
| P143 - Batch #1 - PCR product - 5.9 ng/µl | P143 - Batch #2 - PCR product - 40.5 ng/µl | P143 - Batch #3 - PCR product - 12.4 ng/µl | P192 - Batch #1 - PCR product - 39.7 ng/µl | P192 - Batch #2 - PCR product - 23.1 ng/µl | P192 - Batch #3 - PCR product - 82.5 ng/µl |
- As we can see, the highest concentration of PCR product seems to be in P143 - Batch #2 and P192 - Batch #3.
- Further on an analytic Gelelectrophoresis was performed to be sure that the expected fragment was amplified.
| DNA ladder 1kb | P143 - Batch #1 - PCR product | P143 - Batch #2 - PCR product | P143 - Batch #3 - PCR product | P192 - Batch #1 - PCR product | P192 - Batch #2 - PCR product | P192 - Batch #3 - PCR product |
- As we can see, the fragment has the expected length of about 1600 BP referring to the PCR product of P143 and about XXX BP referring to the product of P192.
- Further on the analytical gel confirms the output of the nano drop.
Preparative digestion of PCR reaction batch #2 of P143 (pActin) XbaI/PstI and PCR reaction batch #3 of P192 with XbaI/AgeI
Investigator: Andi
Aim of the experiment: Preparative digestion of PCR reaction batch #2 of P143 (pActin) XbaI/PstI and PCR reaction batch #3 of P192 with XbaI/AgeI
Procedure:
- Batch for preparative digestion of PCR reaction batch #2 of P143 (pActin) with XbaI/PstI.
| volume | reagent |
| 24 µl | PCR reaction batch #2 of P143 |
| 5 µl | CutSmart Buffer |
| 1 µl | PstI #1 (20 U/µl) |
| 1 µl | PstI #2 (20 U/µl) |
| 1 µl | XbaI (20 U/µl) |
| 18 µl | ddH2O |
| =50 µl | TOTAL |
- I used 1 µl of both PstI batches because one seems to be broken
- Incubation for 2.5 h at 37 °C.
- The digested fragment was purified via PCR-purification (Qiagen-kit)
- A yield of 16.9 ng/µl could be measured for the resulting F84
- Batch for preparative digestion of PCR reaction batch #3 of P192 (Thermonuclease) with XbaI/AgeI-HF.
| volume | reagent |
| 24 µl | PCR reaction batch #2 of P143 |
| 5 µl | CutSmart Buffer |
| 1 µl | AgeI-HF (20 U/µl) |
| 1 µl | XbaI (20 U/µl) |
| 19 µl | ddH2O |
| =50 µl | TOTAL |
- Incubation for 2.5 h at 37 °C.
- The digested fragment was purified via PCR-purification (Qiagen-kit)
- A yield of 46.2 ng/µl could be measured for the resulting F85
Ligation of F84(Digestion of PCR product (GC rich buffer) of P143 (pActin) with PstI/XbaI) and F42 (psB1C3 digested with PstI/XbaI) and Ligation of F85(Digestion of PCR product (Herculase II) of P192 (Thermonuclease) with XbaI/AgeI and F59 (psB1C3 digested with AgeI-HF + XbaI)
Investigator: Andi
Aim of the experiment: Ligation of F84(Digestion of PCR product (GC rich buffer) of P143 (pActin) with PstI/XbaI) with F42 (psB1C3 digested with PstI/XbaI) and Ligation of F85(Digestion of PCR product (Herculase II) of P192 (Thermonuclease) with XbaI/AgeI-HF) with F59 (psB1C3 digested with AgeI-HF/XbaI)
Procedure:
- Ligation batch for F84+F42 (psB1C3+pActin)
| volume | reagent |
| 12.6 µl | F84 (16.9 ng/µl, 1500 bp) |
| 1.4 µl | F42 (73.5 ng/µl, 2100 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 4 µl | ddH2O |
| =20 µl | TOTAL |
- Ligation batch for F85+F59 (psB1C3+Thermonuclease)
| volume | reagent |
| 1.6 µl | F85 (46.2 ng/µl, 500 bp) |
| 1.5 µl | F59 (66.7 ng/µl, 2100 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 13.9 µl | ddH2O |
| =20 µl | TOTAL |
- Negative control was also prepared for both batches. Instead of the insert, the same volume of ddH2O was added to the reaction batch.
- The ligation was performed over night at 16°C.
Transformation of E. coli XL1 blue with plasmids P157, P208 and P213
Investigator: Florian
Aim of the experiment: Transformation of E. coli XL1 blue with plasmids P157, P208 and P213.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 10 µl of DNA was added to 200 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of the cell suspension was plated on one chloramphenicol plate.
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellets were resuspended in 100 µl of LB-medium and these concentrated cell suspensions were plated again on new chlorampenicol plates.
Wednesday, June 12th
Transformation of E. coli XL1 blue with F42+F84, F90+F86, F90+F87, F90+F88, F90+F89, F59+F85
Investigator: Rosario
Aim of the experiment: Transformation of E. coli XL1 blue with F42+F84, F90+F86, F90+F87, F90+F88, F90+F89, F59+F85
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 10 µl of DNA was added to 200 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- Centrifugation for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellets were resuspended in 100 µl of LB-medium and these concentrated cell suspensions were plated on new chlorampenicol plates.
Preparative digestion and gelelectrophoresis of P240 (PhyB + Linker) with AgeI/SpeI, P244 (IgKappa) with AgeI/SpeI, P197 (XylE) with NgoMIV/SpeI, P150 (FluA) with NgoMIV/SpeI, P126 (N-TEV) with AgeI/SpeI, P127 (C-TEV) with NgoMIV/SpeI, P168 (PIF3) with NgoMIV/SpeI and P222 (PIF6) with NgoMIV/SpeI
Investigator: Louise, Florian
Aim of the experiment: Preparative digestion and gelelectrophoresis of P240 (PhyB + Linker) with AgeI/SpeI, P244 (IgKappa) with AgeI/SpeI, P197 (XylE) with NgoMIV/SpeI, P150 (FluA) with NgoMIV/SpeI, P126 (N-TEV) with AgeI/SpeI, P127 (C-TEV) with NgoMIV/SpeI, P168 (PIF3) with NgoMIV/SpeI and P222 (PIF6) with NgoMIV/SpeI
Procedure:
- Batch for preparative digestion of P240 (PhyB + Linker) with AgeI/SpeI
| volume | reagent |
| 20 µl | P240 |
| 4 µl | CutSmart Buffer |
| 1 µl | AgeI(20 U/µl) |
| 1 µl | SpeI-HF (20 U/µl) |
| 14 µl | ddH2O |
| =40 µl | TOTAL |
- Batch for preparative digestion of P244 (IgKappa) with AgeI/SpeI
| volume | reagent |
| 20 µl | P244 |
| 4 µl | CutSmart Buffer |
| 1 µl | AgeI(20 U/µl) |
| 1 µl | SpeI-HF (20 U/µl) |
| 14 µl | ddH2O |
| =40 µl | TOTAL |
- Batch for preparative digestion of P197 (XylE) with NgoMIV/SpeI
| volume | reagent |
| 20 µl | P197 |
| 4 µl | CutSmart Buffer |
| 1 µl | NgoMIV(20 U/µl) |
| 1 µl | SpeI-HF (20 U/µl) |
| 14 µl | ddH2O |
| =40 µl | TOTAL |
- Batch for preparative digestion of P150 (FluA) with NgoMIV/SpeI
| volume | reagent |
| 20 µl | P150 |
| 4 µl | CutSmart Buffer |
| 1 µl | NgoMIV(20 U/µl) |
| 1 µl | SpeI-HF (20 U/µl) |
| 14 µl | ddH2O |
| =40 µl | TOTAL |
- Batch for preparative digestion of P126 (N-TEV) with AgeI/SpeI
| volume | reagent |
| 20 µl | P126 |
| 4 µl | CutSmart Buffer |
| 1 µl | AgeI(20 U/µl) |
| 1 µl | SpeI-HF (20 U/µl) |
| 14 µl | ddH2O |
| =40 µl | TOTAL |
- Batch for preparative digestion of P127 (C-TEV) with NgoMIV/SpeI
| volume | reagent |
| 20 µl | P127 |
| 4 µl | CutSmart Buffer |
| 1 µl | NgoMIV(20 U/µl) |
| 1 µl | SpeI-HF (20 U/µl) |
| 14 µl | ddH2O |
| =40 µl | TOTAL |
- Batch for preparative digestion of P168 (PIF3) with NgoMIV/SpeI
| volume | reagent |
| 20 µl | P168 |
| 4 µl | CutSmart Buffer |
| 1 µl | NgoMIV(20 U/µl) |
| 1 µl | SpeI-HF (20 U/µl) |
| 14 µl | ddH2O |
| =40 µl | TOTAL |
- Batch for preparative digestion of P222 (PIF6) with NgoMIV/SpeI
| volume | reagent |
| 20 µl | P222 |
| 4 µl | CutSmart Buffer |
| 1 µl | NgoMIV(20 U/µl) |
| 1 µl | SpeI-HF (20 U/µl) |
| 14 µl | ddH2O |
| =40 µl | TOTAL |
- Incubation for 2.5 h at 37 °C.
- 4 µl of DNA loading buffer (10x) were added to the 40 µl reaction batches after digestion and were loaded on a 1% agarose gel for preparative gelelectrophoresis
- Preparative gelelectrophoresis was performed at 90 V for 60 min.
| 1 kbp DNA ladder | digestion of P126 (N-TEV) with AgeI/SpeI | digestion of P127 (C-TEV) with NgoMIV/SpeI | digestion of P150 (FluA) with NgoMIV/SpeI |
| as expected, upper band was cut out | as expected, lower band was cut out | as expected, lower band was cut out |
| 1 kbp DNA ladder | digestion of P168 (PIF3) with NgoMIV/SpeI | digestion of P197 (XylE) with NgoMIV/SpeI | digestion of P222 (PIF6) with NgoMIV/SpeI |
| as expected, lower band was cut out | as expected, lower band was cut out | as expected, lower band was cut out |
| 100 bp DNA ladder (Generuler) | digestion of P244 (IgKappa) with AgeI/SpeI | digestion of P240 (PhyB + Linker) with AgeI/SpeI | 1 kbp DNA ladder |
| as expected, upper band was cut out | as expected, upper band was cut out |
- the DNA was extracted from the gel with QIAquick Gel Extraction Kit, QIAGEN.
- the fragments received the following names:
| P126 (N-TEV) | F93 |
| P127 (C-TEV) | F94 |
| P150 (FluA) | F95 |
| P168 (PIF3) | F96 |
| P197 (XylE) | F97 |
| P222 (PIF6) | F98 |
| P240 (PhyB + Linker) | F91 |
| P244 (IgKappa) | F92 |
Transformation of E. coli XL1 blue with plasmids P126 (N-TEV), P127 (C-TEV), P222 (PIF6), P240 (PhyB+Linker) and P244 (IgKappa)
Investigator: Florian
Aim of the experiment: Transformation of E. coli XL1 blue with plasmids P126 (N-TEV), P127 (C-TEV), P222 (PIF6), P240 (PhyB+Linker) and P244 (IgKappa).
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 10 µl of DNA was added to 200 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of the cell suspension were plated on chloramphenicol plates.
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellets were resuspended in 100 µl of LB-medium and these concentrated cell suspensions were plated again on new chlorampenicol plates.
Picking of P157, P208, P213, F59 + F81 (Thermonuclease in pSB1C3) and F59 + F83 (Laccase in pSB1C3)
Investigator: Jeff
Aim of the study: Picking of P157, P208, P213, F59 + F81 (Thermonuclease in pSB1C3) and F59 + F83 (Laccase in pSB1C3).
Operational sequence:
- Picking and overnight culture after standard laboratory's protocol. (4 µl CamR and 4 ml LB-medium)
- 2 colonies were picked for every ligation product and 1 colony was picked for every transformation product.
Ligation of E. coli XL1 blue with F92+F89, F92+F87, F92+F86, F92+F88, F92+F97, F90+F97, F91+F94, F93+F79 and their negative controls
Investigator: Jeff, Flo, Louise, Christopher
Aim of the experiment: Ligation of E. coli XL1 blue with F92+F89, F92+F87, F92+F86, F92+F88, F92+F97, F90+F97, F91+F94, F93+F79 and their negative controls
Procedure:
- Ligation batch for F92+F89 :
| volume | reagent |
| 0.86 µl | F92 (116.7 ng/µl, 2153 bp) |
| 16.14 µl | F89 (6.8 ng/µl, 39 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 0.0 µl | ddH2O |
| =20 µl | TOTAL |
- Ligation batch for F92+F87 :
| volume | reagent |
| 0.86 µl | F92 (116.7 ng/µl, 2153 bp) |
| 7.0 µl | F87 (10.2 ng/µl, 510 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 9.14 µl | ddH2O |
| =20 µl | TOTAL |
- Ligation batch for F92+F86:
| volume | reagent |
| 0.86 µl | F92 (116.7 ng/µl, 2153 bp) |
| 5.7 µl | F86 (31.0 ng/µl, 1260 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 10.44 µl | ddH2O |
| =20 µl | TOTAL |
- Ligation batch for F92+F88 :
| volume | reagent |
| 0.86 µl | F92 (116.7 ng/µl, 2153 bp) |
| 5.4 µl | F88 (8.7 ng/µl, 339 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 10.74 µl | ddH2O |
| =20 µl | TOTAL |
- Ligation batch for F92+F97:
| volume | reagent |
| 0.86 µl | F92 (116.7 ng/µl, 2153 bp) |
| 6.6 µl | F97 (19.4 ng/µl, 918 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 9.5 µl | ddH2O |
| =20 µl | TOTAL |
- Ligation batch for F90+F97:
| volume | reagent |
| 6.5 µl | F90 (15.3 ng/µl, 2144 bp) |
| 6.6 µl | F97 (19.4 ng/µl, 918 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 3.9 µl | ddH2O |
| =20 µl | TOTAL |
- Ligation batch for F91+F94:
| volume | reagent |
| 0.63 µl | F91 (159 ng/µl, 4129 bp) |
| 4.6 µl | F94 (5.6 ng/µl, 354 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 11.77 µl | ddH2O |
| =20 µl | TOTAL |
- Ligation batch for F93+F79:
| volume | reagent |
| 3.6 µl | F93 (27.4 ng/µl, 2440 bp) |
| 3.5 µl | F79 (3.8 ng/µl, 108 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 9.9 µl | ddH2O |
| =20 µl | TOTAL |
- Negative control was also prepared. Instead of the insert, the same volume of ddH2O was added to the reaction batch.
- The ligation was performed at room temperature for 1 hour
Transformation of E. coli XL1 blue with F92+F89, F92+F87, F92+F86, F92+F88, F92+F97, F90+F97, F91+F94, F93+F79 and their negative controls
Investigator: Jeff, Florian
Aim of the experiment: Transformation of E. coli XL1 blue with F92+F89, F92+F87, F92+F86, F92+F88, F92+F97, F90+F97, F91+F94, F93+F79 and their negative controls.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 10 µl of DNA was added to 200 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- Centrifugation for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellets were resuspended in 100 µl of LB-medium and these concentrated cell suspensions were plated on new chloramphenicol plates.
Thursday, June 13th
QuikChange mutagenesis to remove forbidden restriction sites - SDMIV - of P250, P251, P219 (Laccase), P163 (NPT-Cassette)
Investigator: Andi
Aim of the experiment: Removal of forbidden restriction site from P251 (Laccase) and P163 (NPT-Cassette).
Procedure: Quickchange - PCR
Reaction batch for P219 (Laccase)
| volume | reagent | Additional information |
| 2.5 µl | 10x Pfu Ultra II buffer | |
| 1 µl | Plasmid template (P251 - 1:20 dilution) | need to get between 2.5 ng and 25 ng for a reaction batch of 25 µl |
| 1 µl | 1:10 dilution of O74 (10 µM) | |
| 1 µl | 1:10 dilution of O75 (10 µM) | |
| 18 µl | ddH2O | |
| 1 µl | dNTP mix 50 mM | |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U/µl) |
Reaction batch for P163 (NPT-Cassette)
| volume | reagent | Additional information |
| 2.5 µl | 10x Pfu Ultra II buffer | |
| 1 µl | Plasmid template (P163 - 1:60 dilution) | need to get between 2.5 ng and 25 ng for a reaction batch of 25 µl |
| 1 µl | 1:10 dilution of O70 (10 µM) | |
| 1 µl | 1:10 dilution of O71 (10 µM) | |
| 18 µl | ddH2O | |
| 1 µl | dNTP mix 50 mM | |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U/µl) |
Reaction batch for P250 (Laccase)
| volume | reagent | Additional information |
| 2.5 µl | 10x Pfu Ultra II buffer | |
| 1 µl | Plasmid template (P250 - 1:20 dilution) | need to get between 2.5 ng and 25 ng for a reaction batch of 25 µl |
| 1 µl | 1:10 dilution of O74 (10 µM) | |
| 1 µl | 1:10 dilution of O75 (10 µM) | |
| 18 µl | ddH2O | |
| 1 µl | dNTP mix 50 mM | |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U/µl) |
Reaction batch for P217 (Laccase)
| volume | reagent | Additional information |
| 2.5 µl | 10x Pfu Ultra II buffer | |
| 1 µl | Plasmid template (P217 - 1:100 dilution) | need to get between 2.5 ng and 25 ng for a reaction batch of 25 µl |
| 1 µl | 1:10 dilution of O74 (10 µM) | |
| 1 µl | 1:10 dilution of O75 (10 µM) | |
| 18 µl | ddH2O | |
| 1 µl | dNTP mix 50 mM | |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U/µl) |
PCR cycling parameters - P251 (Laccase) and P163 (NPT-Cassette)
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 30 s |
| 2 | 19 | 95 °C | 30 s |
| 52 °C | 1 min | ||
| 68 °C | 4 min | ||
| 4 °C | hold |
- After the PCR, the Quickchange product was stored in the fridge.
PCR of AlcR (P61, pAutoRex8) and IRES (P61, pAutoRex8) and analytical gel electrophoresis
Investigator: Louise, Johanna, Christopher
Aim of the experiment: PCR of AlcR (P61, pAutoRex8) and IRES (P61, pAutoRex8).
Procedure:
Operational sequence:
- PCR reaction mixture for AlcR
| volume | reagent |
| 10 µl | 5x OneTaq Standard Reaction Buffer |
| 1 µl | 10 mM dNTPs |
| 1 µl | 10 µM Forward Primer (O78 (AlcR_fw)) |
| 1 µl | 10 µM Reverse Primer (O79 (AlcR_rv)) |
| 0.25 µL | OneTaq Hot Start DNA Polymerase (Finally: 1.25 units/50 µL) |
| 1 µl | Plasmid DNA (P61) |
| 35.75 µL | ddH2O Water |
| =50 µL | TOTAL |
- PCR reaction mixture for IRES
| volume | reagent |
| 10 µl | 5x OneTaq Standard Reaction Buffer |
| 1 µl | 10 mM dNTPs |
| 1 µl | 10 µM Forward Primer (O68 (IRES_fw)) |
| 1 µl | 10 µM Reverse Primer (O69 (IRES_rv)) |
| 0.25 µL | OneTaq Hot Start DNA Polymerase (Finally: 1.25 units/50 µL) |
| 1 µl | Plasmid DNA (P61) |
| 35.75 µL | ddH2O Water |
| =50 µL | TOTAL |
- Mix with pipette
- The gradient PCR program was performed after following scheme with following conditions (Tm=56 °C; ΔG=4 °C; AlcR in row 11(=60 °C); IRES in row 1 (=52.0 °C)):
| Initial denaturation | 94 °C | 30 s |
| 30 cycles | 94 °C | 30 s |
| Tm=56 °C; ΔG=4 °C | 60 s | |
| 68 °C | 160 s | |
| Final extension | 68 °C | 5 min |
| Hold | 4 °C | infinite |
- After PCR, the product was purified with QIAquick PCR Purification Kit, Qiagen.
- Resulting products was labeled as F99 (AlcR) and F100 (IRES).
Analytical gelelectrophoresis of F99 (AlcR) and F100 (IRES)
Investigator: Louise, Christopher
Aim of the experiment: Analytical gelelectrophoresis of F99 (AlcR) and F100 (IRES).
Procedure:
- 4 µl of F99 and F100 were mixed with 0.44 µl DNA loading buffer (10x)
- Analytical gelelectrophoresis was performed at 90 V for 60 min on a 1% agarose gel.
| 1 kbp DNA ladder | F100 | F99 |
| as expected | as expected |
Miniprep of overnight cultures of transformated E. coli XL1 blue with P157, P208, P213, F59+F81(Thermonuclease), F59+F83 (Laccase)
Investigator: Rosario
Aim of the experiment:Miniprep of overnight cultures of transformated E. coli XL1 blue with P157, P208, P213, F59+F81(Thermonuclease), F59+F83 (Laccase)
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
- The resulting DNA were eluted in tubes labeled as follows:
| Content | New Tube |
| P157 | P245 |
| P208 | P246 |
| P213 | P247 |
| F59+F81 I | P248 |
| F59+F81 II | P249 |
| F59+F83 I | P250 |
| F59+F83 II | P251 |
Analytical digestion and gelelectrophoresis of P248-P249 (Thermonuclease) and P250-251 (Laccase) with EcoRI and SpeI
Investigator: Rosario
Aim of the experiment: Analytical digestion and gelelectrophoresis of P248-P249 (Thermonuclease) and P250-251 (LAccase) with EcoRI and SpeI
Procedure:
- Batch
| volume | reagent |
| 2 µl | CutSmart Buffer (10x) |
| 0.25 µl | EcoRI |
| 0.25 µl | SpeI |
| 15 µl | ddH2O |
| 2.5 µl | Plasmid DNA (P248-P251) |
| =20 µl | TOTAL |
- Incubation for 60 min at 37 °C.
- Analytical gelelectrophoresis was performed at 90 V for 60 min
- For P248-P251 a 1% agarose gel was used.
| Lane: | DNA marker 1kb | Digestion of P248 with EcoRI & SpeI | Digestion of P249 with EcoRI & SpeI | Digestion of P250 with EcoRI & SpeI | empty | Digestion of P251 with EcoRI & SpeI |
| Result: | - | as expected | as expected | no insert (Laccase) | - | no insert (Laccase) |
Transformation of E. coli XL1 blue with P204
Investigator: Johanna
Aim of the experiment: Transformation of E. coli XL1 blue with P204
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 2 µl of DNA was added to 200 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- Centrifugation for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellets were resuspended in 100 µl of LB-medium and these concentrated cell suspensions were plated on new chloramphenicol plates.
Sequencing of P61 (pSH21/pAutoRex8)
Investigator: Rosario, Katrin
Aim of the study: Sequencing of P61 (pSH21/pAutoRex8)
Procedure:
Eurofins SmartSeq Kit protocol (15 µl plasmid DNA (50 - 100 ng), 2 µl sequencing primer).
| # | P61 |
| Name | pSH 21 (AlcR/IRES) |
| O88 (primer 1) | FR02305828 |
| O89 (primer 2) | FR02305829 |
Picking of F92+F89, F92+F87, F92+F86, F92+F88, F92+F97, F90+F97, F91+F94, F93+F79, P126 (N-TEV), P127 (C-TEV), P222 (PIF6), P240 (PhyB+Linker) and P244 (IgKappa), F42+F84, F90+F86, F90+F87, F90+F88, F90+F89, F59+F85
Investigator: Rosario, Katrin
Aim of the study: Picking of F92+F89, F92+F87, F92+F86, F92+F88, F92+F97, F90+F97, F91+F94, F93+F79, P126 (N-TEV), P127 (C-TEV), P222 (PIF6), P240 (PhyB+Linker) and P244 (IgKappa), F42+F84, F90+F86, F90+F87, F90+F88, F90+F89, F59+F85
Operational sequence:
- Picking and overnight culture after standard laboratory's protocol. (4 µl CamR and 4 ml LB-medium)
- There were no colonies on plates F42+F84 and xy
- There were a lot of colonies on NK 90 but there were significantly more colonies on the plates with ligation products
- 2 colonies were picked for every ligation product.
Friday, June 14th
Picking of transformed Plasmid E. coli XL1 blue with P204, F92+F86 and F42+F84
Investigator: Johanna
Aim of the study: Picking of P204, F92+F86 and F42+F84
Operational sequence:
- Picking and overnight culture after standard laboratory's protocol. (4 µl CamR, 4 ml LB-medium)
- 3 colonies were picked for every transformation product.
Mass of Minipreps of overnight cultures of transformated E. coli XL1 blue
Investigator: Rosario, Johanna, Andi
Aim of the experiment:Mass of Minipreps of overnight cultures of transformated E. coli XL1 blue.
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
- The resulting DNA were eluted in tubes labeled as follows:
| Content | New Tube |
| F59+F85 | P252 |
| F59+F85 | P253 |
| F90+F86 | P254 |
| F90+F86 | P255 |
| F90+F87 | P256 |
| F90+F87 | P257 |
| F90+F88 | P258 |
| F90+F88 | P259 |
| F90+F89 | P260 |
| F90+F89 | P261 |
| F90+F97 | P262 |
| F90+F97 | P263 |
| F91+F94 | P264 |
| F91+F94 | P265 |
| F92+F87 | P266 |
| F92+F87 | P267 |
| F92+F97 | P268 |
| F92+F97 | P269 |
| F92+F88 | P270 |
| F92+F88 | P271 |
| F92+F89 | P272 |
| F92+F89 | P273 |
| F93+F79 | P274 |
| F93+F79 | P275 |
| P222 | P276 |
| P222 | P277 |
| P240 | P278 |
| P240 | P279 |
| P126 | P280 |
| P126 | P281 |
| P244 | P282 |
| P244 | P283 |
| P127 | P284 |
| P127 | P285 |
Transformation of E. coli XL1 blue with P250, P251, P219 (Laccase), P163 (NPT-Cassette)
Investigator: Rosario
Aim of the experiment: Transformation of E. coli XL1 blue with P250, P251, P219 (Laccase), P163 (NPT-Cassette)
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 2 µl of DNA was added to 200 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- Centrifugation for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellets were resuspended in 100 µl of LB-medium and these concentrated cell suspensions were plated on new chloramphenicol plates.
Sequencing of P276 (Retrafo of P222, fw), P282 (Retrafo of P244, fw), P278 (Retrafo of P240, fw+rv) and P217 (fw+rv)
Investigators: Louise
Aim of the experiment: Sequencing of P276(Retrafo of P222, fw), P282(Retrafo of P244, fw), P278 (Retrafo of P240, fw+rv) and P217 (fw+rv).
Procedure: Peparation of DNA for sequencing according to the Eurofins SmartSeq Kit protocol (15 µl of plasmid DNA (50 - 100 ng/µl) and 2 µl sequencing primer 10 µM).
The different genes received the following barcodes:
| Label | Name | Barcode1 | Barcode2 |
|---|---|---|---|
| P276 | PIF6 | FR02305830 (O3) | - |
| P282 | IgKappa-SigP in pSB1C3 | FR02305831 (O3) | |
| P278 | PhyB-36AALinker in pSB1C3 | FR02305832 (O3) | FR02305833 (O4) |
| P217 | Laccase QCIV | FR02305826 (O3) | FR02305827 (O4) |
Analytical digestion and gelelectrophoresis of P252-P275 with EcoRI and SpeI
Investigator: Rosario, Johanna, Louise
Aim of the experiment: Analytical digestion and gelelectrophoresis of P252-P275 with EcoRI and SpeI
Procedure:
- Batches for every Plasmid of P252-P275 contained
| volume | reagent |
| 2 µl | CutSmart Buffer (10x) |
| 0.25 µl | EcoRI |
| 0.25 µl | SpeI |
| 15 µl | ddH2O |
| 2.5 µl | Plasmid DNA (P252-P275) |
| =20 µl | TOTAL |
- The batches for the Quickchangeproduct were prepared with 6 µl of Quickchangeproduct and 2 µl DNA loading buffer.
- Incubation for 60 min at 37 °C.
- Analytical gelelectrophoresis was performed at 90 V for 60 min
- For P252-P275 a 1% agarose gel was used.
| Lane: | DNA marker 1kb | Digestion of P252 with EcoRI & SpeI | Digestion of P253 with EcoRI & SpeI | Digestion of P254 with EcoRI & SpeI | Digestion of P255 with EcoRI & SpeI | Digestion of P256 with EcoRI & SpeI | Digestion of P257 with EcoRI & SpeI | Digestion of P258 with EcoRI & SpeI |
| Result: | - | corrupt | as expected | corrupt | corrupt | corrupt | as expected | corrupt |
| Lane: | DNA marker 1kb | Digestion of P259 with EcoRI & SpeI | Digestion of P260 with EcoRI & SpeI | Digestion of P261 with EcoRI & SpeI | Digestion of P262 with EcoRI & SpeI | Digestion of P263 with EcoRI & SpeI | Digestion of P264 with EcoRI & SpeI | Digestion of P265 with EcoRI & SpeI |
| Result: | - | as expected | as expected (sequencing necessary) | as expected (sequencing necessary) | bad miniprep | as expected | not sure (has to be repeated with control P278) | not sure (has to be repeated with control P278) |
| Lane: | DNA marker 1kb | Digestion of P266 with EcoRI & SpeI | Digestion of P267 with EcoRI & SpeI | Digestion of P268 with EcoRI & SpeI | Digestion of P269 with EcoRI & SpeI | Digestion of P270 with EcoRI & SpeI | Digestion of P271 with EcoRI & SpeI | Digestion of P272 with EcoRI & SpeI |
| Result: | - | as expected | as expected | as expected | as expected | as expected | as expected | as expected (has to be sequenced) |
| Lane: | DNA marker 1kb | Digestion of P273 with EcoRI & SpeI | Digestion of P274 with EcoRI & SpeI | Digestion of P275 with EcoRI & SpeI | Digestion of QC I product of P251 with DpnI | Digestion of QC I product of P163I with DpnI | Digestion of QC V product of P163II with DpnI | Digestion of QC V product of P217 with DpnI |
| Result: | - | as expected (has to be sequenced) | as expected (has to be sequnced) | as expected (has to be sequnced) | corrupt, no QC product | corrupt, no QC product | corrupt, no QC product | as expected, QC product |
Preparative Digestion of IRES (F100) and AlcR (F99) with EcoRI and SpeI
| volume | reagent |
| 4 µl | CutSmart Buffer (10x) |
| 1 µl | EcoRI |
| 1 µl | SpeI |
| 9 µl | ddH2O |
| 25 µl | Fragment DNA (F99 or F100) |
| =40 µl | TOTAL |
- Incubation for 150 min at 37 °C
- Purification by QIAGEN PCR Purification Kit
- new Names: F99 -> F101 and F100 -> F102
Oligohybridization of Strep-TEV (O90 + O91)
Investigator: Rosario
Aim of the experiment: Oligohybridization of Strep-TEV (O90 + O91)
Procedure:
- 25 µL of 100 pmol/µl of Strep-TEV_fw and 25 µL of 100 pmol/µl Strep-TEV_rev
- Heating up to 95 °C for 5 min
- Cooling at RT in a styropor box overnight
Saturday, June 15th
Miniprep of overnight cultures of transformated E. coli XL1 blue with P204 (IgKappa-SigP in pSB1C3), F92+F82 (IgKappa-SigP_EreB in pSB1C3)
Investigator: Jeff
Aim of the experiment: Miniprep of overnight cultures of transformated E. coli XL1 blue with P204 (IgKappa-SigP in pSB1C3), F92+F82 (IgKappa-SigP_EreB in pSB1C3).
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
- The resulting DNA were eluted in tubes labeled as follows:
| Content | New Tube | Concentration in ng/µl |
| ReTraFo of P204 (IgKappa-SigP in pSB1C3) | P286 | 105.7 |
| ReTraFo of P204 (IgKappa-SigP in pSB1C3) | P287 | 303.6 |
| ReTraFo of P204 (IgKappa-SigP in pSB1C3) | P288 | 268.1 |
| F92+F86 (IgKappa-SigP_EreB in pSB1C3) | P289 | 121.2 |
| F92+F86 (IgKappa-SigP_EreB in pSB1C3) | P290 | 402.3 |
| F92+F86 (IgKappa-SigP_EreB in pSB1C3) | P291 | 405.2 |
Ligation of F41+F101 (AlcR) and F41+F102 (IRES of poliovirus)
Investigator: Jeff
Aim of the experiment: Ligation of F41+F101 (AlcR) and F41+F102 (IRES of poliovirus).
Procedure:
- Ligation batch for F41+F101 (AlcR), 50 ng vector used, 1:1:
| volume | reagent |
| 0.56 µl | F41 (89.9 ng/µl, 2086 bp) |
| 5.23 µl | F101 (11.3 ng/µl, 2466 bp) |
| 11.21 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 0.5 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- Ligation batch for F41+F102 (IRES of poliovirus), 100 ng vector used, 3:1:
| volume | reagent |
| 1.11 µl | F41 (89.9 ng/µl, 2086 bp) |
| 7.92 µl | F102 (11.4 ng/µl, 628 bp) |
| 7.97 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 0.5 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- Negative control was also prepared. Instead of the insert, the same volume of ddH2O was added to the reaction batch.
- The ligation was performed for 1 hour at room temperature.
Analytical digestion and gelelectrophoresis of P289-P291 (IgKappa-SigP_EreB in pSB1C3) with EcoRI & SpeI
Investigator: Jeff
Aim of the experiment: Analytical digestion and gelelectrophoresis of P289-P291 (IgKappa-SigP_EreB in pSB1C3) with EcoRI & SpeI.
Procedure:
- Batch
| volume | reagent |
| 2 µl | CutSmart Buffer (10x) |
| 0.25 µl | EcoRI-HF |
| 0.25 µl | SpeI-HF |
| 15 µl | ddH2O |
| 2.5 µl | Plasmid DNA (P289-P291, P245) |
| =20 µl | TOTAL |
- Incubation for 60 min at 37 °C.
- Analytical gelelectrophoresis was performed at 90 V for 60 min on a 1% agarose gel.
| Lane: | DNA ladder 100bp | Digestion of P289 (IgKappa-SigP_EreB in pSB1C3) with EcoRI & SpeI, clone 1 | Digestion of P290 (IgKappa-SigP_EreB in pSB1C3) with EcoRI & SpeI, clone 2 | Digestion of P291 (IgKappa-SigP_EreB in pSB1C3) with EcoRI & SpeI, clone 3 | Digestion of P245 (EreB in pSB1C3) with EcoRI & SpeI |
| Result: | as expected | as expected | as expected | as expected (control) |
Transformation of E. coli XL1 blue with F41+F101 (AlcR in pSB1C3) and F41+F102 (IRES in pSB1C3)
Investigator: Jeff
Aim of the experiment: Transformation of E. coli XL1 blue with F41+F101 (AlcR in pSB1C3) and F41+F102 (IRES in pSB1C3).
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 10 µl of ligation product was added to 200 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of the cell suspension was plated on one chloramphenicol plate.
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on a new chlorampenicol plate.
Sunday, June 16th
QuikChange mutagenesis to remove forbidden restriction sites - SDMIV - of P217 (Laccase), P163 (NPT-Cassette)
Investigator: Andi
Aim of the experiment: Removal of forbidden restriction site from P217 (Laccase) and P163 (NPT-Cassette).
Procedure: Quikchange - PCR
Reaction batch for P217 (Laccase)
| volume | reagent | Additional information |
| 2.5 µl | 10x Pfu Ultra II buffer | |
| 1 µl | Plasmid template (P217 - 1:100 dilution) | need to get between 2.5 ng and 25 ng for a reaction batch of 25 µl |
| 1 µl | 1:10 dilution of O74 (10 µM) | |
| 1 µl | 1:10 dilution of O75 (10 µM) | |
| 18 µl | ddH2O | |
| 1 µl | dNTP mix 50 mM | |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U/µl) |
Reaction batch for P163 (NPT-Cassette)
| volume | reagent | Additional information |
| 2.5 µl | 10x Pfu Ultra II buffer | |
| 1 µl | Plasmid template (P163 - 1:50 dilution) | need to get between 2.5 ng and 25 ng for a reaction batch of 25 µl |
| 1 µl | 1:10 dilution of O70 (10 µM) | |
| 1 µl | 1:10 dilution of O71 (10 µM) | |
| 18 µl | ddH2O | |
| 1 µl | dNTP mix 50 mM | |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U/µl) |
PCR cycling parameters - P251 (Laccase) and P163 (NPT-Cassette)
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 30 s |
| 2 | 19 | 95 °C | 30 s |
| 52 °C | 1 min | ||
| 68 °C | 5 min | ||
| 4 °C | hold |
- After the PCR, the Quickchange product was stored in the fridge.
Picking of transformed Plasmid E. coli XL1 blue with QC of P217, P163, F41+F101 and F41+F102
Investigator: Andi, Jeff
Aim of the study: Picking of QC of P217, P163, F41+F101 and F41+F102
Operational sequence:
- Picking and overnight culture after standard laboratory's protocol. (4 µl Cam, 4 ml LB-medium)
- 1 colonies were picked for every transformation product.
Ligation of F59+F74 (SERK-SigP) and F59+F103 (Strep-TEV)
Investigator: Andi, Jeff
Aim of the experiment: Ligation of F59+F74 (SERK-SigP) and F59+F103 (Strep-TEV).
Procedure:
- Ligation batch for F59+F74 (SERK-SigP), 100 ng vector used, insert:vector = 3:1
| volume | reagent |
| 1.5 µl | F59 (66.7 ng/µl, 2086 bp) |
| 3 µl | F74 (4.8 ng/µl, 100 bp) |
| 12.5 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- Ligation batch for F59+F103 (Strep-TEV), 100 ng vector used, insert:vector = 3:1
| volume | reagent |
| 1.5 µl | F59 (66.7 ng/µl, 2086 bp) |
| 15.5 µl | F103 (504.9 ng/µl, 54 bp) |
| 7.97 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- Negative control was prepared by replacing the volume of the insert by ddH2O.
- The ligation was performed for 1 hour at room temperature.
Transformation of E. coli XL1 blue with P217 (QC of 16.05.2013), P163 (QC of 16.05.2013), P7 (Retrafo of PhyB) and ligation products F59+F74 (SERK-SigP in pSB1C3) and F59+F103 (Strep-TEV in pSB1C3)
Investigator: Andi, Jeff
Aim of the experiment: Transformation of E. coli XL1 blue with P217 (QC of 16.05.2013) and P163 (QC of 16.05.2013), P7 (Retrafo of PhyB) and ligation products F59+F74 (SERK-SigP in pSB1C3) and F59+F103 (Strep-TEV in pSB1C3).
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 10 µl of ligation product/ 2 µl of plasmid (P7) was added to 200 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- The cell suspension were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on a new chlorampenicol plate.
Week 9
Monday, June 17th
Miniprep of overnight cultures of transformated E. coli XL1 blue with QC I of P163 (NPT-casette), QC V of P217 (laccase), ligation products F41+F101 (AlcR) and F41+F102 (IRES)
Investigator: Rosario
Aim of the experiment: Miniprep of overnight cultures of transformated E. coli XL1 blue with QC I of P163 (NPT-casette), QC V of P217 (laccase), ligation products F41+F101 (AlcR) and F41+F102 (IRES) Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
- The resulting DNA were eluted in tubes labeled as follows:
| Content | New Tube |
| QC I of P163 | P292 |
| QC V of P217 | P293 |
| F41+F101 I | P294 |
| F41+F101 II | P295 |
| F41+F101 III | P296 |
| F41+F102 I | P297 |
| F41+F102 II | P298 |
| F41+F102 III | P299 |
Picking of transformed Plasmid E. coli XL1 blue with P217 (QC of 16.05.2013), P163 (QC of 16.05.2013), P7 (Retrafo of PhyB) and ligation products F59+F74 (SERK-SigP in pSB1C3) and F59+F103 (Strep-TEV in pSB1C3)
Investigator: Rosario
Aim of the study: Picking of transformed Plasmid E. coli XL1 blue with P217 (QC of 16.05.2013), P163 (QC of 16.05.2013), P7 (Retrafo of PhyB) and ligation products F59+F74 (SERK-SigP in pSB1C3) and F59+F103 (Strep-TEV in pSB1C3)
Operational sequence:
- Picking and overnight culture after standard laboratory's protocol. (4 µl Cam, 4 ml LB-medium)
- 1 colony was picked for every transformation product.
Preparative digestion and gelelectrophoresis of P264 (PhyBlinker+cTev) with AgeI/SpeI, P299 (IRES) with NgoMIV/SpeI, P274 (nTevlinker)with AgeI/SpeI, P111 (TEV) with AgeI/SpeI and P248 (Nuclease) with NgoMIV/SpeI
Investigator: Johanna, Flo
Aim of the experiment: Preparative digestion and gelelectrophoresis of P264 (PhyBlinker+cTev) with AgeI/SpeI, P299 (IRES) with NgoMIV/SpeI, P274 (nTevlinker)with AgeI/SpeI, P111 (TEV) with AgeI/SpeI and P248 (Nuclease) with NgoMIV/SpeI
Procedure:
- Batch for preparative digestion of P264 (PhyBlinker+cTev) with AgeI/SpeI
| volume | reagent |
| 20 µl | P264 |
| 4 µl | CutSmart Buffer |
| 1 µl | AgeI(20 U/µl) |
| 1 µl | SpeI-HF (20 U/µl) |
| 14 µl | ddH2O |
| =40 µl | TOTAL |
- Batch for preparative digestion of P299 (IRES) with NgoMIV/SpeI
| volume | reagent |
| 20 µl | P299 |
| 4 µl | CutSmart Buffer |
| 1 µl | NgoMIV(20 U/µl) |
| 1 µl | SpeI-HF (20 U/µl) |
| 14 µl | ddH2O |
| =40 µl | TOTAL |
- Batch for preparative digestion of P274 (nTevlinker)with AgeI/SpeI
| volume | reagent |
| 20 µl | P274 |
| 4 µl | CutSmart Buffer |
| 1 µl | AgeI(20 U/µl) |
| 1 µl | SpeI-HF (20 U/µl) |
| 14 µl | ddH2O |
| =40 µl | TOTAL |
- Batch for preparative digestion of P111 (TEV) with AgeI/SpeI
| volume | reagent |
| 20 µl | P111 |
| 4 µl | CutSmart Buffer |
| 1 µl | AgeI(20 U/µl) |
| 1 µl | SpeI-HF (20 U/µl) |
| 14 µl | ddH2O |
| =40 µl | TOTAL |
- Batch for preparative digestion of P248 (Nuclease) with NgoMIV/SpeI
| volume | reagent |
| 20 µl | P248 |
| 4 µl | CutSmart Buffer |
| 1 µl | NgoMIV(20 U/µl) |
| 1 µl | SpeI-HF (20 U/µl) |
| 14 µl | ddH2O |
| =40 µl | TOTAL |
- Incubation for 2.5 h at 37 °C.
- 4 µl of DNA loading buffer (10x) were added to the 40 µl reaction batches after digestion and were loaded on a 1% agarose gel for preparative gelelectrophoresis.
- Preparative gelelectrophoresis was performed at 90 V for 60 min.
| Lane: | Digestion of P111 with AgeI & SpeI | 1 kbp DNA ladder | Digestion of P248 with NgoMIV & SpeI | Digestion of P264 with AgeI & SpeI |
| Result: | as expected | as expected | as expected |
| Lane: | Digestion of P274 with AgeI & SpeI | 1 kbp DNA ladder | Digestion of P299 with NgoMIV & SpeI |
| Result: | low concentration, discarded | not digested, discarded |
- The DNA was extracted from the gel with QIAquick Gel Extraction Kit, QIAGEN
Analytical digestion and gelelectrophoresis of P293-P299 with EcoRI & SpeI and P292 with EcoRI & PstI
Investigator: Rosario
Aim of the experiment: Analytical digestion and gelelectrophoresis of P293-P299 with EcoRI & SpeI and P292 with EcoRI & PstI Procedure:
- Batch
| volume | reagent |
| 2 µl | CutSmart Buffer (10x) |
| 0.25 µl | EcoRI-HF |
| 0.25 µl | SpeI-HF (PstI-HF for P292) |
| 15 µl | ddH2O |
| 2.5 µl | Plasmid DNA (P292-P299) |
| =20 µl | TOTAL |
- Incubation for 60 min at 37 °C.
- Analytical gelelectrophoresis was performed at 90 V for 60 min on a 1% agarose gel.
| Lane: | 1 kbp DNA ladder | Digestion of P292 (QC I P163 (NPT-casette)) with EcoRI & PstI | Digestion of P293 (QC V P217 (Laccase)) with EcoRI & SpeI | Digestion of P294 (F41+F101 (AlcR)) with EcoRI & SpeI, clone 1 | Digestion of P295 (F41+F101 (AlcR)) with EcoRI & SpeI, clone 2 | Digestion of P296 (F41+F101 (AlcR)) with EcoRI & SpeI, clone 3 | Digestion of P297 (F41+F102 (IRES)) with EcoRI & SpeI, clone 1 | Digestion of P298 (F41+F102 (IRES)) with EcoRI & SpeI, clone 2 | Digestion of P299 (F41+F102 (IRES)) with EcoRI & SpeI, clone 3 | 1 kbp DNA ladder |
| Result: | corrupt | corrupt | as expected | as expected | as expected | as expected | as expected | as expected |
DpnI digestion of P163QCI and P217QCV
Investigator: Flo, Jeff
Aim of the experiment: DpnI digestion of P163QCI and P217QCV.
Procedure:
- 1 µl of DpnI was added to the QuikChange products of P163QCI and P217QCV.
- The mixtures were incubated for 1 h at 37 °C; afterwards they were ready for transformation.
Analytical gelelectrophoresis of DpnI digestion of P163QCI and P217QCV
Investigator: Jeff, Flo
Aim of the experiment: Analysis of the DpnI digestion of P163QCI and P217QCV
Procedure:
- 4.5 µl of the QuikChange products of P163 and P217, named P163QCI and P217QCV were mixed together with 0.5 µl of DNA loading buffer (10x).
- Analytical gelelectrophoresis was performed at 90 V for 30 min.
| Lane: | 1 kbp DNA ladder | DpnI digestion of P163QCI.1 | DpnI digestion of P163QCI.2 | DpnI digestion of P217QCV.1 | DpnI digestion of P217QCV.2 |
| Result: | empty | empty | empty | empty |
Ligation of F77+F105 (NLS in pSB1C3 + mature thermonuclease)
Investigator: Jeff, Flo
Aim of the experiment: Ligation of F77+F105 (NLS in pSB1C3 + mature thermonuclease).
Procedure:
- Ligation batch for F77+F105 (NLS in pSB1C3 + mature thermonuclease), 100 ng vector used, insert:vector = 3:1
| volume | reagent |
| 1.78 µl | F77 (56.2 ng/µl, 2143 bp) |
| 5.22 µl | F105 (12 ng/µl, 447 bp) |
| 10 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- Negative control was prepared by replacing the volume of the insert by ddH2O.
- The ligation was performed overnight at 16 °C.
Transformation of E. coli XL1 blue with P217 (QC of 16.05.2013), P163 (QC of 16.05.2013) and P160, P264, P274 and P275 for retransformations
Investigator: Flo
Aim of the experiment: Transformation of E. coli XL1 blue with P217 (QC of 16.05.2013), P163 (QC of 16.05.2013) and P160, P264, P274 and P275 for retransformations.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 10 µl of QuikChange product/2 µl of plasmid DNA was added to 200 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- The cell suspension were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on a new chlorampenicol plate.
Sequencing of P248 (Nuc A, fw), P275 (N-TEV_36AAlinker, fw), P299 (IRES, fw), P292 (QC I of P163 NPT-casette,fw+rv), P293 (QC V of P217 Laccase,fw+rv) and P290 (IgKappa_EreB,fw+rv)
Investigators: Rosario
Aim of the experiment: Sequencing of P248 (Nuc A, fw), P275 (N-TEV_36AAlinker, fw), P299 (IRES, fw), P292 (QC I of P163 NPT-casette,fw+rv), P293 (QC V of P217 Laccase,fw+rv) and P290 (IgKappa_EreB,fw+rv).
Procedure: Peparation of DNA for sequencing according to the Eurofins SmartSeq Kit protocol (15 µl of plasmid DNA (50 - 100 ng/µl) and 2 µl sequencing primer 10 µM).
The different genes received the following barcodes:
| Label | Name | Barcode1 | Barcode2 |
|---|---|---|---|
| P248 | NucA | FR02305834 (O3) | - |
| P275 | N-TEV_36AAlinker | FR02305835 (O3) | |
| P299 | IRES | FR02305825 (O3) | - |
| P292 | QC I NPT-casette | FR02305824 (O3) | FR02305823 (O4) |
| P293 | QC V Laccase | FR02305822 (O3) | FR02305821 (O4) |
| P290 | IgKappa_EreB | FR02305820 (O3) | FR02305819 (O4) |
Tuesday, June 18th
Picking of E. coli XL1 blue transformated with P160, P264, P274 and P275
Investigator: Rosario
Aim of the study: Picking of E. coli XL1 blue transformated with P160, P264, P274 and P275.
Operational sequence:
- Picking and overnight culture after standard laboratory's protocol. (4 µl Cam, 4 ml LB-medium)
- 2 colonies were picked for every transformation product.
Transformation of E. coli XL1 blue with F77+F105 (NLS in pSB1C3 + mature thermonuclease) and P299 Retrafo
Investigator: Rosario
Aim of the experiment: Transformation of E. coli XL1 blue with P217 (QC of 16.05.2013), P163 (QC of 16.05.2013) and P160, P264, P274 and P275 for retransformations.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 2 µl of plasmid DNA was added to 200 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- The cell suspension were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on a new chlorampenicol plate.
Miniprep of overnight cultures of transformated E. coli XL1 blue with F59+F103 (Strep-TEV-Linker in PsB1C3) and F59+F74 (Serk-Sigp in PsB1C3) and P7 (PhyB)
Investigator: Louise
Aim of the experiment: Miniprep of overnight cultures of transformated E. coli XL1 blue with F59+F103 (Strep-TEV-Linker in PsB1C3) and F59+F74 (Serk-Sigp in PsB1C3) and P7 (PhyB).
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
- The resulting DNA were eluted in tubes labeled as follows:
| Content | New Tube |
| P7 (PhyB) clone 1 | P300 |
| P7 (PhyB) clone 2 | P301 |
| F59+F74 (Serk-Sigp in PsB1C3) clone 1 | P302 |
| F59+F74 (Serk-Sigp in PsB1C3) clone 2 | P303 |
| F59+F103 (Strep-TEV-Linker in PsB1C3) clone 1 | P304 |
| F59+F103 (Strep-TEV-Linker in PsB1C3) clone 2 | P305 |
Analytical digestion and gelelectrophoresis of P300-P301 (PhyB), P302-P303 (Serk-Sigp in PsB1C3) and P304-P305 (Strep-TEV-Linker in PsB1C3) with XbaI and AgeI
Investigator: Louise, Rosario, Florian
Aim of the experiment: Analytical digestion and gelelectrophoresis of P300-P301 (PhyB), P302-P303 (Serk-Sigp in PsB1C3) and P304-P305 (Strep-TEV-Linker in PsB1C3) with XbaI and AgeI.
Procedure:
- Batch
| volume | reagent |
| 2 µl | CutSmart Buffer (10x) |
| 0.25 µl | AgeI |
| 0.25 µl | XbaI |
| 15 µl | ddH2O |
| 2.5 µl | Plasmid DNA (P300-P305) |
| =20 µl | TOTAL |
- Incubation for 60 min at 37 °C.
- Analytical gelelectrophoresis was performed at 90 V for 60 min
| Lane: | Digestion of P300 with XbaI/AgeI | Digestion of P301 with XbaI/AgeI | Digestion of P302 with XbaI/AgeI | Digestion of P303 with XbaI/AgeI | Digestion of P304 with XbaI/AgeI | Digestion of P305 with XbaI/AgeI | 1 kb DNA Ladder | 100 bp DNA Ladder |
| Result: | as expected | as expected | as expected | as expected | as expected | as expected |
Preparative digestion and gelelectrophoresis of P212 (SERK-TMD) with NgoMIV/SpeI, P238 (SERK-TMD_Linker_GFP) with NgoMIV/SpeI, P303 (SERK-SigP) with AgeI/SpeI and P304 (StrepTEV-Linker) with AgeI/SpeI
Investigator: Louise, Flo, Jeff, Rosario
Aim of the experiment: Preparative digestion and gelelectrophoresis of P212 (SERK-TMD) with NgoMIV/SpeI, P238 (SERK-TMD_Linker_GFP) with NgoMIV/SpeI, P303 (SERK-SigP) with AgeI/SpeI and P304 (StrepTEV-Linker) with AgeI/SpeI.
Procedure:
- Batch for preparative digestion of P212 (SERK-TMD) with NgoMIV/SpeI
| volume | reagent |
| 20 µl | P212 |
| 4 µl | CutSmart Buffer |
| 1 µl | NgoMIV(20 U/µl) |
| 1 µl | SpeI-HF (20 U/µl) |
| 14 µl | ddH2O |
| =40 µl | TOTAL |
- Batch for preparative digestion of P238 (SERK-TMD_Linker_GFP) with NgoMIV/SpeI
| volume | reagent |
| 20 µl | P238 |
| 4 µl | CutSmart Buffer |
| 1 µl | NgoMIV(20 U/µl) |
| 1 µl | SpeI-HF (20 U/µl) |
| 14 µl | ddH2O |
| =40 µl | TOTAL |
- Batch for preparative digestion of P303 (SERK-SigP) with AgeI/SpeI
| volume | reagent |
| 20 µl | P303 |
| 4 µl | CutSmart Buffer |
| 1 µl | AgeI(20 U/µl) |
| 1 µl | SpeI-HF (20 U/µl) |
| 14 µl | ddH2O |
| =40 µl | TOTAL |
- Batch for preparative digestion of P304 (StrepTEV-Linker) with AgeI/SpeI
| volume | reagent |
| 20 µl | P304 |
| 4 µl | CutSmart Buffer |
| 1 µl | AgeI(20 U/µl) |
| 1 µl | SpeI-HF (20 U/µl) |
| 14 µl | ddH2O |
| =40 µl | TOTAL |
- Incubation for 2.5 h at 37 °C.
- 4 µl of DNA loading buffer (10x) were added to the 40 µl reaction batches after digestion and P238, P303 and P304 were loaded on a 1% agarose gel, while P212 was loaded on a 2% agarose gel for preparative gelelectrophoresis.
- Preparative gelelectrophoresis was performed at 90 V for 60 min.
| Lane: | Digestion of P238 (SERK-TMD_Linker_GFP) with NgoMIV/SpeI | 1 kbp DNA ladder | Digestion of P303 (SERK-SigP) with AgeI/SpeI | Digestion of P304 (StrepTEV-Linker) with AgeI/SpeI |
| Result: | as expected | as expected | as expected |
| Lane: | 1 kbp DNA ladder | Digestion of P212 (SERK-TMD) with NgoMIV/SpeI | 100 bp DNA Ladder |
| Result: | as expected |
- The DNA was extracted from the gel with QIAquick Gel Extraction Kit, QIAGEN
Ligation of F109+F86 (SERK-SigP + EreB), F109+F87 (SERK-SigP + NanoLuc), F109+F88 (SERK-SigP + SpyCatcher), F109+F89 (SERK-SigP + SpyTag), F109+F97 (SERK-SigP + XylE), F109+F107 (SERK-SigP + SERK-TMD) and F110+F108 (Strep-tag-TEV-site-linker + SERK-TMD_8AAlinker_GFPmut1)
Investigator: Jeff, Flo
Aim of the experiment: Ligation of F109+F86 (SERK-SigP + EreB), F109+F87 (SERK-SigP + NanoLuc), F109+F88 (SERK-SigP + SpyCatcher), F109+F89 (SERK-SigP + SpyTag), F109+F97 (SERK-SigP + XylE), F109+F107 (SERK-SigP + SERK-TMD) and F110+F108 (Strep-tag-TEV-site-linker + SERK-TMD_8AAlinker_GFPmut1).
Procedure:
- Ligation batch for F109+F86 (SERK-SigP + EreB), 100 ng vector used, insert:vector = 3:1
| volume | reagent |
|---|---|
| 0.92 µl | F109 (108.2 ng/µl, 2177 bp) |
| 5.57 µl | F86 (31.0 ng/µl, 1254 bp) |
| 10.51 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- Ligation batch for F109+F87 (SERK-SigP + NanoLuc), 100 ng vector used, insert:vector = 3:1
| volume | reagent |
|---|---|
| 0.92 µl | F109 (108.2 ng/µl, 2177 bp) |
| 6.89 µl | F87 (10.2 ng/µl, 510 bp) |
| 9.19 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- Ligation batch for F109+F88 (SERK-SigP + SpyCatcher), 100 ng vector used, insert:vector = 3:1
| volume | reagent |
|---|---|
| 0.92 µl | F109 (108.2 ng/µl, 2177 bp) |
| 5.37 µl | F88 (8.7 ng/µl, 339 bp) |
| 10.71 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- Ligation batch for F109+F89 (SERK-SigP + SpyTag), 100 ng vector used, insert:vector = 3:1
| volume | reagent |
|---|---|
| 0.92 µl | F109 (108.2 ng/µl, 2177 bp) |
| 12.0 µl | F89 (6.8 ng/µl, 39 bp) |
| 4.08 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- Ligation batch for F109+F97 (SERK-SigP + XylE), 100 ng vector used, insert:vector = 3:1
| volume | reagent |
|---|---|
| 0.92 µl | F109 (108.2 ng/µl, 2177 bp) |
| 6.52 µl | F97 (19.4 ng/µl, 918 bp) |
| 9.56 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- Ligation batch for F109+F107 (SERK-SigP + SERK-TMD), 100 ng vector used, insert:vector = 3:1
| volume | reagent |
|---|---|
| 0.92 µl | F109 (108.2 ng/µl, 2177 bp) |
| 5.38 µl | F107 (4.8 ng/µl, 186 bp) |
| 10.70 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- Ligation batch for F110+F108 (Strep-tag-TEV-site-linker + SERK-TMD_8AAlinker_GFPmut1), 100 ng vector used, insert:vector = 3:1
| volume | reagent |
|---|---|
| 1.38 µl | F110 (72.4 ng/µl, 2143 bp) |
| 5.53 µl | F108 (23.6 ng/µl, 933 bp) |
| 10.09 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- Negative control was prepared by replacing the volume of the insert by ddH2O.
- The ligation was performed for 1 hour at room temperature.
QuikChange mutagenesis to remove forbidden restriction sites - SDMV of P217 (Laccase RFC10) and SDMI of P162 (NPT-Cassette RFC10)
Investigator: Jeff
Aim of the experiment: Removal of the last existing forbidden restriction site from P217 (Laccase) and the first one from P162 (NPT-Cassette).
Procedure: Quikchange - PCR
Reaction batch for P217 (Laccase)
| volume | reagent | Additional information |
| 2.5 µl | 10x Pfu Ultra II buffer | |
| 0.5 µl | Plasmid template (P217 - 1:100 dilution) | need to get between 2.5 ng and 25 ng for a reaction batch of 25 µl |
| 1 µl | 1:10 dilution of O74 (=10 µM) | |
| 1 µl | 1:10 dilution of O75 (=10 µM) | |
| 18.5 µl | ddH2O | |
| 1 µl | dNTP mix 50 mM | |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U/µl) |
Reaction batch for P163 (NPT-Cassette)
| volume | reagent | Additional information |
| 2.5 µl | 10x Pfu Ultra II buffer | |
| 0.5 µl | Plasmid template (P162 - 1:50 dilution) | need to get between 2.5 ng and 25 ng for a reaction batch of 25 µl |
| 1 µl | 1:10 dilution of O70 (10 µM) | |
| 1 µl | 1:10 dilution of O71 (10 µM) | |
| 18.5 µl | ddH2O | |
| 1 µl | dNTP mix 50 mM | |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U/µl) |
PCR cycling parameters - P251 (Laccase) and P163 (NPT-Cassette)
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 30 s |
| 2 | 19 | 95 °C | 30 s |
| 52 °C | 1 min | ||
| 68 °C | 4 min | ||
| 4 °C | hold |
- After the PCR, the Quickchange products were mixed with 1 µl of DpnI (10 U/µl) and incubated at 37 °C for 1 h.
Analytical gelelectrophoresis of the DpnI digested products of the performed QuikChange of SDMV of P217 (Laccase RFC10) and SDMI of P162 (NPT-Cassette RFC10)
Investigator: Jeff
Aim of the experiment:
Procedure:
- 4.5 µl of each of the DpnI digested QuikChange products were mixed with 1 µl of DNA loading buffer (10x).
- Analytical gelelectrophoresis was performed at 90 V for 40 min at an 1% agarose gel.
| Lane: | 1 kbp DNA ladder | DpnI digestion of P163QCI | DpnI digestion of P217QCV |
| Result: | as expected | as expected |
Transformation of E. coli XL1-blue with ligation products F109+F86 (SERK-SigP + EreB), F109+F87 (SERK-SigP + NanoLuc), F109+F88 (SERK-SigP + SpyCatcher), F109+F89 (SERK-SigP + SpyTag), F109+F97 (SERK-SigP + XylE), F109+F107 (SERK-SigP + SERK-TMD) and F110+F108 (Strep-tag-TEV-site-linker + SERK-TMD_8AAlinker_GFPmut1), with DpnI digested QuikChange products P162QCI and P217QCV, and P212 (ReTraFo), P238 (ReTraFo)
Investigator: Jeff
Aim of the experiment: Transformation of E. coli XL1-blue with ligation products F109+F86 (SERK-SigP + EreB), F109+F87 (SERK-SigP + NanoLuc), F109+F88 (SERK-SigP + SpyCatcher), F109+F89 (SERK-SigP + SpyTag), F109+F97 (SERK-SigP + XylE), F109+F107 (SERK-SigP + SERK-TMD) and F110+F108 (Strep-tag-TEV-site-linker + SERK-TMD_8AAlinker_GFPmut1), with DpnI digested QuikChange products P162QCI and P217QCV, and P212 (ReTraFo), P238 (ReTraFo).
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 10 µl of QuikChange products/10 µl of ligation products/2 µl of plasmid DNA was added to 200 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- The cell suspension were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on a new chlorampenicol plate.
Sequencing of P294 (AlcR)
Investigators: Rosario
Aim of the experiment: Sequencing of P294 (AlcR).
Procedure: Peparation of DNA for sequencing according to the Eurofins SmartSeq Kit protocol (15 µl of plasmid DNA (50 - 100 ng/µl) and 2 µl sequencing primer 10 µM).
The different genes received the following barcodes:
| Label | Name | Barcode1 | Barcode2 |
|---|---|---|---|
| P294 | AlcR | FR02305817 (O3) | FR02305818(O4) |
Wednesday, June 19th
QuikChange mutagenesis to remove forbidden restriction sites - SDMI of P294 (AlcR)
Investigator: Florian - Master of QuikChange 2.0
Aim of the experiment: Removal of the first forbidden restriction site from P294 (AlcR).
Procedure: Quikchange - PCR
Reaction batch for P294 (AlcR)
| volume | reagent | Additional information |
| 2.5 µl | 10x Pfu Ultra II buffer | |
| 0.5 µl | Plasmid template (P294 - 1:50 dilution) | need to get between 2.5 ng and 25 ng for a reaction batch of 25 µl |
| 1 µl | 1:10 dilution of O80 (=10 µM) | |
| 1 µl | 1:10 dilution of O81 (=10 µM) | |
| 18.5 µl | ddH2O | |
| 1 µl | dNTP mix 50 mM | |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U/µl) |
PCR cycling parameters - P294 (AlcR)
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 30 s |
| 2 | 19 | 95 °C | 30 s |
| 52 °C | 1 min | ||
| 68 °C | 5 min | ||
| 4 °C | hold |
- After the PCR, the Quickchange products were mixed with 1 µl of DpnI (10 U/µl) and incubated at 37 °C for 1 h.
Analytical gelelectrophoresis of the DpnI digested products of the performed QuikChange of SDMI of P294 (AlcR)
Investigator: Florian
Aim of the experiment: Analytical gelelectrophoresis of the DpnI digested products of the performed QuikChange of SDMI of P294 (AlcR).
Procedure:
- 4 µl of each of the DpnI digested QuikChange products were mixed with 1 µl of DNA loading buffer (10x).
- Analytical gelelectrophoresis was performed at 90 V for 50 min at an 1% agarose gel.
| Lane: | 1 kbp DNA ladder | DpnI digestion of P294QCI |
|---|---|---|
| Result: | only Primers |
Transformation of E. coli XL1-blue with DpnI digested QuikChange product P294QCI and P294 (ReTrafo of FluA)
Investigator: Florian
Aim of the experiment: Transformation of E. coli XL1-blue with DpnI digested QuikChange product P294QCI and P294 (ReTrafo of FluA).
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 10 µl of QuikChange products/2 µl of plasmid DNA was added to 200 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- The cell suspension were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on a new chlorampenicol/ampicillin plate.
Sequencing of P308 (ReTrafo of P264, PhyB_36AALinker_C-TEV in pSB1C3), P310 (ReTrafo of P274, N-TEV_36AALinker in pSB1C3), P303 (SERK-SigP in pSB1C3), P304 (Strep-TEV-Linker in pSB1C3), P305 (Strep-TEV-Linker in pSB1C3) and P301 (ReTrafo of P7, Phytotchrome B)
Investigators: Rosario
Aim of the experiment: Sequencing of P308 (ReTrafo of P264, PhyB_36AALinker_C-TEV in pSB1C3), P310 (ReTrafo of P274, N-TEV_36AALinker in pSB1C3), P303 (SERK-SigP in pSB1C3), P304 (Strep-TEV-Linker in pSB1C3), P305 (Strep-TEV-Linker in pSB1C3) and P301 (ReTrafo of P7, Phytotchrome B).
Procedure: Peparation of DNA for sequencing according to the Eurofins SmartSeq Kit protocol (15 µl of plasmid DNA (50 - 100 ng/µl) and 2 µl sequencing primer 10 µM).
The different genes received the following barcodes:
| Label | Name | Barcode1 | Barcode2 |
|---|---|---|---|
| P308 | PhyB_36AALinker_C-TEV in pSB1C3 | FR02305816 (O3) | FR02305815 (O4) |
| P310 | N-TEV_36AALinker in pSB1C3 | FR02305814 (O3) | |
| P303 | SERK-SigP in pSB1C3 | FR02305813 (O3) | - |
| P304 | Strep-TEV-Linker in pSB1C3 | FR02305812 (O3) | - |
| P305 | Strep-TEV-Linker in pSB1C3 | FR02305811 (O3) | - |
| P301 | Phytotchrome B | FR02305810 (O3) | FR02305809 (O4) |
Miniprep of overnight cultures of transformated E. coli XL1 blue with ReTrafo of P160 (SV40 NLS), ReTrafo of P264 (PhyB_36AALinker_C-TEV in pSB1C3), ReTrafo of P274 (N-TEV_36AALinker in pSB1C3) and ReTrafo of P275 (N-TEV_36AALinker in pSB1C3)
Investigator: Rosario
Aim of the experiment: Miniprep of overnight cultures of transformated E. coli XL1 blue with ReTrafo of P160 (SV40 NLS), ReTrafo of P264 (PhyB_36AALinker_C-TEV in pSB1C3), ReTrafo of P274 (N-TEV_36AALinker in pSB1C3) and ReTrafo of P275 (N-TEV_36AALinker in pSB1C3).
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
- The resulting DNA were eluted in tubes labeled as follows:
| Content | New Tube |
| P160 I | P306 |
| P160 II | P307 |
| P264 I | P308 |
| P264 II | P309 |
| P274 I | P310 |
| P274 II | P311 |
| P275 I | P312 |
| P275 II | P313 |
Pouring of CamR gel plates
Investigator: Louise, Volker
Aim of the experiment: Pouring of CamR gel plates
Procedure: Preparation of 5 l of LB-Medium with agar was performed after standard recipe
Picking of QC I of P162 (npt-casette in pSB1C3), QC V of P217 (Laccase), ReTrafo of P212, ReTrafo of P238, ReTrafo of P299. F77+F105 (NucA+NLS), F110+F108 (Strep-tag-TEV-site-linker+SERK-TMD_8AAlinker_GFPmut1), F109+F86 (SERK-SigP+EreB), F109+F87 (SERK-SigP+NanoLuc), F109+F88 (SERK-SigP+SpyCatcher), F109+F89 (SERK-SigP+SpyTag), F109+F97 (SERK-SigP+xylE), F109+F107 (SERK-SigP+SERK-TMD)
Investigator: Rosario
Aim of the experiment: Picking of QC I of P162 (npt-casette in pSB1C3), QC V of P217 (Laccase), ReTrafo of P212, ReTrafo of P238, ReTrafo of P299. F77+F105 (NucA+NLS), F110+F108 (Strep-tag-TEV-site-linker+SERK-TMD_8AAlinker_GFPmut1), F109+F86 (SERK-SigP+EreB), F109+F87 (SERK-SigP+NanoLuc), F109+F88 (SERK-SigP+SpyCatcher), F109+F89 (SERK-SigP+SpyTag), F109+F97 (SERK-SigP+xylE), F109+F107 (SERK-SigP+SERK-TMD)
Procedure:
- Retransformations (P212, P238, P299) 1 clone each
- Quickchanges (QCI P162, QCV P217) 2 clones each
- Ligation F77+F105 4 clones because of the high number of colonies at F77 negative ctrl
- All other Ligations (F110+F108, F109+F86, F109+F87, F109+F88, F109+F89, F109+F97, F109+F107) 2 clones each
Thursday, June 20th
Miniprep of overnight cultures of transformated E. coli XL1 blue with QC I of P162 (npt-casette in pSB1C3), QC V of P217 (Laccase), ReTrafo of P212, ReTrafo of P238, ReTrafo of P299. F77+F105 (NucA+NLS), F110+F108 (Strep-tag-TEV-site-linker+SERK-TMD_8AAlinker_GFPmut1), F109+F86 (SERK-SigP+EreB), F109+F87 (SERK-SigP+NanoLuc), F109+F88 (SERK-SigP+SpyCatcher), F109+F89 (SERK-SigP+SpyTag), F109+F97 (SERK-SigP+xylE), F109+F107 (SERK-SigP+SERK-TMD)
Investigator: Rosario
Aim of the experiment: Miniprep of overnight cultures of transformated E. coli XL1 blue with QC I of P162 (npt-casette in pSB1C3), QC V of P217 (Laccase), ReTrafo of P212, ReTrafo of P238, ReTrafo of P299. F77+F105 (NucA+NLS), F110+F108 (Strep-tag-TEV-site-linker+SERK-TMD_8AAlinker_GFPmut1), F109+F86 (SERK-SigP+EreB), F109+F87 (SERK-SigP+NanoLuc), F109+F88 (SERK-SigP+SpyCatcher), F109+F89 (SERK-SigP+SpyTag), F109+F97 (SERK-SigP+xylE), F109+F107 (SERK-SigP+SERK-TMD).
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
- The resulting DNA were eluted in tubes labeled as follows:
| Content | New Tube |
|---|---|
| QC I P162 I | P315 |
| QC I P162 II | P316 |
| QC V P217 I | P317 |
| QC V P217 II | P318 |
| P212 ReTrafo | P319 |
| P238 ReTrafo | P320 |
| P299 ReTrafo | P321 |
| F77+F105 I | P322 |
| F77+F105 II | P323 |
| F77+F105 III | P324 |
| F77+F105 IV | P325 |
| F110+F108 I | P326 |
| F110+F108 II | P327 |
| F109+F86 I | P328 |
| F109+F86 II | P329 |
| F109+F87 I | P330 |
| F109+F87 II | P331 |
| F109+F88 I | P332 |
| F109+F88 II | P333 |
| F109+F89 I | P334 |
| F109+F89 II | P335 |
| F109+F97 I | P336 |
| F109+F97 II | P337 |
| F109+F107 I | P338 |
| F109+F107 II | P339 |
Retransformation of E. coli XL1 blue with P170, P111, P314, P160, P275, P244, P240, P212, P208, P114, P217, P204, P213 and P222
Investigator: Johanna, Andi
Aim of the experiment: Retransformation of E. coli XL1 blue with P170, P111, P314, P160, P275, P244, P240, P212, P208, P114, P217, P204, P213 and P222.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 2 µl of ddH2O was added to empty tubes
- 2 µl of DNA was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- The transformated cells were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated on chlorampenicol (P170, P111, P160, P275, P244, P240, P212, P208, P217, P204, P213, P222) and ampicillin plates (P314, P114).
Sequencing of P320 (ReTrafo of P238,SERK-TMD_8AAlinker_GFPmut1)
Investigators: Rosario
Aim of the experiment: Sequencing of P320 (ReTrafo of P238,SERK-TMD_8AAlinker_GFPmut1).
Procedure: Peparation of DNA for sequencing according to the Eurofins SmartSeq Kit protocol (15 µl of plasmid DNA (50 - 100 ng/µl) and 2 µl sequencing primer 10 µM).
The different genes received the following barcodes:
| Label | Name | Barcode1 | Barcode2 |
|---|---|---|---|
| P308 | SERK-TMD_8AAlinker_GFPmut1 | FR02305808 (O3) | FR02305807 (O4) |
Preparation of media for competent cells
Investigator : Johanna, Louise
Aim of the experiment:Preparation of media for competent cells
Procedure:
- Picking of one colony of overnight stem culture of non competent XL1-Blue E.Coli in 50 ml LB-Medium without antibiotics (under the sterile bench)
- this was incubate overnight at 37 °C
- Batch for 1l of a 0.1 M MgCl2-solution
| amount | reagent |
| 20.33 g | MgCl2 (M=203.3 g/mol) |
| 1 l | ELGA H2O |
- Batch for 500ml of a 50 mM CaCl2-solution
| amount | reagent |
| 3.67 g | CaCl2 (M=147.02 g/mol) |
| 500 ml | ELGA H2O |
- Batch for 50ml of a 50 mM CaCl2-solution, 15% v/v Glycerin
| amount | reagent |
| 0.368 g | CaCl2 (M=147.02 g/mol) |
| 0.458 g | Glycerin (density=1.26) |
| 50 ml | ELGA H2O |
- Autoclave each bottle of solution and leave each in a 4°C room.
QuikChanges QC pActin and QC1 AlcR
Investigator : Andi
Aim of the experiment: QuikChanges QC pActin (P114) and QC1 AlcR (P294)
Procedure: Quickchange - PCR
Reaction batch 1 for P114 (pActin) with Pfu Ultra II
| volume | reagent | Additional information |
| 2.5 µl | 10x Pfu Ultra II buffer | |
| 1 µl | Plasmid template (P114 undiluted) | 4.5 ng in 25 µl batch |
| 1 µl | 1:10 dilution of O76 (=10 µM) | 0.5 µl Primer + 4.5 µl ddH2O |
| 1 µl | 1:10 dilution of O77 (=10 µM) | 0.5 µl Primer + 4.5 µl ddH2O |
| 18 µl | ddH2O | |
| 1 µl | dNTP mix 50 mM | |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U/µl) |
PCR cycling parameters - P114 (pActin)
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 30 s |
| 2 | 20 | 95 °C | 30 s |
| 3 | 52 °C | 1 min | |
| 4 | 68 °C | 6.5 min | |
| 5 | 4 °C | hold |
Reaction batch 2 for P114 (pActin) with Q5 Polymerase Mastermix
| volume | reagent | Additional information |
| 12.5 µl | 2x QS Polymerase Mastermix | |
| 1 µl | Plasmid template (P114 - 1:10 dilution) | 0.45 ng in 25 µl batch |
| 1.25 µl | 1:10 dilution of O76 (=10 µM) | 0.5 µl Primer + 4.5 µl ddH2O |
| 1.25 µl | 1:10 dilution of O77 (=10 µM) | 0.5 µl Primer + 4.5 µl ddH2O |
| 9 µl | ddH2O |
PCR cycling parameters - P114 (pActin) with Q5 Polymerase
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 30 s |
| 2 | 20 | 95 °C | 30 s |
| 3 | 55 °C | 1 min | |
| 4 | 72 °C | 3.25 min | |
| 5 | 4 °C | hold |
Reaction batch for P294 (AlcR) with Pfu Ultra II
| volume | reagent | Additional information |
| 2.5 µl | 10x Pfu Ultra II buffer | |
| 0.5 µl | Plasmid template (P294 dilutsion 1:100) | 2.79 ng in 25 µl batch |
| 1 µl | 1:10 dilution of O76 (=10 µM) | 0.5 µl Primer + 4.5 µl ddH2O |
| 1 µl | 1:10 dilution of O77 (=10 µM) | 0.5 µl Primer + 4.5 µl ddH2O |
| 18.5 µl | ddH2O | |
| 1 µl | dNTP mix 50 mM | |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U/µl) |
PCR cycling parameters - P294 (AlcR)
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 30 s |
| 2 | 20 | 95 °C | 30 s |
| 3 | 52 °C | 1 min | |
| 4 | 68 °C | 6.5 min | |
| 5 | 4 °C | hold |
- After the PCR, the Quickchange products of P114 (pActin) with Q5 Mastermix and P294 (AlcR)were mixed with 1 µl of DpnI (10 U/µl) and incubated at 37 °C for 1 h.
- After the PCR, the Quickchange products of P114 (pActin) with PfU II Polymerase was stored in the Cycler at 4 °C over night
Transformation of E. coli XL1-blue with DpnI digested QuikChange product P114 (pActin) and P294 (AlcR)
Investigator: Master of Quickchange, Gel-Penetrator
Aim of the experiment: Transformation of E. coli XL1-blue with DpnI digested QuikChange product P294 (AlcR) QCI and P114 (pActin).
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 10 µl of QuikChange products/2 µl of plasmid DNA was added to 200 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- The cell suspension were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on a new chlorampenicol/ampicillin plate.
PCR of P317 (Laccase) with Q5 Mastermix and Herculase Polymerase
Investigator: Andi, Johanna
Aim of the experiment: PCR of P317 (Laccase) with Q5 Mastermix and Herculase Polymerase
Procedure:
Operational sequence:
- Reaction batch with Herculase Polymerase
| volume | reagent |
| 10 µl | 5x Herculase Fusion Polymerase reaction buffer |
| 2.5 µl | 10 mM dNTPs |
| 1.25 µl | 10 µM Forward Primer O24 (Bpul_for) |
| 1.25 µl | 10 µM Reverse Primer O25 (Bpul_rev) |
| 1 µL | Herculase II Fusion Polymerase |
| 1.5 µl | Plasmid DNA (P317)- 1:500 dilution |
| 0.5 µl | DMSO |
| 32 µL | ddH2O Water |
| =50 µL | TOTAL |
- Reaction batch with Q5 Mastermix
| volume | reagent |
| 25 µl | 2x Q5 Master Mix |
| 2.5 µl | 10 µM Forward Primer O24 (Bpul_for) |
| 2.5 µl | 10 µM Reverse Primer O25 (Bpul_rev) |
| 1 µl | Plasmid DNA (P317)- 1:500 dilution |
| 19 µL | ddH2O Water |
| =50 µL | TOTAL |
- The content has been mixed with a pipette
- The PCR program was performed after following scheme:
| Initial denaturation | 94 °C | 30 s |
| 30 cycles | 94 °C | 30 s |
| 51 °C | 60 s | |
| 68 °C | 90 s | |
| Final extension | 68 °C | 5 min |
| Hold | 4 °C | infinite |
- After the PCR has finished, the product was hold at 4 °C in the Cycler
Analytical gelelectrophoresis of the DpnI digested products of the performed QuickChange of SDMI of P294 (AlcR)and P114 (pActin)
Investigator: Andi, Johanna
Aim of the experiment: Analytical gelelectrophoresis of the DpnI digested products of the performed QuikChange of SDMI of P294 (AlcR)and P114 (pActin.)
Procedure:
- 9 µl of each of the DpnI digested QuikChange products were mixed with 1 µl of DNA loading buffer (10x).
- Analytical gelelectrophoresis was performed at 90 V for 50 min at an 1% agarose gel.
| Lane: | 100 bp DNA ladder | DpnI digestion of P294QCI | DpnI digestion of P114QCI | 100 bp DNA ladder |
|---|---|---|---|---|
| Result: | expected result | only Primers |
Analytical digestion and gelelectrophoresis of P315 and P316 (QC I P162 NPT-casette), P162 (npt-casette in pSB1C3), P317 and P318 (QC V P217 Laccase), P217 (Laccase), P322 (F77+F105 NucA+NLS), P248 (Thermonuc in pSB1C3), P320 (SERK-TMD_8AAlinker_GFPmut1), P326+P327 (Strep-tag-TEV-site-linker+SERK-TMD_8AAlinker_GFPmut1), P328+P329 (SERK-SigP+EreB), P157 (EreB), P330+P331 (SERK-SigP+NanoLuc), P208 (NanoLuc), P332+P333 (SERK-SigP+SpyCatcher), P213 (SpyCatcher), P334+335 (SERK-SigP+SpyTag), P170 (SpyTag), P336+P337 (SERK-SigP+xylE), P197 (xylE), P338+P339 (SERK-SigP+SERK-TMD), P212(SERK-TMD)
Investigator: Rosario, Katrin
Aim of the experiment: Analytical digestion and gelelectrophoresis of P315 and P316 (QC I P162 NPT-casette), P162 (npt-casette in pSB1C3), P317 and P318 (QC V P217 Laccase), P217 (Laccase), P322 (F77+F105 NucA+NLS), P248 (Thermonuc in pSB1C3), P320 (SERK-TMD_8AAlinker_GFPmut1), P326+P327 (Strep-tag-TEV-site-linker+SERK-TMD_8AAlinker_GFPmut1), P328+P329 (SERK-SigP+EreB), P157 (EreB), P330+P331 (SERK-SigP+NanoLuc), P208 (NanoLuc), P332+P333 (SERK-SigP+SpyCatcher), P213 (SpyCatcher), P334+335 (SERK-SigP+SpyTag), P170 (SpyTag), P336+P337 (SERK-SigP+xylE), P197 (xylE), P338+P339 (SERK-SigP+SERK-TMD), P212(SERK-TMD)
Procedure:
- Batch
| volume | reagent |
| 2 µl | CutSmart Buffer (10x) |
| 0.25 µl | EcoRI |
| 0.25 µl | PstI |
| 15 µl | ddH2O |
| 2.5 µl | Plasmid DNA |
| =20 µl | TOTAL |
- Incubation for 90 min at 37 °C.
- Analytical gelelectrophoresis was performed at 90 V for 60 min
| 1 kb DNA Ladder | P315 | P316 | P162 (Control) | P317 | P318 | P217 (Control) | P322 | P323 | P324 | P325 | P248 (Control) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| as expected | as expected | as expected | as expected | as expected | as expected | corrupt | corrupt | corrupt | corrupt | as expected |
| 1 kb DNA Ladder | P334 | P335 | P170 (Control) | P336 | P337 | P197 (Control) | P338 | P339 | P212 (Control) | P320 |
|---|---|---|---|---|---|---|---|---|---|---|
| as expected | as expected | as expected | as expected | as expected | as expected | as expected | as expected | as expected | is repeated on other gel |
| 1 kb DNA Ladder | P320 (Control) | P326 | P327 |
|---|---|---|---|
| as expected | as expected | as expected |
| 1 kb DNA Ladder | P328 | P329 | P157 (Control) | P330 | P331 | P208 (Control) | P332 | P333 | P213 (Control) | P326 | P327 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| as expected | as expected | as expected | as expected | as expected | corrupt | as expected | as expected | corrupt | is repeated on other gel | is repeated on other gel |
Friday, June 21st
Analytical gelelectrophoresis of the PCR product of P317 (Laccse) with PfU II Polymerase, Herculase Polymerase and the DpnI digested products of the performed QuickChange of P114 (pActin)
Investigator: Master of QuikChange
Aim of the experiment: Analytical gelelectrophoresis of the PCR product of P317 (Laccse) with PfU II Polymerase, Herculase Polymerase and the DpnI digested products of the performed QuickChange of P114 (pActin)
Procedure:
- 9 µl of each of the PCR products were mixed with 1 µl of DNA loading buffer (10x).
- 9 µl of each of the DpnI digested QuikChange product was mixed with 1 µl of DNA loading buffer (10x).
- Analytical gelelectrophoresis was performed at 90 V for 50 min at an 1% agarose gel.
| Lane: | 1000 bp DNA ladder | PCR product of P317 with Q5 Master Mix | PCR product of P317 with Herculase Polymerase | DpnI digestion of P114 |
|---|---|---|---|---|
| Result: | expected result | expected result | expected result |
Preparation of competent cells according to the CaCl 2-method (Cohen et al.,1972)
Investigator: Johanna, Louise
Aim of the experiment: Preparation of competent cells according to the CaCl 2-method (Cohen et al.,1972)
Procedure:
- Add Tetracyclin (ratio: 1:1000) to the 2YT-medium
- Measure the OD550 of it until OD550=0.5
- Put the culture in Falcon tubes (leaving on ice)
- Centrifuge the falcon tubes by 5000 rpm, 4°C, 10 min
- Resuspend the falcon tubes each with 40 ml 0,1 M MgCl2 solution after removing the supernatants
- Centrifuge the falcon tubes by 5000 rpm, 4°C, 10 min
- Resuspend the falcon tubes each with 20 ml 50mM CaCl2 solution after removing the supernatants
- Incubate the falcon tubes on ice for 30 min
- Centrifuge the falcon tubes by 5000 rpm, 4°C, 10 min
- Resuspend the falcon tubes each with 2 ml 50mM CaCl2, 15 % v/v Glycerin solution after removing the supernatants. From this step everything needs to be carry out in a 4°C room.
- Mix the solutions of each falcon tube and transfer the content in eppis (each with 100 and accordingly 150 µl)
- Store the boxes of eppis at -80 °C
Preparative Digestion of P308 (PhyB-Linker-C-TEV), P299 (IRES), P111 (TEV), P326 (TEV-Linker-TM-GFP), P328 (SERKSigP_EreB), P330 (SERK-SigP_NLuc), P332 (SERK-SigP_SpyCatcher), P334 (SERK-SigP_SpyTag), P336 (SERK-SigP_XylE), P338 (SERK-SigP_SERK-TMD), P8 (pSB1C3) and PCR product of Laccase (F112)
Investigator: Katrin
Aim of the experiment: Peparative Digestion of P308 (PhyB-Linker-C-TEV), P299 (IRES), P111 (TEV), P326 (TEV-Linker-TM-GFP), P328 (SERKSigP_EreB), P330 (SERK-SigP_NLuc), P332 (SERK-SigP_SpyCatcher), P334 (SERK-SigP_SpyTag), P336 (SERK-SigP_XylE), P338 (SERK-SigP_SERK-TMD) and P8 (PhyB)
Procedure:
Batch for preparative digestion of effectors (P328, P330, P332, P334, P336, P338) with AgeI and PstI) (7x Mastermix)
| volume | reagent |
| 28 µl | Buffer 4 (10x) |
| 7 µl | AgeI (20 U/µl) |
| 7 µl | PstI (20 U/µl) |
| 98 µl | ddH2O |
| =140 µl | TOTAL |
- 20 µl Mastermix + 20 µl Plasmid each
Batches for preparative digestions:
| volume | reagent |
| 20 µl | Plasmid |
| 4 µl | Buffer 4 (10x) |
| 1 µl | Enzyme 1 |
| 1 µl | Enzyme 2 |
| 14 µl | ddH2O |
| =40 µl | TOTAL |
| Plasmid | Name | Enzyme 1 | Enzyme 2 |
| P308 | PhyB-Linker-C-TEV | SpeI | PstI |
| P299 | IRES | XbaI | PstI |
| P111 | TEV | SpeI | PstI |
| P326 | TEV-Linker-TM-GFP | NgoMIV | PstI |
| P8 | pSB1C3 | XbaI | AgeI |
| F112 | Laccase | XbaI | AgeI |
- Incubation for 2.5 h at 37 °C.
- 4 µl of DNA loading buffer (10x) were added to the 40 µl reaction batches after digestion and they were loaded on a 1% agarose gel
- Preparative gelelectrophoresis was performed at 90 V for 60 min.
| Name: | P308 dig. SpeI + PstI | 1kb DNA ladder | P299 dig. XbaI + PstI | P111 dig. SpeI + PstI |
| cut out: | Backbone | Insert (lower band) | Backbone |
| Name: | P326 dig. NgoMIV + PstI | 1kb DNA ladder | P328 dig. AgeI + Pst | P330 dig. AgeI + PstI |
| cut out: | Insert (lower band) | Backbone | Backbone |
| Name: | P332 dig. AgeI + PstI | 1kb DNA ladder | P334 dig. AgeI + PstI | P336 dig. AgeI + PstI |
| cut out: | Backbone | Backbone | Backbone |
| Name: | P338 dig. AgeI + PstI | 1kb DNA ladder | P8 dig. AgeI + XbaI | |
| cut out: | Backbone | Backbone (lower band) |
- Extracted via QIAGEN gel extraction kit, digested PCR product purified via PCR Purification Kit
Ligation of F116+F117 (TEV-Linker-TM-GFP+SERKSigP_EreB), F116+F118 (TEV-Linker-TM-GFP+SERK-SigP_NLuc), F116+F119 (TEV-Linker-TM-GFP+SERK-SigP_SpyCatcher), F116+F120 (TEV-Linker-TM-GFP+SERK-SigP_SpyTag), F116+F121 (TEV-Linker-TM-GFP+SERK-SigP_XylE), F123+F124 (Laccase+pSB1C3)
Investigator: Katrin
Aim of the experiment: Ligation of F116+F117 (TEV-Linker-TM-GFP+SERKSigP_EreB), F116+F118 (TEV-Linker-TM-GFP+SERK-SigP_NLuc), F116+F119 (TEV-Linker-TM-GFP+SERK-SigP_SpyCatcher), F116+F120 (TEV-Linker-TM-GFP+SERK-SigP_SpyTag), F116+F121 (TEV-Linker-TM-GFP+SERK-SigP_XylE), F123+F124 (Laccase+pSB1C3)
Procedure:
- Ligation batch for F116+F117 (TEV-Linker-TM-GFP+SERKSigP_EreB)
| volume | reagent |
|---|---|
| 3.52 µl | F117 (28.4 ng/µl, 3413 bp) |
| 4.81 µl | F116 (18.2 ng/µl, 996 bp) |
| 8.67 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- Ligation batch for F116+F118 (TEV-Linker-TM-GFP+SERK-SigP_NLuc)
| volume | reagent |
|---|---|
| 1.89 µl | F118 (52.8 ng/µl, 2663 bp) |
| 6.16 µl | F116 (18.2 ng/µl, 996 bp) |
| 8.95nbsp;µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- Ligation batch for F116+F119 (TEV-Linker-TM-GFP+SERK-SigP_SpyCatcher)
| volume | reagent |
|---|---|
| 2.57 µl | F119 (38.0 ng/µl, 2492 bp) |
| 6.59 µl | F116 (18.2 ng/µl, 996 bp) |
| 7.84nbsp;µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- Ligation batch for F116+F120 (TEV-Linker-TM-GFP+SERK-SigP_SpyTag)
| volume | reagent |
|---|---|
| 0.8 µl | F120 (62.5 ng/µl, 2192 bp) |
| 3.75 µl | F116 (18.2 ng/µl, 996 bp) |
| 3.96nbsp;µl | ddH2O |
| 1 µl | T4 ligase buffer (10x) |
| 0.5 µl | T4 ligase (1 U/µl) |
| =10 µl | TOTAL |
- Ligation batch for F116+F121 (TEV-Linker-TM-GFP+SERK-SigP_XylE)
| volume | reagent |
|---|---|
| 0.73 µl | F121 (68.3 ng/µl, 3071 bp) |
| 2.56 µl | F116 (18.2 ng/µl, 996 bp) |
| 5.95nbsp;µl | ddH2O |
| 1 µl | T4 ligase buffer (10x) |
| 0.5 µl | T4 ligase (1 U/µl) |
| =10 µl | TOTAL |
- Ligation batch for F123+F124 (Laccase+pSB1C3)
| volume | reagent |
|---|---|
| 2.03 µl | F123 (49.3 ng/µl, 2086 bp) |
| 3.80 µl | F124 (58.9 ng/µl, 1555 bp) |
| 11.17nbsp;µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- Negative control was prepared by replacing the volume of the insert by ddH2O.
- The ligation was performed for 1 hour at room temperature.
Transformation of E. coli XL1-blue with ligation products F116+F117 (TEV-Linker-TM-GFP+SERKSigP_EreB), F116+F118 (TEV-Linker-TM-GFP+SERK-SigP_NLuc), F116+F119 (TEV-Linker-TM-GFP+SERK-SigP_SpyCatcher), F116+F120 (TEV-Linker-TM-GFP+SERK-SigP_SpyTag), F116+F121 (TEV-Linker-TM-GFP+SERK-SigP_XylE), F123+F124 (Laccase+pSB1C3) and negative controls
Investigator: Jeff, Katrin
Aim of the experiment: Transformation of E. coli XL1-blue with ligation products F116+F117 (TEV-Linker-TM-GFP+SERKSigP_EreB), F116+F118 (TEV-Linker-TM-GFP+SERK-SigP_NLuc), F116+F119 (TEV-Linker-TM-GFP+SERK-SigP_SpyCatcher), F116+F120 (TEV-Linker-TM-GFP+SERK-SigP_SpyTag), F116+F121 (TEV-Linker-TM-GFP+SERK-SigP_XylE), F123+F124 (Laccase+pSB1C3), QCI(Insertion) of pActin and negative controls
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 10 µl of QuikChange products/2 µl of plasmid DNA was added to 150 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- The cell suspension were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on a new chlorampenicol/ampicillin plate.
Sequencing of P303 (SERK-SigP in PsB1C3, fw), P310 (N-TEV_36AALinker in pSB1C3, rev), P248 (Thermonuc in pSB1C3, fw), P312 (N-TEV_36AALinker in pSB1C3, fw), P316 (npt-casette after QCI, fw,rev), P318 (Laccase after QCV, still RFC10, fw, rev), P320 (SERK-TMD_8AAlinker_GFPmut1, fw, rev), P326 (Strep-tag-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1, fw, rev), P328 (SERK-SigP_EreB, fw, rev), P330 (SERK-SigP_NanoLuc, fw), P332 (SERK-SigP_SpyCatcher, fw), P334 (SERK-SigP_SpyTag, fw), P336 (SERK-SigP_XylE, fw, rev), P338 (SERK-SigP_SERK-TMD, fw)
Investigators: Louise, Katrin
Aim of the experiment: Sequencing of P303 (SERK-SigP in PsB1C3, fw), P310 (N-TEV_36AALinker in pSB1C3, rev), P248 (Thermonuc in pSB1C3, fw), P312 (N-TEV_36AALinker in pSB1C3, fw), P316 (npt-casette after QCI, fw,rev), P318 (Laccase after QCV, still RFC10, fw, rev), P320 (SERK-TMD_8AAlinker_GFPmut1, fw, rev), P326 (Strep-tag-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1, fw, rev), P328 (SERK-SigP_EreB, fw, rev), P330 (SERK-SigP_NanoLuc, fw), P332 (SERK-SigP_SpyCatcher, fw), P334 (SERK-SigP_SpyTag, fw), P336 (SERK-SigP_XylE, fw, rev), P338 (SERK-SigP_SERK-TMD, fw).
Procedure: Peparation of DNA for sequencing according to the Eurofins SmartSeq Kit protocol (15 µl of plasmid DNA (50 - 100 ng/µl) and 2 µl sequencing primer 10 µM).
The different genes received the following barcodes:
| Label | Name | Barcode1 | Barcode2 |
|---|---|---|---|
| P303 | SERK-SigP | FR02305805 (O3) | |
| P310 | N-TEV_36AAlinker | FR02305806 (O4) | |
| P248 | Thermonuc. | FR02305804 (O3) | |
| P312 | NTEV_36aaLinker | FR02305803 (O3) | |
| P316 | npt-casette QCI | FR02305802 (O3) | FR02305801 (O4) |
| P318 | Laccase QCV | FR02305800 (O3) | FR02305799 (O4) |
| P320 | SERK-TM_8aaLinker_GFPmut1 | FR02305798 (O3) | FR02305797 (O4) |
| P326 | Strep-tag-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1 | FR02305796 (O3) | FR02305795 (O4) |
| P328 | SERK-SigP_EreB | FR02305794 (O3) | FR02305793 (O4) |
| P330 | SERK-SigP_Nanoluc | FR02305792 (O3) | |
| P332 | SERK-SigP_Spycatcher | FR02305791 (O3) | |
| P334 | SERK-SigP_Spy-tag | FR02305790 (O3) | |
| P336 | SERK-SigP_XylE | FR02305789 (O3) | FR02305788 (O4) |
| P338 | SERK-SigP_SERK-TM | FR02305787 (O3) |
PCR of P61 to get the IRES of poliovirus 1 Mahoney strain
Investigator: Jeff
Aim of the experiment: PCR of P61 to get the IRES of poliovirus 1 Mahoney strain.
Procedure:
Operational sequence:
- Reaction batch with Q5 Mastermix:
| volume | reagent |
|---|---|
| 25 µl | 2x Q5 Master Mix |
| 2.5 µl | 10 µM Forward Primer O68 (IRES_polio_fw) |
| 2.5 µl | 10 µM Reverse Primer O69 (IRES_polio_rv) |
| 1 µl | Plasmid DNA (P61)- 1:1000 dilution (desired c between 1 pg and 1 ng) |
| 19 µL | ddH2O Water |
| =50 µL | TOTAL |
- The content has been mixed with a pipette
- The PCR program was performed after following scheme:
| Initial denaturation | 98 °C | 30 s |
|---|---|---|
| 30 cycles | 98 °C | 10 s |
| 63 °C | 30 s | |
| 72 °C | 20 s | |
| Final extension | 72 °C | 2 min |
| Hold | 4 °C | infinite |
- After PCR, the product was purified with QIAquick PCR Purification Kit, Qiagen.
Picking of Re-Transformations P111 (TEV in pSB1C3, Re-Trafo), P160 (Sv40 NLS in pSB1C3), P114 (pActin), P314 (FluA), Glutathione S-transferase (GST, BBa_K620000), QCI AlcR
Investigator: Katrin
Aim of the experiment: Picking of Re-Transformations P111 (TEV in pSB1C3, Re-Trafo), P160 (Sv40 NLS in pSB1C3), P114 (pActin), P314 (FluA), Glutathione S-transferase (GST, BBa_K620000), QCI AlcR.
Procedure:
- Retransformations 1 clone each
- Resuspendsion in 4 ml LB-medium with 4 µl chloramphenicol (P111, P160) or ampicillin (P114, P314)
Saturday, June 22nd
QuikChange of npt-casette (P316, QCII) and of AlcR (P342, QCII)
Investigator: Jeff
Aim of the experiment: QuikChange of npt-casette (P316, QCII) and of AlcR (P342, QCII).
Procedure:
Reaction batch for P316 (npt-casette after QCI)
| volume | reagent | Additional information |
|---|---|---|
| 2.5 µl | 10x Pfu Ultra II buffer | |
| 1 µl | Plasmid template (P316 - 1:100 dilution) | need to get between 2.5 ng and 25 ng for a reaction batch of 25 µl |
| 1 µl | 1:10 dilution of O72 (=10 µM) | |
| 1 µl | 1:10 dilution of O73 (=10 µM) | |
| 19 µl | ddH2O | |
| 1 µl | dNTP mix 50 mM | |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U/µl) |
Reaction batch for P342 (AlcR after QCI):
| volume | reagent | Additional information |
|---|---|---|
| 2.5 µl | 10x Pfu Ultra II buffer | |
| 1 µl | Plasmid template (P342 - 1:100 dilution) | need to get between 2.5 ng and 25 ng for a reaction batch of 25 µl |
| 1 µl | 1:10 dilution of O82 (10 µM) | |
| 1 µl | 1:10 dilution of O83 (10 µM) | |
| 19 µl | ddH2O | |
| 1 µl | dNTP mix 50 mM | |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U/µl) |
PCR cycling parameters - P316 (npt-casette after QCI) and P342 (AlcR after QCI):
| Segment | Cycles | Temperature | Time |
|---|---|---|---|
| 1 | 1 | 95 °C | 30 s |
| 2 | 20 | 95 °C | 30 s |
| 52 °C | 1 min | ||
| 68 °C | 5 min | ||
| 3 | 4 °C | hold |
Miniprep of overnight cultures of transformated E. coli XL1 blue with P111(Retrafo), QCI AlcR, P114(Retrafo), P314 (Retrafo), P160 (Retrafo), Glutathione S-transferase (GST, BBa_K620000)
Investigator: Katrin
Aim of the experiment: Miniprep of overnight cultures of transformated E. coli XL1 blue with P111(Retrafo), QCI AlcR, P114(Retrafo), P314 (Retrafo), P160 (Retrafo), Glutathione S-transferase (GST, BBa_K620000).
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
- The resulting DNA were eluted in tubes labeled as follows:
Analytical digestion and gelelectrophoresis of QCII AlcR (P341, P342) and control P294 with EcoRI and PstI, gelelectrophoresis of PCR of IRES
Investigator: Katrin
Aim of the experiment: Analytical digestion and gelelectrophoresis of QCII AlcR (P341, P342) and control P294 with EcoRI and PstI
Procedure:
- Batch
| volume | reagent |
| 2 µl | CutSmart Buffer (10x) |
| 0.25 µl | EcoRI |
| 0.25 µl | PstI |
| 15 µl | ddH2O |
| 2.5 µl | Plasmid DNA (P341, P342; P249) |
| =20 µl | TOTAL |
- Incubation for 60 min at 37 °C.
- Analytical gelelectrophoresis was performed at 90 V for 60 min
| Lane: | 1 kb DNA Ladder | PCR of IRES | Digestion of P249 with EcoRI and PstI (control) | Digestion of P341 with EcoRI and PstI | Digestion of P342 with EcoRI and PstI |
| Result: | as expected | as expected | as expected | as expected |
Preparative Digestion of P310 (N-Tev-linker), P249 (Thermonuclease), P160 (SV40 NLS), PCR of IRES
Investigator: Katrin
Aim of the experiment: Preparative Digestion of P310 (N-Tev-linker), P249 (Thermonuclease), P160 (SV40 NLS), PCR of IRES
Procedure:
Preparative digestion of P310 (N-Tev-linker)
| volume | reagent |
| 20 µl | P310 |
| 4 µl | CutSmart Buffer (10x) |
| 1 µl | AgeI (20 U/µl) |
| 1 µl | SpeI (20 U/µl) |
| 14 µl | ddH2O |
| =40 µl | TOTAL |
Preparative digestion of P160 (SV40 NLS)
| volume | reagent |
| 20 µl | P160 |
| 4 µl | CutSmart Buffer (10x) |
| 1 µl | AgeI (20 U/µl) |
| 1 µl | PstI (20 U/µl) |
| 14 µl | ddH2O |
| =40 µl | TOTAL |
Preparative digestion of P249 (Thermonuclease)
| volume | reagent |
| 20 µl | P249 |
| 4 µl | CutSmart Buffer (10x) |
| 1 µl | NgoMIV (20 U/µl) |
| 1 µl | PstI (20 U/µl) |
| 14 µl | ddH2O |
| =40 µl | TOTAL |
Preparative digestion of PCR of IRES)
| volume | reagent |
| 25 µl | PCR product |
| 5 µl | CutSmart Buffer (10x) |
| 1 µl | EcoRI (20 U/µl) |
| 1 µl | SpeI (20 U/µl) |
| 18 µl | ddH2O |
| =50 µl | TOTAL |
- Incubation for 2.5 h at 37 °C.
- 4 µl of DNA loading buffer (10x) were added to the 40 µl reaction batches after digestion and they were loaded on a 1% agarose gel
- Preparative gelelectrophoresis was performed at 90 V for 60 min.
| Name: | P249 dig. NgoMIV + PstI | 1kb DNA ladder | P160 dig. AgeI + PstI | P111 dig. SpeI + AgeI |
| cut out: | Insert(lower band) | Backbone | Backbone |
- Extracted via QIAGEN gel extraction kit, digested PCR product purified via PCR Purification Kit
Ligation of F126+F127 (NLS in pSB1C3+Thermonuclease), F125+F96 (N-TEV-linker in pSB1C3+PIF3), F125+F98 (N-TEV-linker in pSB1C3+PIF6), F124+F41 (IRES+pSB1C3)
Investigator: Katrin
Aim of the experiment: Ligation of F126+F127 (NLS in pSB1C3+Thermonuclease), F125+F96 (N-TEV-linker in pSB1C3+PIF3), F125+F98 (N-TEV-linker in pSB1C3+PIF6), F124+F41 (IREs+pSB1C3)
Procedure:
- Ligation batch for F126+F127 (NLS in pSB1C3+Thermonuclease
| volume | reagent |
|---|---|
| 7.2 µl | F126 (13.9 ng/µl, 2143 bp) |
| 4.1 µl | F127 (15.2 ng/µl, 447 bp) |
| 5.7 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- Ligation batch for F125+F96 (N-TEV-linker in pSB1C3+PIF3)
| volume | reagent |
|---|---|
| 6.9 µl | F125 (14.5 ng/µl, 2554 bp) |
| 2.3 µl | F96 (15.1 ng/µl, 297 bp) |
| 7.8nbsp;µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- Ligation batch for F124+F41 (IREs+pSB1C3)
| volume | reagent |
|---|---|
| 1.3 µl | F124 (70.8 ng/µl, 628 bp) |
| 1.2 µl | F41 (81.1 ng/µl, 2086 bp) |
| 14.5nbsp;µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- Negative control was prepared by replacing the volume of the insert by ddH2O.
- The ligation was performed for 1 hour at room temperature.
Transformation of E. coli XL1-blue with ligation products F126+F127 (NLS in pSB1C3+Thermonuclease), F125+F96 (N-TEV-linker in pSB1C3+PIF3), F125+F98 (N-TEV-linker in pSB1C3+PIF6), F124+F41 (IRES+pSB1C3) and negative controls
Investigator: Volker
Aim of the experiment: Transformation of E. coli XL1-blue with ligation products F126+F127 (NLS in pSB1C3+Thermonuclease), F125+F96 (N-TEV-linker in pSB1C3+PIF3), F125+F98 (N-TEV-linker in pSB1C3+PIF6), F124+F41 (IRES+pSB1C3) and negative controls
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 10 µl of QuikChange products/2 µl of plasmid DNA was added to 150 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- The cell suspension were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on a new chlorampenicol plate.
Picking of F116+F121, F116+F117, F123+F124, F116+F119, F116+F120, F116+F118, QCI (insertion of P114 (pActin)
Investigator: Katrin Aim of the experiment: Picking of F116+F121, F116+F117, F123+F124, F116+F119, F116+F120, F116+F118, QCI (insertion of P114 (pActin)
Procedure:
- 2 clones were picked from each plate
- Resuspendsion in 4 ml LB-medium with 4 µl chloramphenicol or ampicillin (QCI (insertion of P114 (pActin))
Sunday, June 23rd
QuikChange II of pActin (P359, P360)
Investigator: Katrin
Aim of the experiment: QuikChangeII of pActin (P359, P360) Procedure:
Reaction batch
| volume | reagent | Additional information |
|---|---|---|
| 2.5 µl | 10x Pfu Ultra II buffer | |
| 1 µl | Plasmid template (P359, P360) | need to get between 2.5 ng and 25 ng for a reaction batch of 25 µl |
| 1 µl | 1:10 dilution of O76 (=10 µM) | |
| 1 µl | 1:10 dilution of O77 (=10 µM) | |
| 19 µl | ddH2O | |
| 1 µl | dNTP mix 50 mM | |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U/µl) |
PCR cycling parameters
| Segment | Cycles | Temperature | Time |
|---|---|---|---|
| 1 | 1 | 95 °C | 30 s |
| 2 | 20 | 95 °C | 30 s |
| 52 °C | 1 min | ||
| 68 °C | 6 min | ||
| 3 | 4 °C | hold |
Miniprep of overnight cultures of transformated E. coli XL1 blue F123+F124, F116+F117, F116+F119, F116+F118, F116+F121, F116+F120, QCI pActin (P114)
Investigator: Katrin
Aim of the experiment: Miniprep of overnight cultures of transformated E. coli XL1 blue F123+F124, F116+F117, F116+F119, F116+F118, F116+F121, F116+F120, QCI pActin (P114)
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
Analytical digestion and gelelectrophoresis of P347-P358
Investigator: Katrin
Aim of the experiment: Analytical digestion and gelelectrophoresis of P347-P358
Procedure:
| volume | reagent |
| 2 µl | CutSmart Buffer (10x) |
| 0.25 µl | EcoRI |
| 0.25 µl | PstI |
| 15 µl | ddH2O |
| 2.5 µl | Plasmid DNA (P347-P358) |
| =20 µl | TOTAL |
- Incubation for 60 min at 37 °C.
- Analytical gelelectrophoresis was performed at 90 V for 60 min
| Lane: | 1 kb DNA Ladder | Digestion of P347 with EcoRI and PstI | Digestion of P348 with EcoRI and PstI | Digestion of P349 with EcoRI and PstI | Digestion of P350 with EcoRI and PstI | Digestion of P351 with EcoRI and PstI | Digestion of P352 with EcoRI and PstI | Digestion of P353 with EcoRI and PstI | Digestion of P354 with EcoRI and PstI |
| Result: |
| Lane: | 1 kb DNA Ladder | Digestion of P355 with EcoRI and PstI | Digestion of P356 with EcoRI and PstI | Digestion of P357 with EcoRI and PstI | Digestion of P358 with EcoRI and PstI |
| Result: |
Dpn1 Digestion and transformation of E. coli XL1-blue with QCII of AlcR and QCII of npt-cassette
Investigator: Katrin
Aim of the experiment: Dpn1 Digestion and transformation of E. coli XL1-blue with QCII of AlcR and QCII of npt-cassette
Procedure:
- 1 µl of Dpn1 was added to the reaction, the batch was incubated for one hour at 37°C
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 10 µl of QuikChange products was added to 150 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- The cell suspension were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on a new chlorampenicol plate.
Picking of F125+F98, F125+F96, F128+F41
Investigator: Katrin Aim of the experiment:Picking of F125+F98, F125+F96, F128+F41
Procedure:
- 2 clones were picked from each plate
- Resuspendsion in 4 ml LB-medium with 4 µl chloramphenicol
- F126+F127 was put back into the incubator because there were no colonies visible
Sequencing of P35, P143, P359, P360, P249
Investigators: Katrin
Aim of the experiment: Sequencing of P35, P143, P359, P360, P249
Procedure: The DNA was prepared for sequencing according to the Eurofins SmartSeq Kit protocol (15 µl of plasmid DNA (50 - 100 ng) and 2 µl sequencing primer).
The different genes we sequenced received the following barcodes:
| Label | Barcode1 | Barcode2 |
|---|---|---|
| P35 | FR02305786 (O4) | |
| P143 | FR02305785 (O92) | FR02305784 (O93) |
| P359 | FR02305783 (O62) | FR02305782 (O63) |
| P360 | FR02305781 (O62) | FR02305780 (O63) |
| P249 | FR02305779 (O3) | FR02305778 (O4) |
Week 10
Monday, June 24th
Miniprep of ligations F125+F98 (N-TEV-linker in pSB1Cs + PIF6), F125+F96 (N-TEV-linker in pSB1Cs + PIF3) and F128+F41 (PCR of IRES + pSB1C3 backbone) + analytical gel
Investigator: Johanna
Aim of the experiment: Preparation of ligation products F125+F98, F125+F96 and F128+F41
Procedure:
- Miniprep according to QIAGEN QIAprep Kit
| label | content | conc. in ng/µl |
| P361 | N-TEV-linker (in pSB1C3) + PIF6 | 163.2 |
| P362 | N-TEV-linker in (in pSB1C3) + PIF6 | 183.5 |
| P363 | N-TEV-linker in (in pSB1C3) + PIF3 | 159.6 |
| P364 | N-TEV-linker in (in pSB1C3) + PIF3 | 200.1 |
| P365 | PCR of IRES + (in pSB1C3) backbone | 136.0 |
| P366 | PCR of IRES + (in pSB1C3) backbone | 153.7 |
- Analytical digestion with EcoRI and PstI
| reagent | volume |
| CutSmart buffer | 2 µl |
| EcoRI | 0.25 µl |
| PstI | 0.25 µl |
| ddH2O | 15 µl |
| plasmid DNA | 2.5 µl |
| Total | 20 µl |
- 60 min incubation, 37 °C
- Gel elektrophoresis on 1% agarose gel
| 1 kbp DNA ladder | P361 | P362 | P363 | P364 | P365 | P366 |
| as expected | as expected | as expected | as expected | as expected | as expected |
Dpn1 digestion and transformation of QCII of pActin (insertion) + analytical Gel
Investigator: Jeff
Aim of the experiment: Dpn1 digestion and transformation of QCII of pActin (insertion)
Procedure:
- 1 µl of Dpn1 (10 U/µl) was added to the reaction, the batch was incubated for one hour at 37°C
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 8 µl of QuikChange products was added to 150 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- The cell suspension were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on a new ampicillin plate.
- 0.5 µl of loading buffer were added to 4.5 µl of Dpn1 digested plasmid
- Electrophoresis on 1% agarose gel
| 1 kb dna ladder | P359 | P360 |
| successful | successful |
Sequencing of P266 (IgKappa-SigP_Nanoluc), P268 (IgKappa-SigP_XylE), P270 (IgKappa-SigP_SpyCatcher), P272 (IgKappa-SigP_SpyTag), P361 (N-TEV-linker+PIF6), P364 (N-TEV-36AAlinker-PIF3) and P366 (IRES)
Investigator: Florian
Aim of the experiment: Sequencing of P266 (IgKappa-SigP_Nanoluc), P268 (IgKappa-SigP_XylE), P270 (IgKappa-SigP_SpyCatcher), P272 (IgKappa-SigP_SpyTag), P361 (N-TEV-linker+PIF6), P364 (N-TEV-36AAlinker-PIF3) and P366 (IRES)
Procedure: DNA samples were prepared according to the Eurofins SmartSeq Kit protocol (15 µl of plasmid DNA (50 - 100 ng) and 2 µl sequencing primer).
The samples received the following barcodes:
| Label | Barcode1 | Barcode2 |
|---|---|---|
| P266 | FR02305766 fw | FR02305767 rv |
| P268 | FR02305768 fw | FR02305769 rv |
| P270 | FR02305770 fw | |
| P272 | FR02305771 fw | |
| P361 | FR02305772 fw | FR02305773 rv |
| P364 | FR02305774 fw | FR02305775 rv |
| P366 | FR02305776 fw | FR02305777 rv |
Retransformation of P270 (IgKappa-SigP_SpyCatcher), P272 (IgKappa-SigP_SpyTag), P308 (PhyB_36AALinker_C-TEV) and P366 (IRES)
Investigator: Johanna
Aim of the experiment: Retransformation of P270 (IgKappa-SigP_SpyCatcher in pSB1C3), P272 (IgKappa-SigP_SpyTag in pSB1C3), P308 (PhyB_36AALinker_C-TEV in pSB1C3) and P366 (IRES in pSB1C3)
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 1.5 µl of plasmid DNA was added to 150 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of cell suspension were plated on chlorampenicol agar
- The rest of the cell suspension were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium; the concentrated cell suspension was plated on another chlorampenicol agar plate.
Ligation of F92 + F130 (IgKappa-SigP + Laccase), F109 + F130 (SERK-SigP + Laccase), F113 + F131 (PhyB_36AA-Linker_C-TEV + IRES), F115 + F131 (TEV + IRES)
Investigator: Florian, Jeff
Aim of the experiment: Ligation of F92 + F130 (IgKappa-SigP + Laccase), F109 + F130 (SERK-SigP + Laccase), F113 + F131 (PhyB_36AA-Linker_C-TEV + IRES), F115 + F131 (TEV + IRES).
Procedure:
- Ligation batch for F92 + F130 (IgKappa-SigP + Laccase)
| volume | reagent |
|---|---|
| 0.86 µl | F92 (116.7 ng/µl, 2144 bp) |
| 7.55 µl | F130 (28.3 ng/µl, 1527 bp) |
| 8.59 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- Ligation batch for F109 + F130 (SERK-SigP + Laccase)
| volume | reagent |
|---|---|
| 0.92 µl | F109 (108.2 ng/µl, 2177 bp) |
| 7.44 µl | F130 (28.3 ng/µl, 1527 bp) |
| 8.64 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- Ligation batch for F113 + F131 (PhyB_36AA-Linker_C-TEV + IRES)
| volume | reagent |
|---|---|
| 0.82 µl | F113 (122.2 ng/µl, 5281 bp) |
| 2.17 µl | F131 (16.4 ng/µl, 627 bp) |
| 14.01 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- Ligation batch for F115 + F131 (TEV + IRES)
| volume | reagent |
|---|---|
| 1.76 µl | F115 (56.9 ng/µl, 2794 bp) |
| 4.1 µl | F131 (16.4 ng/µl, 627 bp) |
| 11.14 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- Negative control was prepared by replacing the volume of the insert by ddH2O.
- The ligation was performed for 1 hour at room temperature.
Transformation of E. coli XL1-blue with ligation products F92 + F130 (IgKappa-SigP + Laccase), F109 + F130 (SERK-SigP + Laccase), F113 + F131 (PhyB_36AA-Linker_C-TEV + IRES), F115 + F131 (TEV + IRES) and negative controls and BioBrick BBa_K325909 (lucABCDEG under pBAD)
Investigator: Florian, Jeff
Aim of the experiment: Transformation of E. coli XL1-blue with ligation products F92 + F130 (IgKappa-SigP + Laccase), F109 + F130 (SERK-SigP + Laccase), F113 + F131 (PhyB_36AA-Linker_C-TEV + IRES), F115 + F131 (TEV + IRES) and negative controls and BioBrick BBa_K325909 (lucABCDEG under pBAD).
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 10 µl of QuikChange products/2 µl of plasmid DNA was added to 150 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- The cell suspension were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on a new chlorampenicol plate.
Preparative digestion of P308 (PhyB_36AAlinker_C-TEV), P348 (Laccase RFC25) and P366 (polioviral IRES)
Investigator: Flo, Jeff
Aim of the experiment: Preparative digestion of P308 (PhyB_36AAlinker_C-TEV), P348 (Laccase RFC25) and P366 (polioviral IRES).
Procedure:
| volume | reagent |
|---|---|
| 14 µl | P308 |
| 4 µl | CutSmart Buffer (10x) |
| 1 µl | SpeI-HF (20 U/µl) |
| 1 µl | PstI-HF (20 U/µl) |
| 20 µl | ddH2O |
| =40 µl | TOTAL |
| volume | reagent |
|---|---|
| 14 µl | P348 |
| 4 µl | CutSmart Buffer (10x) |
| 1 µl | NgoMIV (20 U/µl) |
| 1 µl | SpeI-HF (20 U/µl) |
| 20 µl | ddH2O |
| =40 µl | TOTAL |
| volume | reagent |
|---|---|
| 20 µl | P366 |
| 4 µl | CutSmart Buffer (10x) |
| 1 µl | XbaI (20 U/µl) |
| 1 µl | PstI-HF (20 U/µl) |
| 14 µl | ddH2O |
| =40 µl | TOTAL |
- Reaction batches were incubated for 2.5 h at 37 °C.
- 4 µl DNA loading buffer (10x) were added to the reaction batches after the incubation time and were loaded on a 1% agaorose gel.
- Preparative geleletrophoresis was performed at 90 V for 60 min.
| Lane | Digestion of P308 with SpeI & PstI | 1 kbp DNA ladder | Digestion of P348 with NgoMIV & SpeI | Digestion of P366 with XbaI & PstI |
|---|---|---|---|---|
| result | 2 further unexpected bands; seems that PstI is contaminated with a prefix restriction enzyme; used PstI enzyme batch is discareded; upper band was cut out | as expected; lower band was cut out | as expected; lower band was cut out |
Picking of F126+F127 (NLS+ThermoNuc), AlcR QCII and npt-cassette QCII
Investigator: Jeff
Aim of the experiment: Picking of F126+F127 (NLS+ThermoNuc), AlcR QCII and npt-cassette QCII for plasmid preparation
Procedure:
- F126+F127 (NLS+ThermoNuc) 7 clones due to high number of colonies on negative ctrl
- AlcR QCII and npt-cassette QCII 2 clones each
Tuesday, June 25th
Sequencing of P344 pASK-Flua(Trippelmutante)
Investigator: Jeff
Aim of the experiment: Sequencing of P344. Concentration was determined by Nanodop to 132 ng/µl
Procedure:
DNA samples were prepared according to the Eurofins SmartSeq Kit protocol (15 µl of plasmid DNA (50 - 100 ng) and 2 µl sequencing primer). The samples received the following barcodes:
| Label | Barcode1 |
|---|---|
| P344 with F83 (primer from Skerra lab! not fragment 83 from iGEM!) | FR02305765 |
Retransformation of P35 (Viability sensor with RFP) and P249 (Thermonuc in pSB1C3)
Investigator: Jeff, Rosario
Aim of the experiment: Retransformation of P35 (Viability sensor with RFP) and P249 (Thermonuc in pSB1C3)
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- empty tube was washed with 2 µl water and the volume was added to 150 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of cell suspension were plated on chlorampenicol agar
- The rest of the cell suspension were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium; the concentrated cell suspension was plated on another chlorampenicol agar plate.
Miniprep of overnight cultures of transformated E. coli XL1 blue with QCII of P316 (npt-casette in pSB1C3), QCII of P342 (AlcR in pSB1C3) and F126+F127 (NLS in pSB1C3 + Thermonuclease)
Investigator: Louise
Aim of the experiment: Miniprep of overnight cultures of transformated E. coli XL1 blue with QCII of P316 (npt-casette in pSB1C3), QCII of P342 (AlcR in pSB1C3) and F126+F127 (NLS in pSB1C3 + Thermonuclease).
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
- The resulting DNA were eluted in tubes labeled as follows:
| Content | New Tube |
|---|---|
| F126+F127 clone 1 | P367 |
| F126+F127 clone 2 | P368 |
| F126+F127 clone 3 | P369 |
| F126+F127 clone 4 | P370 |
| F126+F127 clone 5 | P371 |
| F126+F127 clone 6 | P372 |
| F126+F127 clone 7 | P373 |
| QCII npt clone 1 | P374 |
| QCII npt clone 2 | P375 |
| QCII AlcR clone 1 | P376 |
| QCII AlcR clone 2 | P377 |
Analytical digestion and gelelectrophoresis of P367-P373 (NLS in pSB1C3 + Thermonuclease) with EcoRI and PstI, P374-P375 (QCII of npt casette) with XbaI and PstI, P376-P377 (QCII of AlcR) with XbaI and SpeI
Investigator: Louise, Rosario, Florian
Aim of the experiment:Analytical digestion and gelelectrophoresis of P367-P373 (NLS in pSB1C3 + Thermonuclease) with EcoRI and PstI, P374-P375 (QCII of npt casette) with XbaI and PstI, P376-P377 (QCII of AlcR) with XbaI and SpeI.
Procedure:
- Master-Mix for P367-P373 and control P249
| volume | reagent |
| 20 µl | CutSmart Buffer (10x) |
| 2.5 µl | EcoRI |
| 2.5 µl | PstI |
| 150 µl | ddH2O |
| =175 µl | TOTAL |
- Master-Mix for P374-P375 and control P316
| volume | reagent |
| 8 µl | CutSmart Buffer (10x) |
| 1 µl | XbaI |
| 1 µl | PstI |
| 60 µl | ddH2O |
| =70 µl | TOTAL |
- Master-Mix for P376-P377 and control P342
| volume | reagent |
| 8 µl | CutSmart Buffer (10x) |
| 1 µl | XbaI |
| 1 µl | SpeI |
| 60 µl | ddH2O |
| =70 µl | TOTAL |
- Batch for analytical digestion of P367-P377 and controls
| volume | reagent |
| 17.5 µl | Master-Mix |
| 2.5 µl | Plasmid DNA |
| =20 µl | TOTAL |
- Incubation for 60 min at 37 °C.
- Analytical gelelectrophoresis was performed at 90 V for 60 min
| Lane | 1 kbp DNA ladder | Digestion of P367 with EcoRI and PstI | Digestion of P368 with EcoRI and PstI | Digestion of P369 with EcoRI and PstI | Digestion of P370 with EcoRI and PstI | Digestion of P371 with EcoRI and PstI | Digestion of P372 with EcoRI and PstI | Digestion of P373 with EcoRI and PstI | Digestion of P249 with EcoRI and PstI |
|---|---|---|---|---|---|---|---|---|---|
| result | as expected | as expected | not as expected | not as expected | as expected | as expected | not as expected | as expected |
| Lane | 1 kbp DNA ladder | Digestion of P316 with XbaI and PstI | Digestion of P374 with XbaI and PstI/SpeI | Digestion of P374 with XbaI and PstI/SpeI | Digestion of P375 with XbaI and PstI/SpeI | Digestion of P375 with XbaI and PstI/SpeI |
|---|---|---|---|---|---|---|
| Digestion of P376 with XbaI and SpeI | Digestion of P377 with XbaI and PstI/SpeI | Digestion of P377 with XbaI and PstI/SpeI | Digestion of P342 with XbaI and SpeI | |||
| result | not as expected, maybe digestion with EcoRI instead of XbaI, has to be repeated | not as expected, maybe digestion with EcoRI instead of XbaI, has to be repeated | not as expected, maybe digestion with EcoRI instead of XbaI, has to be repeated | not as expected, maybe digestion with EcoRI instead of XbaI, has to be repeated | not as expected, maybe digestion with EcoRI instead of XbaI, has to be repeated | |
| as expected | not fully digested | not fully digested | as expected |
Preparative digestion and gelelectrophoresis of P26 (Dnb1c Promotor) and P35 (RD29 Promotor) with EcoRI and KpnI
Investigator: Louise, Florian
Aim of the experiment: Preparative digestion and gelelectrophoresis of P26 (Dnb1c Promotor) and P35 (RD29 Promotor) with EcoRI and KpnI .
Procedure:
- Batch for P26
| volume | reagent |
| 20 µl | Plasmid DNA |
| 4 µl | CutSmart Buffer |
| 1 µl | EcoRI |
| 1 µl | KpnI |
| 14 µl | ddH2O |
| =40 µl | TOTAL |
- Batch for P35
| volume | reagent |
| 7 µl | Plasmid DNA |
| 4 µl | CutSmart Buffer |
| 1 µl | EcoRI |
| 1 µl | KpnI |
| 27 µl | ddH2O |
| =40 µl | TOTAL |
- Incubation for 2.5 h at 37 °C.
- 4 µl of DNA loading buffer (10x) were added to the 40 µl reaction batches after digestion and the samples were loaded on a 1% agarose gel.
- Preparative gelelectrophoresis was performed at 90 V for 60 min.
| Lane | Digestion of P26 with EcoRI and KpnI | 100 bp DNA ladder | Digestion of P26 with EcoRI and KpnI | 1 kbp DNA ladder |
|---|---|---|---|---|
| result | not as expected, no band was cut out | as expected; upper band was cut out, F132 |
Picking of ligation products F92 + F130 (IgKappa-SigP + Laccase), F109 + F130 (SERK-SigP + Laccase), F113 + F131 (PhyB_36AA-Linker_C-TEV + IRES), F115 + F131 (TEV + IRES) and BioBrick BBa_K325909 (lucABCDEG under pBAD) and Retransformation of P270 (IgKappa-SigP_SpyCatcher), P272 (IgKappa-SigP_SpyTag), P308 (PhyB_36AALinker_C-TEV) and P366 (IRES)
Investigator: Rosario
Aim of the experiment: Picking of ligation products F92 + F130 (IgKappa-SigP + Laccase), F109 + F130 (SERK-SigP + Laccase), F113 + F131 (PhyB_36AA-Linker_C-TEV + IRES), F115 + F131 (TEV + IRES) and BioBrick BBa_K325909 (lucABCDEG under pBAD) and Retransformation of P270 (IgKappa-SigP_SpyCatcher), P272 (IgKappa-SigP_SpyTag), P308 (PhyB_36AALinker_C-TEV) and P366 (IRES).
Procedure:
- 2 clones were picked for each ligation product and for the biobrick
- 1 clone was picked for each ReTrafo
Wednesday, June 26th
Miniprep of overnight cultures of transformated E. coli XL1 blue with P270, F113+F131, F109+F130, P308, F115+F131, F92+F130, F115+F131, P272, P366, lucBB
Investigator: Ingmar
Aim of the experiment: Miniprep of overnight cultures of transformated E. coli XL1 blue with P270, F113+F131, F109+F130, P308, F115+F131, F92+F130, F115+F131, P272, P366 and lucBB.
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
- The resulting DNA were eluted in tubes labeled as follows:
| Content | New Tube |
|---|---|
| P270 Retrafo | P379 |
| F113+F131 | P380 |
| F109+F130 | P381 |
| P308 Retrafo | P382 |
| F115+F131 | P383 |
| F92+F130 | P384 |
| F113+F131 | P385 |
| F115+F131 | P386 |
| F92+F130 | P387 |
| P272 Retrafo | P388 |
| P366 | P389 |
| luc BB | P390 |
| F109+F130 | P391 |
| luc BB | P392 |
Analytical digestion and gelelectrophoresis of P374-P375 (QC II of npt-casette) and P316 (QC I of npt-casette) as a reference with XbaI and PstI
Investigator: Rosario
Aim of the experiment: Analytical digestion and gelelectrophoresis of P374-P375 (QC II of npt-casette) and P316 (QC I of npt-casette) as a reference with XbaI and PstI
Procedure:
| volume | reagent |
| 2 µl | CutSmart Buffer (10x) |
| 0.25 µl | XbaI |
| 0.25 µl | PstI |
| 15 µl | ddH2O |
| 2.5 µl | Plasmid DNA (P316,P374,P375) |
| =20 µl | TOTAL |
- Incubation for 60 min at 37 °C.
- Analytical gelelectrophoresis was performed at 90 V for 60 min
| Lane: | 1000 bp DNA ladder | Digestion of P316 with XbaI and PstI | Digestion of P374 with XbaI and PstI | Digestion of P375 with XbaI and PstI |
|---|---|---|---|---|
| Result: | corrupt | corrupt | corrupt |
Picking of Retransformation of P35 (Viability sensor with RFP) and P249 (Thermonuc in pSB1C3)
Investigator: Rosario
Aim of the experiment: Picking of Retransformation of P35 (Viability sensor with RFP) and P249 (Thermonuc in pSB1C3).
Procedure:
- 1 clone was picked for each ReTrafo
Sequencing of P367 (NLS-Thermonuclease), P376 (AlcR QC II), P374 (npt-casette QC II), P348 (Laccase), P350 (SERK-SigP_EreB_Strep-tag-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1), P352 (SERK-SigP_SpyCatcher_Strep-tag-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1), P354 (SERK-SigP_NanoLuc_Strep-tag-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1), P356 (SERK-SigP_XylE_Strep-tag-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1), P358 (SERK-SigP_SpyTag_Strep-tag-TEV-site-linker_SERK-TMD-8AAlinker_GFPmut1), P26 (DREB1C promoter)
Investigator: Rosario
Aim of the experiment: Sequencing of P367 (NLS-Thermonuclease), P376 (AlcR QC II), P374 (npt-casette QC II), P348 (Laccase), P350 (SERK-SigP_EreB_Strep-tag-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1), P352 (SERK-SigP_SpyCatcher_Strep-tag-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1), P354 (SERK-SigP_NanoLuc_Strep-tag-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1), P356 (SERK-SigP_XylE_Strep-tag-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1), P358 (SERK-SigP_SpyTag_Strep-tag-TEV-site-linker_SERK-TMD-8AAlinker_GFPmut1), P26 (DREB1C promoter).
Procedure: The DNA was prepared for sequencing according to the Eurofins SmartSeq Kit protocol (15 µl of plasmid DNA (50 - 100 ng) and 2 µl sequencing primer).
The different genes we sequenced received the following barcodes:
| Label | Barcode1 | Barcode2 |
|---|---|---|
| P367 | FR02305740 (O3) | |
| P376 | FR02305741 (O3) | FR02305742 (O4) |
| P374 | FR02305743 (O3) | FR02305744 (O4) |
| P348 | FR02305745 (O3) | FR02305746 (O4) |
| P350 | FR02305747 (O3) | FR02305748 (O4) |
| P352 | FR02305749 (O3) | FR02305750 (O4) |
| P354 | FR02305751 (O3) | FR02305752 (O4) |
| P356 | FR02305753 (O3) | FR02305754 (O4) |
| P358 | FR02305755 (O3) | FR02305756 (O4) |
| P26 | FR02305757 (O4) |
Thursday, June 27th
Sequencing of P387 (IgkKappa-SigP_Laccase (QCV, RFC25)), P391 (SERK-SigP_Laccase (QCV, RFC25))and P355 (SERK-SigP_XylE_Strep-tag-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1 clone 2)
Investigator: Ingmar
Aim of the experiment: Sequencing of P387 (IgkKappa-SigP_Laccase (QCV, RFC25)), P391 (SERK-SigP_Laccase (QCV, RFC25))and P355 (SERK-SigP_XylE_Strep-tag-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1 clone 2).
Procedure: The DNA was prepared for sequencing according to the Eurofins SmartSeq Kit protocol (15 µl of plasmid DNA (50 - 100 ng) and 2 µl sequencing primer).
The different genes we sequenced received the following barcodes:
| Label | Barcode1 | Barcode2 |
|---|---|---|
| P387 | FR02305758 (O3) | FR02305759 (O4) |
| P391 | FR02305760 (O3) | |
| P355 | FR02305761 (O3) | FR02305762 (O4) |
Re-Transformation of E. coli XL1-blue with P170(SpyTag), P165(t35s), P308(PhyB_36AALinker_C-TEV), P348(Laccase QCV RFC25), P326(Strep-tag-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1), P361(N-TEV-36AAlinker-PIF6), P364(N-TEV-36AAlinker-PIF3), P266(IgKappa-SigP_Nanoluc)
Investigator: Katrin
Aim of the experiment: Re-Transformation of E. coli XL1-blue with P170(SpyTag), P165(t35s), P308(PhyB_36AALinker_C-TEV), P348(Laccase QCV RFC25), P326(Strep-tag-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1), P361(N-TEV-36AAlinker-PIF6), P364(N-TEV-36AAlinker-PIF3), P266(IgKappa-SigP_Nanoluc) Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 150 µl of competent cells were added to the tubes containing about 1 µl of plasmid DNA and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- The cell suspension were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on a new chlorampenicol plate.
PCR of P143 (pLuc-Actin, Amplification of pActin)
Investigator: Rosario, Flo
Aim of the experiment: PCR of P143 (pLuc-Actin, Amplification of pActin).
Procedure:
Operational sequence:
- Reaction batch with Q5 Mastermix:
| volume | reagent |
|---|---|
| 25 µl | 2x Q5 Master Mix |
| 2.5 µl | 10 µM Forward Primer O96 (pActin_Oryza_fw) |
| 2.5 µl | 10 µM Reverse Primer O97 (pActin_Oryza_rv) |
| 1 µl | Plasmid DNA (P143)- 1:1000 dilution (desired c between 1 pg and 1 ng) |
| 19 µL | ddH2O Water |
| =50 µL | TOTAL |
- The content has been mixed with a pipette
- The PCR program was performed after following scheme:
| Initial denaturation | 98 °C | 30 s |
|---|---|---|
| 30 cycles | 98 °C | 10 s |
| 62 °C | 30 s | |
| 72 °C | 45 s | |
| Final extension | 72 °C | 2 min |
| Hold | 4 °C | infinite |
- After PCR, the product was purified with QIAquick PCR Purification Kit, Qiagen.
Analytical gelelectrophoresis of F133
Investigator: Jeff
Procedure: Analytical gelelectrophoresis of F133 (PCR product of F143 with O96 & O97) to check the quality of the performed PCR.
Procedure:
- 4.5 µl of F133 were mixed together 0.5 µl of DNA loading buffer (10x)
- Analytical gelelectrophoresis was performed on a 1% agarose gel at 90 V for 40 min.
| Lane: | 1000 bp DNA ladder | F133 |
|---|---|---|
| Result: | as expected; but byproduct visible |
Analytical digestion and gelelectrophoresis of P381, P391, P347, P384, P387, P380, P385, P309, P383, P386, P390, P392
Investigator: Rosario
Aim of the experiment: Analytical digestion and gelelectrophoresis of P381, P391, P347, P384, P387, P380, P385, P309, P383, P386, P390, P392 with EcoRI and PstI
| Volume | Reagent |
|---|---|
| 2 µl | CutSmart Buffer (10x) |
| 0.25 µl | EcoRI |
| 0.25 µl | PstI |
| 15 µl | ddH2O |
| 2.5 µl | Plasmid DNA |
| =20 µl | TOTAL |
- Incubation for 60 min at 37 °C.
- Analytical gelelectrophoresis was performed at 90 V for 60 min
| Lane: | 1000 bp DNA ladder | Digestion of P381 with EcoRI and PstI | Digestion of P391 with EcoRI and PstI | Digestion of P347 with EcoRI and PstI | Digestion of P384 with EcoRI and PstI | Digestion of P387 with EcoRI and PstI |
|---|---|---|---|---|---|---|
| Result: | as expected | as expected | not fully digested | as expected | as expected |
| Lane: | 1000 bp DNA ladder | Digestion of P380 with EcoRI and PstI | Digestion of P385 with EcoRI and PstI | Digestion of P309 with EcoRI and PstI | Digestion of P383 with EcoRI and PstI | Digestion of P386 with EcoRI and PstI | Digestion of P390 with EcoRI and PstI | Digestion of P392 with EcoRI and PstI |
|---|---|---|---|---|---|---|---|---|
| Result: | as expected | as expected | as expected | as expected | as expected | as expected | as expected |
Preparative digestion and gelelectrophoresis of P391 (SERK-SigP_Laccase), P326 (Strep-tag-TEV-linker_SERK-TMD_8AAlinker_GFPmut1), P385 (PhyB_36AAlinker_C-TEV_IRES), P361 (N-TEV_36AAlinker_PIF6), P364 (N-TEV_36AAlinker_PIF3), P266 (IgKappa-SigP_NanoLuc), P388 (IgKappa-SigP_SpyTag), P247 (SpyCatcher), P379 (IgKappa-SigP_SpyCatcher), P374 (npt-casette) and P165 (t35S)
Investigator: Flo, Rosario
Aim of the experiment: Preparative digestion and gelelectrophoresis of P391 (SERK-SigP_Laccase), P326 (Strep-tag-TEV-linker_SERK-TMD_8AAlinker_GFPmut1), P385 (PhyB_36AAlinker_C-TEV_IRES), P361 (N-TEV_36AAlinker_PIF6), P364 (N-TEV_36AAlinker_PIF3), P266 (IgKappa-SigP_NanoLuc), P388 (IgKappa-SigP_SpyTag), P247 (SpyCatcher), P379 (IgKappa-SigP_SpyCatcher), P374 (npt-casette) and P165 (t35S).
Procedure:
Batch for preparative digestion of effectors (P328, P330, P332, P334, P336, P338) with AgeI and PstI)
| volume | reagent |
|---|---|
| 20 µl | Plasmid DNA |
| 4 µl | CutSmart Buffer (10x) |
| 1 µl | Enzyme 1 (20 U/µl) |
| 1 µl | Enzyme 2 (20 U/µl) |
| 14 µl | ddH2O |
| =40 µl | TOTAL |
| Plasmid | Name | Enzyme 1 | Enzyme 2 |
|---|---|---|---|
| P391 | SERK-SigP_Laccase | AgeI | PstI |
| P326 | Strep-tag-TEV-linker_SERK-TMD_8AAlinker_GFPmut1 | NgoMIV | PstI |
| P385 | PhyB_36AAlinker_C-TEV_IRES | SpeI | PstI |
| P361 | N-TEV_36AAlinker_PIF6 | XbaI | PstI |
| P364 | N-TEV_36AAlinker_PIF3 | XbaI | PstI |
| P266 | IgKappa-SigP_NanoLuc | EcoRI | AgeI |
| P388 | IgKappa-SigP_SpyTag | AgeI | PstI |
| P247 | SpyCatcher | EcoRI | NgoMIV |
| P379 | IgKappa-SigP_SpyCatcher | AgeI | PstI |
| P374 | npt-casette | EcoRI | XbaI |
| P165 | t35S | EcoRI | SpeI |
- Incubation for 2.5 h at 37 °C.
- 4 µl of DNA loading buffer (10x) were added to the 40 µl reaction batches after digestion and they were loaded on a 1% agarose gel
- Preparative gelelectrophoresis was performed at 90 V for 60 min.
| Lane | Digestion of P391 with AgeI & PstI | 1kb DNA ladder | Digestion of P326 with NgoMIV & PstI | Digestion of P385 with SpeI & PstI |
|---|---|---|---|---|
| Results | sideproducts?!; upper band was cut out | as expected, lower band was cut out | as expected |
| Lane | Digestion of P361 with XbaI & PstI | 1kb DNA ladder | Digestion of P364 with XbaI & PstI | Digestion of P266 with EcoRI & AgeI |
|---|---|---|---|---|
| Results | as expected, lower band was cut out | as expected, lower band was cut out | as expected, lower band was cut out |
| Lane | Digestion of P388 with AgeI & PstI | 1kb DNA ladder | Digestion of P382 with NgoMIV & PstI | Digestion of P247 with EcoRI & NgoMIV |
|---|---|---|---|---|
| Results | as expected | not expected, wrong plasmid | as expected |
| Lane | Digestion of P379 with AgeI & PstI | 1kb DNA ladder | Digestion of P374 with EcoRI & XbaI | Digestion of P165 with EcoRI & SpeI |
|---|---|---|---|---|
| Results | as expected | as expected | as expected, but only little DNA |
- Extracted via QIAGEN gel extraction kit, digested PCR product purified via PCR Purification Kit
Ligation of F134 + F135 (SERK-SigP-Laccase + Strep-tag-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1), F136 + F137 (PhyB_36AAlinker_C-TEV_IRES + N-TEV_36AAlinker_PIF6), F136 + F138 (PhyB_36AAlinker_C-TEV_IRES + N-TEV_36AAlinker_PIF3) and F143 + F144 (npt-csette + t35S)
Investigator: Flo, Rosario
Aim of the experiment: Ligation of F134 + F135 (SERK-SigP-Laccase + Strep-tag-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1), F136 + F137 (PhyB_36AAlinker_C-TEV_IRES + N-TEV_36AAlinker_PIF6), F136 + F138 (PhyB_36AAlinker_C-TEV_IRES + N-TEV_36AAlinker_PIF3) and F143 + F144 (npt-csette + t35S).
Procedure:
- Ligation batch for F134 + F135 (SERK-SigP-Laccase + Strep-tag-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1) (vector:insert = 1:3):
| volume | reagent |
|---|---|
| 0.83 µl | F134 (120.6 ng/µl, 1630 bp) |
| 10.29 µl | F135 (17.8 ng/µl, 996 bp) |
| 5.88 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- Ligation batch for F136 + F137 (PhyB_36AAlinker_C-TEV_IRES + N-TEV_36AAlinker_PIF6) (vector:insert = 1:3):
| volume | reagent |
|---|---|
| 0.5 µl | F136 (201.2 ng/µl, 5916 bp) |
| 4.6 µl | F137 (8.5 ng/µl, 771 bp) |
| 11.9 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- Ligation batch for F136 + F138 (PhyB_36AAlinker_C-TEV_IRES + N-TEV_36AAlinker_PIF3) (vector:insert = 1:3):
| volume | reagent |
|---|---|
| 0.5 µl | F136 (201.2 ng/µl, 5916 bp) |
| 4.07 µl | F138 (9.6 ng/µl, 771 bp) |
| 12.43 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- Ligation batch for F143 + F144 (npt-csette + t35S) (vector:insert = 1:3):
| volume | reagent |
|---|---|
| 0.53 µl | F143 (188.8 ng/µl, 3587 bp) |
| 6.05 µl | F144 (3.0 ng/µl, 217 bp) |
| 10.42 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- Negative control was prepared by replacing the volume of the insert by ddH2O.
- The ligation was performed overnight at 16 °C.
Friday, June 28th
Transformation of E. coli XL1-blue with P338(Ig-Kappa-SigP_SpyTag, Retrafo), F135+F134(SERK-SigP_Laccase+Strep-TEV_linker_SERK-TMD_linker_GFP), F136+F138(PhyB_linker_C-TEV_IRES+N-TEV_linker_PIF3), F136+F137(PhyB_linker_C-TEV_IRES+N-TEV_linker_PIF6), F143+F144(npt-Cassette+Terminator) and negative controls (F135, F136, F143)
Investigator: Katrin
Aim of the experiment: Transformation of E. coli XL1-blue with P338(Ig-Kappa-SigP_SpyTag, Retrafo), F135+F134(SERK-SigP_Laccase+Strep-TEV_linker_SERK-TMD_linker_GFP), F136+F138(PhyB_linker_C-TEV_IRES+N-TEV_linker_PIF3), F136+F137(PhyB_linker_C-TEV_IRES+N-TEV_linker_PIF6), F143+F144(npt-Cassette+Terminator) and negative controls (F135, F136, F143)
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 8 µl of DNA were added to the tubes containing 150 µl of competent cells and gently mixed, for re-transformation, 1 µl of P388 was added to 150 µl of competent cells
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- The cell suspension were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated on a new chlorampenicol plate.
QuikChange III of AlcR (P376)
Investigator: Andi, Johanna
Aim of the experiment: QuikChange III of AlcR (P376)
Reaction batch
| volume | reagent | Additional information |
|---|---|---|
| 2.5 µl | 10x Pfu Ultra II buffer | |
| 1 µl | Plasmid template (P376), (1:120 diluted) | need to get between 2.5 ng and 25 ng for a reaction batch of 25 µl |
| 1 µl | 1:10 dilution of O84 (=10 µM) | |
| 1 µl | 1:10 dilution of O85 (=10 µM) | |
| 19 µl | ddH2O | |
| 1 µl | dNTP mix 50 mM | |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U/µl) |
PCR cycling parameters
| Segment | Cycles | Temperature | Time |
|---|---|---|---|
| 1 | 1 | 95 °C | 30 s |
| 2 | 20 | 95 °C | 30 s |
| 52 °C | 1 min | ||
| 68 °C | 5 min | ||
| 3 | 4 °C | hold |
Preparative digestion and gelelectrophoresis of P388 (Ig-Kappa-SigP_SpyTag), P246 (Nanoluc), F133 (PCR of pActin)
Investigator: Katrin
Aim of the experiment: Preparative digestion and gelelectrophoresis of P388 (Ig-Kappa-SigP_SpyTag), P246 (Nanoluc), F133 (PCR of pActin)
Procedure:
reaction batch for P388(Ig-Kappa-SigP_SpyTag)
| volume | reagent |
|---|---|
| 20 µl | P388 |
| 4 µl | CutSmart Buffer (10x) |
| 1 µl | AgeI-HF (20 U/µl) |
| 1 µl | PstI-HF (20 U/µl) |
| 14 µl | ddH2O |
| =40 µl | TOTAL |
reaction batch for P246(Nanoluc)
| volume | reagent |
|---|---|
| 20 µl | P246 |
| 4 µl | CutSmart Buffer (10x) |
| 1 µl | NgoMIV-HF (20 U/µl) |
| 1 µl | PstI-HF (20 U/µl) |
| 14 µl | ddH2O |
| =40 µl | TOTAL |
reaction batch for F133(PCR of pActin)
| volume | reagent |
|---|---|
| 25 µl | F133 |
| 5 µl | CutSmart Buffer (10x) |
| 1 µl | XbaI (20 U/µl) |
| 1 µl | PstI-HF (20 U/µl) |
| 18 µl | ddH2O |
| =50 µl | TOTAL |
- Reaction batches were incubated for 3 h at 37 °C.
- 4 µl DNA loading buffer (10x) were added to the reaction batches after the incubation time and were loaded on a 1% agaorose gel.
- Preparative geleletrophoresis was performed at 90 V for 60 min.
| Lane | Digestion of F133 with XbaI & PstI | 1 kbp DNA ladder | Digestion of P246 with NgoMIV,PstI | Digestion of P388 with AgeI, PstI |
|---|---|---|---|---|
| result | as expected (but smaller fragment visible), upper band was cut out | as expected, lower band was cut out | as expected |
Ligation of F146+F147 (Ig-Kappa-SigP_SpyTag+Nanoluc) and F145+F42 (pActin+pSB1C3)
Investigator: Katrin
Aim of the experiment: Ligation of F146+F147 (Ig-Kappa-SigP_SpyTag+Nanoluc) and F145+F42 (pActin+pSB1C3)
Procedure:
- Ligation batch for F146+F147 (Ig-Kappa-SigP_SpyTag+Nanoluc)
| volume | reagent |
|---|---|
| 0.90 µl | F146 (111.2 ng/µl, 2189 bp) |
| 3.99 µl | F147 (17.5 ng/µl, 510 bp) |
| 12.11 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- Ligation batch for F145+F42 (pActin + pSB1C3) (batch: 30 ng vector!)
| volume | reagent |
|---|---|
| 23.35 µl | F145 (2.5 ng/µl, 1353 bp) |
| 0.41 µl | F42 (73.5 ng/µl, 2086 bp) |
| 2.7 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =27 µl | TOTAL |
- Negative controls were prepared by replacing the volume of the insert by ddH2O.
- The ligation was performed for 1 hour at room temperature.
Picking of ReTrafos of E. coli XL1-blue with P170(SpyTag), P165(t35s), P308(PhyB_36AALinker_C-TEV), P348(Laccase QCV RFC25), P326(Strep-tag-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1), P361(N-TEV-36AAlinker-PIF6), P364(N-TEV-36AAlinker-PIF3), P266(IgKappa-SigP_Nanoluc)
Investigator: Katrin
Aim of the experiment: Picking of ReTrafos of E. coli XL1-blue with P170(SpyTag), P165(t35s), P308(PhyB_36AALinker_C-TEV), P348(Laccase QCV RFC25), P326(Strep-tag-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1), P361(N-TEV-36AAlinker-PIF6), P364(N-TEV-36AAlinker-PIF3), P266(IgKappa-SigP_Nanoluc
Procedure:
- one clone per plate was picked
- 4 ml LB-medium, 4 µl chloramphenicol
Dpn1 digestion and transformation of QCIII of AlcR + analytical Gel
Investigator: Katrin
Aim of the experiment: Dpn1 digestion and transformation of QCIII of AlcR + analytical Gel
Procedure:
- 1 µl of Dpn1 (10 U/µl) was added to the reaction, the batch was incubated for one hour at 37°C
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 8 µl of QuikChange products was added to 150 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- The cell suspension were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellchloramphenicol plate.
- 0.5 µl of loading buffer were added to 4.5 µl of Dpn1 digested plasmid
- Electrophoresis on 1% agarose gel
| Lane | 1 kbp DNA ladder | QCIII product of AlcR |
|---|---|---|
| Results | as expected |
Transformation of E. coli XL1-blue with ligation products F146+F147 (Ig-Kappa-SigP_SpyTag+Nanoluc) and F145+F42 (pActin+pSB1C3) and negative controls F146 and F42
Investigator: Katrin
Aim of the experiment: Transformation of E. coli XL1-blue with ligation products F146+F147 (Ig-Kappa-SigP_SpyTag+Nanoluc) and F145+F42 (pActin+pSB1C3) and negative controls F146 and F42
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 8 µl of DNA were added to the tubes containing 150 µl of competent cells and gently mixed
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- The cell suspension were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated on a new chlorampenicol plate.
Oligohybridization of mMCSII (mMCSII_fw (O98) + mMCSII_rv (O99)) and (GGGGS)x5_TEV (GGGGSx5_TEV_fw (O102) + GGGGSx5_TEV_rv (O103))
Investigator: Jeff
Aim of the experiment: Oligohybridization of mMCSII (mMCSII_fw (O98) + mMCSII_rv (O99)) and (GGGGS)x5_TEV (GGGGSx5_TEV_fw (O102) + GGGGSx5_TEV_rv (O103)).
Procedure:
- 25 µL of 100 pmol/µl of mMCSII_fw (O98) and mMCSII_rv (O99) were pooled and mixed together/25 µL of 100 pmol/µl of GGGGSx5_TEV_fw (O102) and GGGGSx5_TEV_rv (O103) were pooled and mixed together/
- Heating up to 95 °C for 5 min
- Cooling at RT in a styropor box overnight
Saturday, June 29th
Miniprep of Retrafos of P170(SpyTag), P165(t35s), P308(PhyB_36AALinker_C-TEV), P348(Laccase QCV RFC25), P326(Strep-tag-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1), P361(N-TEV-36AAlinker-PIF6), P364(N-TEV-36AAlinker-PIF3), P266(IgKappa-SigP_Nanoluc)
Investigator: Katrin
Aim of the experiment: Miniprep of Retrafos of P170(SpyTag), P165(t35s), P308(PhyB_36AALinker_C-TEV), P348(Laccase QCV RFC25), P326(Strep-tag-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1), P361(N-TEV-36AAlinker-PIF6), P364(N-TEV-36AAlinker-PIF3), P266(IgKappa-SigP_Nanoluc)
Procedure:
- Miniprep according to QIAGEN QIAprep Kit
Transformation of E. coli XL1-blue with ligation product F145+F42 (pActin+pSB1C3, ligation overnight at 16°C) and negative control
Investigator: Katrin
Aim of the experiment: Transformation of E. coli XL1-blue with ligation products F145+F42 (pActin+pSB1C3, ligation overnight at 16°C) and negative control Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 8 µl of DNA were added to the tubes containing 150 µl of competent cells and gently mixed
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- The cell suspension were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated on a new chlorampenicol plate.
Preparative digestion of P402 (Ig-Kappa-SigP_Nanoluc)
Investigator: Katrin
Aim of the experiment: Preparative digestion of P402 (Ig-Kappa-SigP_Nanoluc)
Procedure:
reaction batch
| volume | reagent |
|---|---|
| 20 µl | P402 |
| 4 µl | CutSmart Buffer (10x) |
| 1 µl | AgeI-HF (20 U/µl) |
| 1 µl | SpeI-HF (20 U/µl) |
| 14 µl | ddH2O |
| =40 µl | TOTAL |
- Reaction batches were incubated for 3 h at 37 °C.
Preparative gelelectrophoresis of F148, F149 and digestion product of P402
Investigator: Jeff
Aim of the experiment: Preparative gelelectrophoresis of F148, F149 and digestion product of P402.
Procedure:
- Preparative gelelectrophoresis was performed on a 1% agarose gel at 90 V for 45 min.
| Lane: | 1 kbp DNA ladder | F148 | F149 | Digestion of P402 with AgeI & SpeI |
|---|---|---|---|---|
| Result: | as expected | as expected | as expected (lower band was cut out) |
- Gelpurified products were extracted via QIAquick gel extraction kit, QIAGEN.
PCR of P143 (pLuc-Actin, Amplification of pActin) and P344 (FluA R95KA45IS114T, triple mutant of FluA)
Investigator: Jeff
Aim of the experiment: PCR of P143 (pLuc-Actin, Amplification of pActin) and P344 (FluA R95KA45IS114T, triple mutant of FluA).
Procedure:
Operational sequence:
- Reaction batch with Q5 Mastermix for P143 (pActin amplification):
| volume | reagent |
|---|---|
| 25 µl | 2x Q5 Master Mix |
| 2.5 µl | 10 µM Forward Primer O96 (pActin_Oryza_fw) |
| 2.5 µl | 10 µM Reverse Primer O97 (pActin_Oryza_rv) |
| 1 µl | Plasmid DNA (P143)- 1:1000 dilution (desired c between 1 pg and 1 ng) |
| 19 µL | ddH2O Water |
| =50 µL | TOTAL |
- Reaction batch with Q5 Mastermix for P344 (FluA triple mutant amplification):
| volume | reagent |
|---|---|
| 25 µl | 2x Q5 Master Mix |
| 2.5 µl | 10 µM Forward Primer O104 (FluA_tripmut_fw) |
| 2.5 µl | 10 µM Reverse Primer O105 (FluA_tripmut_rv) |
| 1 µl | Plasmid DNA (P344)- 1:1000 dilution (desired c between 1 pg and 1 ng) |
| 19 µL | ddH2O Water |
| =50 µL | TOTAL |
- The content has been mixed with a pipette
- The PCR program was performed after following scheme for P143 (pActin amplification):
| Initial denaturation | 98 °C | 30 s |
|---|---|---|
| 30 cycles | 98 °C | 10 s |
| 62 °C | 30 s | |
| 72 °C | 45 s | |
| Final extension | 72 °C | 2 min |
| Hold | 4 °C | infinite |
- The PCR program was performed after following scheme for P344 (FluA triple mutant amplification)
| Initial denaturation | 98 °C | 30 s |
|---|---|---|
| 30 cycles | 98 °C | 10 s |
| 69 °C | 30 s | |
| 72 °C | 16 s | |
| Final extension | 72 °C | 2 min |
| Hold | 4 °C | infinite |
- After PCR, the product was purified with QIAquick PCR Purification Kit, Qiagen.
Sunday, June 30th
Miniprep of F134 + F135 (SERK-SigP-Laccase + Strep-tag-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1), F136 + F137 (PhyB_36AAlinker_C-TEV_IRES + N-TEV_36AAlinker_PIF6), F136 + F138 (PhyB_36AAlinker_C-TEV_IRES + N-TEV_36AAlinker_PIF3), F143 + F144 (npt-csette + t35S) and ReTraFo of P388 (IgKappa-SigP_SpyTag)
Investigator: Andi
Aim of the experiment: Miniprep of F134 + F135 (SERK-SigP-Laccase + Strep-tag-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1), F136 + F137 (PhyB_36AAlinker_C-TEV_IRES + N-TEV_36AAlinker_PIF6), F136 + F138 (PhyB_36AAlinker_C-TEV_IRES + N-TEV_36AAlinker_PIF3), F143 + F144 (npt-csette + t35S) and ReTraFo of P388 (IgKappa-SigP_SpyTag).
Procedure:
Miniprep according to QIAGEN QIAprep Kit
Analytical digestion and gelelectrophoresis of P403 – P418
Investigator: Rosario
Aim of the experiment: Analytical digestion and gelelectrophoresis of P403 – P418.
Procedure:
- 23x Mastermix (EcoRI-HF & PstI-HF) for analytical digestion:
| volume | reagent |
|---|---|
| 46 µl | CutSmart Buffer (10x) |
| 5.75 µl | EcoRI-HF (20 U/µl) |
| 5.75 µl | PstI-HF (20 U/µl) |
| 345 µl | ddH2O |
| =402.5 µl | TOTAL |
- 23x Mastermix (EcoRI-HF & PstI-HF) for analytical digestion:
| volume | reagent |
|---|---|
| 17.5 µl | Mastermix for EcoRI & PstI |
| 2.5 µl | Plasmid DNA (P403 – P418) |
| =20 µl | TOTAL |
- Analytical digestion was performed at 37 °C for 1.5 h.
- After incubation 2.22 µl of DNA loading buffer (10x) was added to the reaction batches.
- Analytical gelelectrophoresis was performed at 90 V for 1 h.
| Lane: | 1000 bp DNA ladder | Digestion of P403 with EcoRI & PstI | Digestion of P404 with EcoRI & PstI | Digestion of P405 with EcoRI & PstI | Digestion of P406 with EcoRI & PstI | Digestion of P407 with EcoRI & PstI | Digestion of P408 with EcoRI & PstI | Digestion of P409 with EcoRI & PstI | Digestion of P410 with EcoRI & PstI | Digestion of P374 with EcoRI & PstI | F153 | F154 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Result: | corrupt | as expected | as expected | as expected | corrupt | corrupt | corrupt | corrupt | as expected (control) | no PCR product; corrupt | as expected |
| Lane: | 1000 bp DNA ladder | Digestion of P411 with EcoRI & PstI | Digestion of P412 with EcoRI & PstI | Digestion of P246 with EcoRI & PstI (Control) | Digestion of P413 with EcoRI & PstI | Digestion of P414 with EcoRI & PstI | Digestion of P391 with EcoRI & PstI (Control) |
|---|---|---|---|---|---|---|---|
| Result: | as expected | as expected | as expected | as expected | as expected | as expected |
| Lane: | 1000 bp DNA ladder | Digestion of P415 with EcoRI & PstI | Digestion of P416 with EcoRI & PstI | Digestion of P417 with EcoRI & PstI | Digestion of P418 with EcoRI & PstI | Digestion of P385 with EcoRI & PstI |
|---|---|---|---|---|---|---|
| Result: | as expected | as expected | as expected | as expected | as expected (Control) |
PCR of P143 (pLuc-Actin, Amplification of pActin)
Investigator: Ingmar
Aim of the experiment: PCR of P143 (pLuc-Actin, Amplification of pActin). Procedure:
Operational sequence:
- Reaction batch with Q5 Mastermix for P143 (pActin amplification):
| volume | reagent |
|---|---|
| 25 µl | 2x Q5 Master Mix |
| 2.5 µl | 10 µM Forward Primer O96 (pActin_Oryza_fw) |
| 2.5 µl | 10 µM Reverse Primer O97 (pActin_Oryza_rv) |
| 1 µl | Plasmid DNA (P143)- 1:1000 dilution (desired c between 1 pg and 1 ng) |
| 19 µL | ddH2O Water |
| =50 µL | TOTAL |
- The content has been mixed with a pipette
- The PCR program was performed after following scheme for P143 (pActin amplification):
| Initial denaturation | 98 °C | 30 s |
|---|---|---|
| 30 cycles | 98 °C | 10 s |
| 62 °C | 30 s | |
| 72 °C | 45 s | |
| Final extension | 72 °C | 2 min |
| Hold | 4 °C | infinite |
- The PCR was performed in two different cyclers in parallel to elucidate wheather this might have an influence on the product. Furthermore one sample contained a 100 fold increased template concentration.
- After PCR, the product was purified with QIAquick PCR Purification Kit, Qiagen.
- Analytical gelelectrophoresis was performed on a 1% agarose gel at 90 V for 60 min using NEB 2-Log DNA ladder.
| Lane: | 1 kbp DNA ladder | 1:1000 P143 | 1:1000 P143 | 1:10 P143 |
|---|---|---|---|---|
| Result: | length as expected, but low concentration, small amount of side products | length as expected, but low concentration, small amount of side products | length as expected; higher concentration but increased amount of side products, too. Fragment was labelled F155. |
Preparative digestion and gelelectrophoresis of P367 (NLS+thermonuclease), P416 (PhyB_36AAlinker_C-TEV_IRES_N-TEV_36AAlinker_PIF3/6), P418 (PhyB_36AAlinker_C-TEV_IRES_N-TEV_36AAlinker_PIF3/6 ), P404 (t35S_npt-casette), P395 (SpyTag)and F154 (FluA triple mutant)
Investigator: Jeff
Aim of the experiment: Preparative digestion and gelelectrophoresis of P367 (NLS+thermonuclease), P416 (PhyB_36AAlinker_C-TEV_IRES_N-TEV_36AAlinker_PIF3/6), P418 (PhyB_36AAlinker_C-TEV_IRES_N-TEV_36AAlinker_PIF3/6 ), P404 (t35S_npt-casette), P395 (SpyTag)and F154 (FluA triple mutant).
Procedure:
Batch for preparative digestion of P404 with EcoRI and XbaI, P395 with NgoMIV and SpeI, F154 with XbaI and AgeI, P367 with XbaI and NgoMIV, P416 with SpeI and Pst I and P418 with SpeI and PstI
| volume | reagent |
|---|---|
| 20 µl | Plasmid DNA |
| 4 µl | CutSmart Buffer (10x) |
| 1 µl | Enzyme 1 (20 U/µl) |
| 1 µl | Enzyme 2 (20 U/µl) |
| 14 µl | ddH2O |
| =40 µl | TOTAL |
| volume | reagent |
|---|---|
| 25 µl | PCR product |
| 5 µl | CutSmart Buffer (10x) |
| 1 µl | Enzyme 1 (20 U/µl) |
| 1 µl | Enzyme 2 (20 U/µl) |
| 18 µl | ddH2O |
| =50 µl | TOTAL |
| Plasmid | Name | Enzyme 1 | Enzyme 2 |
|---|---|---|---|
| P404 | t35S_npt-casette | EcoRI | XbaI |
| P395 | SpyTag | NgoMIV | SpeI |
| P418 | PhyB_36AAlinker_C-TEV_IRES_N-TEV_36AAlinker_PIF3/6 | SpeI | PstI |
| P367 | NLS+thermonuclease | XbaI | NgoMIV |
| P416 | PhyB_36AAlinker_C-TEV_IRES_N-TEV_36AAlinker_PIF3/6 | SpeI | PstI |
| F154 | FluA triple mutant | XbaI | AgeI |
- Incubation for 2.5 h at 37 °C.
- 4 µl of DNA loading buffer (10x) were added to the 40 µl reaction batches after digestion and they were loaded on a 1% agarose gel
- Preparative gelelectrophoresis was performed at 90 V for 60 min.
| Lane | Digestion of P367 with XbaI & NgoMIV | 1kb DNA ladder | Digestion of P416 with SpeI & PstI | Digestion of P418 with SpeI & PstI |
|---|---|---|---|---|
| Results | as expected | as expected | as expected |
| Lane | Digestion of P404 with EcoRI & XbaI | 1kb DNA ladder | Digestion of P395 with NgoMIV & SpeI | Digestion of F154 with XbaI & AgeI |
|---|---|---|---|---|
| Results | as expected | as expected; lower band was extracted | as expected |
- Extracted via QIAGEN gel extraction kit, digested PCR product purified via PCR Purification Kit
Ligation of F123 + F152 ((GGGGS)x5-TEV-site-linker in pSB1C3), F156 + F152 ((GGGGS)x5-TEV-site-linker_NLS_Thermonuclease in pSB1C3), F157 + F131 (PhyB_36AAlinker_C-TEV_IRES_N-TEV_36AAlinker_PIF3/6_IRES in pSB1C3), F158 + F131 (PhyB_36AAlinker_C-TEV_IRES_N-TEV_36AAlinker_PIF3/6_IRES in pSB1C3), F159 + F151 (mMCSII_t35S_npt-casette in pSB1C3), F150 + F160 (SERK-SigP_Nanoluc_SpyTag) and F123 + F161 (FluA triple mutant in pSB1C3)
Investigator: Andi
Aim of the experiment: Ligation of F123 + F152 ((GGGGS)x5-TEV-site-linker in pSB1C3), F156 + F152 ((GGGGS)x5-TEV-site-linker_NLS_Thermonuclease in pSB1C3), F157 + F131 (PhyB_36AAlinker_C-TEV_IRES_N-TEV_36AAlinker_PIF3/6_IRES in pSB1C3), F158 + F131 (PhyB_36AAlinker_C-TEV_IRES_N-TEV_36AAlinker_PIF3/6_IRES in pSB1C3), F159 + F151 (mMCSII_t35S_npt-casette in pSB1C3), F150 + F160 (SERK-SigP_Nanoluc_SpyTag) and F123 + F161 (FluA triple mutant in pSB1C3).
Procedure:
- Ligation batch for F123 + F152 ((GGGGS)x5-TEV-site-linker in pSB1C3):
| volume | reagent |
|---|---|
| 2.03 µl | F123 (49.3 ng/µl, 2086 bp) |
| 14.97 µl | F152 (488.2 ng/µl, 102 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- F156 + F152 ((GGGGS)x5-TEV-site-linker_NLS_Thermonuclease in pSB1C3):
| volume | reagent |
|---|---|
| 0.63 µl | F156 (158.3 ng/µl, 2596 bp) |
| 16.37 µl | F152 (488.2 ng/µl, 102 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- F157 + F131 (PhyB_36AAlinker_C-TEV_IRES_N-TEV_36AAlinker_PIF3/6_IRES in pSB1C3):
| volume | reagent |
|---|---|
| 0.67 µl | F157 (148.7 ng/µl, 6711 bp) |
| 1.71 µl | F131 (16.4 ng/µl, 627 bp) |
| 14.62 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- F158 + F131 (PhyB_36AAlinker_C-TEV_IRES_N-TEV_36AAlinker_PIF3/6_IRES in pSB1C3):
| volume | reagent |
|---|---|
| 0.76 µl | F158 (132.4 ng/µl, 6711 bp) |
| 1.71 µl | F131 (16.4 ng/µl, 627 bp) |
| 14.59 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- F159 + F151 (mMCSII_t35S_npt-casette in pSB1C3):
| volume | reagent |
|---|---|
| 1.78 µl | F159 (56.1 ng/µl, 3812 bp) |
| 15.22 µl | F151 (229.4 ng/µl, 37 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- F150 + F160 (SERK-SigP_Nanoluc_SpyTag):
| volume | reagent |
|---|---|
| 0.59 µl | F150 (169.3 ng/µl, 2660 bp) |
| 16.41 µl | F160 (109 ng/µl, 39 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- F123 + F161 (FluA triple mutant in pSB1C3):
| volume | reagent |
|---|---|
| 2.03 µl | F123 (49.3 ng/µl, 2086 bp) |
| 2.58 µl | F161 (29.1 ng/µl, 522 bp) |
| 12.39 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
Week 11
Monday, July 1st
Everyday I'm swaffeling
Investigator: Le swaffleur passive
Preparative digestion and gelelectrophoresis of F155 (pActin PCR product) and P393 (Stress inducible Promoter)
Investigator: Ingmar
Aim of the experiment: Preparative digestion and gelelectrophoresis of F155 (pActin PCR product) and P393 (Stress inducible Promoter).
Procedure:
Batch for preparative digestion of F155 (pActin PCR product) with XbaI and PstI
| volume | reagent |
|---|---|
| 28.5 µl | F155 |
| 4 µl | CutSmart Buffer (10x) |
| 1 µl | XbaI |
| 1 µl | PstI |
| 5.5 µl | ddH2O |
| =40 µl | TOTAL |
Batch for preparative digestion of P393 (Stress inducible Promoter)with EcoRI and SpeI"
| volume | reagent |
|---|---|
| 20 µl | P393 |
| 4 µl | CutSmart Buffer (10x) |
| 1 µl | EcoRI (20 U/µl) |
| 1 µl | SpeI (20 U/µl) |
| 14 µl | ddH2O |
| =40 µl | TOTAL |
- Incubation for 2.5 h at 37 °C.
- 4.5 µl of DNA loading buffer (10x) were added to the 40 µl reaction batches after digestion and they were loaded on a 1% agarose gel
- Preparative gelelectrophoresis was performed at 90 V for 60 min.
| Lane | 1kb DNA ladder | Digestion of F155 with XbaI & PstI | Digestion of P393 with EcoRI & SpeI |
|---|---|---|---|
| Results | as expected, the upper band was cut out | as expected, the lowest band was cut out |
- Extracted via QIAGEN gel extraction kit, digested PCR product purified via PCR Purification Kit
Ligation of F162 (pActin) + F42(pSB1C3 backbone)
Investigator: Ingmar
Aim of the experiment: Ligation of F162 (pActin) + F42(pSB1C3 backbone)
Procedure:
- Ligation batch for ligation of F162 (pActin) + F42(pSB1C3 backbone)
| volume | reagent |
|---|---|
| 1.36 µl | F42 (100 ng) |
| 6.64 µl | F162 |
| 91 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (NEB) |
| =20 µl | TOTAL |
- Negative controls were prepared by replacing the volume of the insert by ddH2O.
- The ligation was performed for 1 hour at room temperature; the ligation batch was transferred to 16 °C afterwads over night.
Transformation of E. coli XL1-blue with ligation product F162+F42 (pActin+pSB1C3, ligation for 1 h at room temperature
Investigator: Ingmar
Aim of the experiment: Transformation of E. coli XL1-blue with ligation product F162+F42 (pActin+pSB1C3, ligation for 1 h at room temperature
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 10 µl of DNA were added to the tubes containing 150 µl of competent cells and gently mixed
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- The cell suspension were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated on a new chlorampenicol plate.
Transformation of E. coli XL1-blue with ligation product F123 + F152 ((GGGGS)x5-TEV-site-linker in pSB1C3), F157 + F131 (PhyB_36AAlinker_C-TEV_IRES_N-TEV_36AAlinker_PIF3/6_IRES in pSB1C3), F158 + F131 (PhyB_36AAlinker_C-TEV_IRES_N-TEV_36AAlinker_PIF3/6_IRES in pSB1C3), F159 + F151 (mMCSII_t35S_npt-casette in pSB1C3) and F123 + F161 (FluA triple mutant in pSB1C3)
Investigator: Rosario
Aim of the experiment: Transformation of E. coli XL1-blue with ligation product F123 + F152 ((GGGGS)x5-TEV-site-linker in pSB1C3), F157 + F131 (PhyB_36AAlinker_C-TEV_IRES_N-TEV_36AAlinker_PIF3/6_IRES in pSB1C3), F158 + F131 (PhyB_36AAlinker_C-TEV_IRES_N-TEV_36AAlinker_PIF3/6_IRES in pSB1C3), F159 + F151 (mMCSII_t35S_npt-casette in pSB1C3) and F123 + F161 (FluA triple mutant in pSB1C3).
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 10 µl of DNA were added to the tubes containing 100 µl of competent cells and gently mixed
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- The cell suspension were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated on a new chlorampenicol plate.
Tuesday, July 2nd
Transformation of E. coli XL1-blue with ligation product F162+F42 (pActin+pSB1C3, ligation over night at 16 °C
Investigator: Ingmar
Aim of the experiment: Transformation of E. coli XL1-blue with ligation product F162+F42 (pActin+pSB1C3, ligation over night at 16 °C.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 10 µl of DNA were added to the tubes containing 150 µl of competent cells and gently mixed
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- The cell suspension were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated on a new chlorampenicol plate.
Picking of F123 + F152, F157 + F131, F158 + F131, F159 + F151 and F123 + F161
Investigator: Louise
Aim of the experiment: Picking of F123 + F152 ((GGGGS)x5-TEV-site-linker in pSB1C3), F157 + F131 (PhyB_36AAlinker_C-TEV_IRES_N-TEV_36AAlinker_PIF3/6_IRES in pSB1C3), F158 + F131 (PhyB_36AAlinker_C-TEV_IRES_N-TEV_36AAlinker_PIF3/6_IRES in pSB1C3), F159 + F151 (mMCSII_t35S_npt-casette in pSB1C3) and F123 + F161 (FluA triple mutant in pSB1C3).
Procedure:
Picked colonies were transfered into 5 ml LB-medium with 5 µl of Chloramphenicol and incubated for at 37 °C in the 180 rpm cell-culture shaker.
Ligation of F162 (pActin) + F42(pSB1C3 backbone) and of F163 (stress inducible Promoter + RFP ) + F159 (t35S-NPT-pSB1C3)
Investigator: Ingmar
Aim of the experiment: As the ligation of the prmoter done on Monday failed, it is repeated with an decreased overall volume resulting in an increased DNA concentration: Ligation of F162 (pActin) + F42(pSB1C3 backbone). Furthermore the stress inducible Promoter cloned to RFP (F163) should be ligated into a pSB1C3 in front og t35S and NPT.
Procedure:
- Ligation batch for ligation of F162 (pActin) + F42(pSB1C3 backbone)
| volume | reagent |
|---|---|
| 1.36 µl | F42 (100 ng) |
| 6.64 µl | F162 |
| 1 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase |
| =10 µl | TOTAL |
- The reaction was prepared twice: one time using a ligase provided by NEB and one time using a ligase providied by Thermo Scientific. Negative controls were prepared by replacing the volume of the insert by ddH2O.
- Ligation batch for ligation of F163 (stress inducible promoter + RFP) + F159(t35S-NPT-pSB1C3 backbone)
| volume | reagent |
|---|---|
| 1.79 µl | F159 (100 ng) |
| 8.61 µl | F163 |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase |
| 6,6 µl | H2O |
| =20 µl | TOTAL |
- The ligation was performed for 2 hours at room temperature; the ligation batch was transferred to 16 °C afterwads over night.
Transformation of E. coli XL1-blue with ligation product F162+F42 (pActin+pSB1C3, ligation batch volume 10 µl, reaction performed at room temperature) and with F159+F163 (stress inducible promoter+RFP+t35S+NPT in pSB1C3)
Investigator: Ingmar
Aim of the experiment: Transformation of E. coli XL1-blue with ligation product F162+F42 (pActin+pSB1C3, ligation batch volume 10 µl, reaction performed at room temperature) and with F159+F163 (stress inducible promoter+RFP+t35S+NPT in pSB1C3).
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 5 µl of DNA were added to the tubes containing 150 µl of competent cells and gently mixed
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- The cell suspension were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated on a new chlorampenicol plate.
PCR of P143 (pLuc-Actin, Amplification of pActin)
Investigator: Ingmar
Aim of the experiment: PCR of P143 (pLuc-Actin, Amplification of pActin). The PCR product was phosphorylated afterwards to test blunt end cloning into a vector. Procedure:
Operational sequence:
- Reaction batch for PCR with Q5 Mastermix for P143 (pActin amplification):
| volume | reagent |
|---|---|
| 25 µl | 2x Q5 Master Mix |
| 2.5 µl | 10 µM Forward Primer O96 (pActin_Oryza_fw) |
| 2.5 µl | 10 µM Reverse Primer O97 (pActin_Oryza_rv) |
| 1 µl | Plasmid DNA (P143)- 1:10 dilution (desired c between 1 pg and 1 ng) |
| 19 µL | ddH2O Water |
| =50 µL | TOTAL |
- The content has been mixed with a pipette
- The PCR program was performed after following scheme for P143 (pActin amplification):
| Initial denaturation | 98 °C | 30 s |
|---|---|---|
| 30 cycles | 98 °C | 10 s |
| 62 °C | 30 s | |
| 72 °C | 45 s | |
| Final extension | 72 °C | 2 min |
| Hold | 4 °C | infinite |
- The PCR was performed in two different cyclers in parallel to elucidate wheather this might have an influence on the product. Furthermore one sample contained a 100 fold increased template concentration.
- After PCR, the product was purified with QIAquick PCR Purification Kit, Qiagen.
- Reaction batch for phosphorylation of thePCR product:
| volume | reagent |
|---|---|
| 15 µl | PCR product |
| 2 µl | 10 x forward buffer A |
| 2 µl | 10 mM ATP |
| 1 µl | Polynucleotide Kinase (Thermo Scientific) |
| =20 µL | TOTAL |
- The reaction was performed for 1 h at 37 °C. Afterwards the mixture was inactivated by heat for 10 min at 75 °C.
Quickchange mutagenesis of P143 (Actin promoter)
Investigator: Ingmar
Aim of the experiment: Deliting the forbidden EcoRI restriction site in the promoter.
Procedure: PCR
Reaction batch 1
| volume | reagent |
|---|---|
| 2.5 µl | 10x Pfu Ultra II buffer |
| 1 µl | Plasmid P143 template |
| 0.5 µl | 1:10 dilution of O106 |
| 19. 5 µl | ddH2O |
| 1 µl | dNTP mix |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
Reaction batch 2
| volume | reagent |
|---|---|
| 2.5 µl | 10x Pfu Ultra II buffer |
| 1 µl | Plasmid P143 template |
| 0.5 µl | O107 |
| 19.5 µl | ddH2O |
| 1 µl | dNTP mix |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
PCR cycling parameters
| Segment | Cycles | Temperature | Time |
|---|---|---|---|
| 1 | 1 | 95 °C | 30 sec |
| 2 | 15 | 95°C | 30 sec |
| 55°C | 1 min | ||
| 67°C | 6,5 min |
- Having completed the PCR cycling parameters listed above both PCR reaction batches were mixed together and the cycling parameters listed above were one time more applied. Furthermore 1 µl Pfu Ultra II DNA polymerase was added.
- Digestion of the parental DNA with DpnI: Addition of 1 µl DpnI to the PCR batch and incubate for 1 h at 37 °C.
Preparative digestion and gelelectrophoresis of P352 (SERK-SigP_SpyCatcher_Strep-tag-II-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1), P358 (SERK-SigP_SpyTag_Strep-tag-II-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1) and P395 (SpyTag)
Investigator: Louise, Jeff
Aim of the experiment: Preparative digestion and gelelectrophoresis of P352 (SERK-SigP_SpyCatcher_Strep-tag-II-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1), P358 (SERK-SigP_SpyTag_Strep-tag-II-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1) and P395 (SpyTag).
- Batch for preparative digestion of P352 (SERK-SigP_SpyCatcher_Strep-tag-II-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1):
| volume | reagent |
|---|---|
| 20 µl | P352 |
| 4 µl | CutSmart Buffer (10x) |
| 1 µl | SpeI |
| 1 µl | PstI-HF |
| 14 µl | ddH2O |
| =40 µl | TOTAL |
- Batch for preparative digestion of P358 (SERK-SigP_SpyTag_Strep-tag-II-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1):
| volume | reagent |
|---|---|
| 20 µl | P358 |
| 4 µl | CutSmart Buffer (10x) |
| 1 µl | SpeI |
| 1 µl | PstI-HF |
| 14 µl | ddH2O |
| =40 µl | TOTAL |
- Batch for preparative digestion of P395 (SpyTag):
| volume | reagent |
|---|---|
| 20 µl | P395 |
| 4 µl | CutSmart Buffer (10x) |
| 1 µl | NgoMIV |
| 1 µl | SpeI |
| 14 µl | ddH2O |
| =40 µl | TOTAL |
- Incubation at 37 °C; 2.5 h
- 4.44 µl of DNA loading buffer (10x) were added to the 40 µl reaction batches, digested with NgoMIV & SpeI (P395), after digestion and they were loaded on a 1% agarose gel.
- Preparative gelelectrophoresis was performed at 90 V for 60 min.
- Reaction batches which were digested with SpeI & PstI were purified with QIAquick PCR purification kit, QIAGEN.
| Lane | 1kb DNA ladder | Digestion of P395 with NgoMIV & SpeI |
|---|---|---|
| Results | as expected; lower band was cut out |
- Extracted via QIAquick gel extraction kit, QIAGEN.
Ligation of F164 + F131 (SERK-SigP_SpyCatcher_Strep-tag-II-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1 in pSB1C3+ IRES) and F165 + F131 (SERK-SigP_SpyTag_Strep-tag-II-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1 in pSB1C3 + IRES)
Investigator: Jeff
Aim of the experiment: Ligation of F164 + F131 (SERK-SigP_SpyCatcher_Strep-tag-II-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1 in pSB1C3+ IRES), F165 + F131 (SERK-SigP_SpyTag_Strep-tag-II-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1 in pSB1C3 + IRES).
Procedure:
- Ligation batch for F164 + F131 (SERK-SigP_SpyCatcher_Strep-tag-II-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1 in pSB1C3+ IRES), vector:insert = 1:3:
| volume | reagent |
|---|---|
| 1.22 µl | F164 (81.7 ng/µl, 3524 bp) |
| 3.25 µl | F131 (16.4 ng/µl, 627 bp) |
| 12.53 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- Ligation batch for F165 + F131 (SERK-SigP_SpyTag_Strep-tag-II-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1 in pSB1C3+ IRES), vector:insert = 1:3:
| volume | reagent |
|---|---|
| 1.65 µl | F165 (60.7 ng/µl, 3224 bp) |
| 3.56 µl | F131 (16.4 ng/µl, 627 bp) |
| 11.79 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- Ligation was performed at RT for 1 h.
Transformation of E. coli XL1-blue with ligation product F164 + F131 (SERK-SigP_SpyCatcher_Strep-tag-II-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1 in pSB1C3+ IRES), F165 + F131 (SERK-SigP_SpyTag_Strep-tag-II-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1 in pSB1C3 + IRES) and retransformations of P352, P358 and P395
Investigator: Jeff
Aim of the experiment: Transformation of E. coli XL1-blue with ligation product F164 + F131 (SERK-SigP_SpyCatcher_Strep-tag-II-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1 in pSB1C3+ IRES), F165 + F131 (SERK-SigP_SpyTag_Strep-tag-II-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1 in pSB1C3 + IRES) and retransformations of P352, P358 and P395.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 10 µl of DNA were added to the tubes containing 150 µl of competent cells and gently mixed
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- The cell suspension were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated on a new chlorampenicol plate.
Ligation of F150 + F166 (IgKappa-SigP_NanoLuc in pSB1C3 + SpyTag)
Investigator: Jeff
Aim of the experiment: Ligation of F150 + F166 (IgKappa-SigP_NanoLuc in pSB1C3 + SpyTag).
Procedure:
- Ligation batch for F150 + F166 (IgKappa-SigP_NanoLuc in pSB1C3 + SpyTag), vector:insert = 1:6:
| volume | reagent |
|---|---|
| 0.59 µl | F150 (169.3 ng/µl, 2660 bp) |
| 2.67 µl | F166 (3.3 ng/µl, 39 bp) |
| 13.79 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- Ligation was performed at 16 °C overnight.
Miniprep of E. coli XL1 blue transformed with F123 + F152 ((GGGGS)x5-TEV-site-linker in pSB1C3), F157 + F131 (PhyB_36AAlinker_C-TEV_IRES_N-TEV_36AAlinker_PIF3/6_IRES in pSB1C3), F158 + F131 (PhyB_36AAlinker_C-TEV_IRES_N-TEV_36AAlinker_PIF3/6_IRES in pSB1C3), F159 + F151 (mMCSII_t35S_npt-casette in pSB1C3) and F123 + F161 (FluA triple mutant in pSB1C3)
Investigator: Ingmar, Jeff
Aim of the experiment: Miniprep of E. coli XL1 blue transformed with F123 + F152 ((GGGGS)x5-TEV-site-linker in pSB1C3), F157 + F131 (PhyB_36AAlinker_C-TEV_IRES_N-TEV_36AAlinker_PIF3/6_IRES in pSB1C3), F158 + F131 (PhyB_36AAlinker_C-TEV_IRES_N-TEV_36AAlinker_PIF3/6_IRES in pSB1C3), F159 + F151 (mMCSII_t35S_npt-casette in pSB1C3) and F123 + F161 (FluA triple mutant in pSB1C3)
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN).
- The resulting tubes were labeled as P420–P432.
Picking of of E. coli XL1 blue transformed with F152 + F156 ((GGGGS)x5-TEV-site-linker + NLS_Thermonuclease in pSB1C3)
Investigator: Jeff
Aim of the experiment: Picking of of E. coli XL1 blue transformed with F152 + F156 ((GGGGS)x5-TEV-site-linker + NLS_Thermonuclease in pSB1C3).
Procedure:
- Picked colonies tips was transferred into cell-culture tubes with air-permeable, sterile cover. Each tube contain 4 mL of LB-medium + 4 µL chloramphenicol(1000x).
- These tubes were transferred in a cell culture shaker at 37 °C and were incubated at 180 rpm overnight
Wednesday, July 3rd
Picking of of E. coli XL1 blue transformed with F162 + F42 (pActin + pSB1C3), F163 + F159 (Stress inducible promotor_RFP + t35S_NPT in pSB1C3, P395, P352, P358, F164+F131 (SERK-SigP_SpyCatcher_Strep-tag-II-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1 in pSB1C3 + IRES), F165+F131 (SERK-SigP_SpyTag_Strep-tag-II-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1 in pSB1C3 + IRES)
Investigator: Ingmar
Aim of the experiment: Picking of of E. coli XL1 blue transformed with F162 + F42 (pActin + pSB1C3), F163 + F159 (Stress inducible promotor_RFP + t35S_NPT in pSB1C3, P395, P352, P358, F164+F131 (SERK-SigP_SpyCatcher_Strep-tag-II-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1 in pSB1C3 + IRES), F165+F131 (SERK-SigP_SpyTag_Strep-tag-II-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1 in pSB1C3 + IRES).
Procedure:
- Picked colonies tips was transferred into cell-culture tubes with air-permeable, sterile cover. Each tube contain 5 mL of LB-medium + 5 µL chloramphenicol(1000x).
- These tubes were transferred in a cell culture shaker at 37 °C and were incubated at 180 rpm overday.
Miniprep of E. coli XL1 blue transformed with F152 + F156 ((GGGGS)x5-TEV-site-linker + NLS_Thermonuclease in pSB1C3)
Investigator: Jeff
Aim of the experiment: Miniprep of E. coli XL1 blue transformed with F152 + F156 ((GGGGS)x5-TEV-site-linker + NLS_Thermonuclease in pSB1C3).
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN).
- The resulting tubes were labeled as P433 – P435.
Transformation of E. coli XL1-blue with ligation product F150 + F166 (IgKappa-SigP_NanoLuc in pSB1C3 + SpyTag)
Investigator: Jeff
Aim of the experiment: Transformation of E. coli XL1-blue with ligation product F150 + F166 (IgKappa-SigP_NanoLuc in pSB1C3 + SpyTag).
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 10 µl of DNA were added to the tubes containing 100 µl of competent cells and gently mixed
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- The cell suspension were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated on a new chlorampenicol plate.
Analytical digestion and gelelectrophoresis of P420 – P435
Investigator: Ingmar
Aim of the experiment: Analytical digestion and gelelectrophoresis of P420 – P435.
Procedure:
| Lane: | 1000 bp DNA ladder | Digestion of P420 with EcoRI & PstI | Digestion of P421 with EcoRI & PstI | Digestion of F123 with EcoRI & PstI | Digestion of P422 with EcoRI & PstI | Digestion of P429 with EcoRI & PstI | Digestion of P430 with EcoRI & PstI | Digestion of P431 with EcoRI & PstI | Digestion of P432 with EcoRI & PstI | 1000 bp DNA ladder |
|---|---|---|---|---|---|---|---|---|---|---|
| Result: | as expected | as expected | as expected | corrupt | corrupt | corrupt | corrupt |
| Lane: | 1000 bp DNA ladder | Digestion of P424 with EcoRI & PstI | Digestion of P425 with EcoRI & PstI | Digestion of P426 with EcoRI & PstI | Digestion of P427 with EcoRI & PstI | Digestion of P428 with EcoRI & PstI | Digestion of P404 with EcoRI & PstI | Digestion of P433 with EcoRI & PstI | Digestion of P434 with EcoRI & PstI | Digestion of P435 with EcoRI & PstI | Digestion of P367 with EcoRI & PstI | 1000 bp DNA ladder |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Result: | as expected | as expected | as expected | corrupt | corrupt | corrupt | corrupt | as expected (control) | no PCR product; corrupt | as expected |
Thursday, July 4th
Quickchange mutagenesis I of P444 (Actin promoter)
Investigator: Louise
Aim of the experiment: Deleting the forbidden EcoRI restriction site in the promoter.
Procedure: PCR
Reaction batch 1
| volume | reagent |
|---|---|
| 2.5 µl | 10x Pfu Ultra II buffer |
| 1 µl | Plasmid P444 template |
| 1.0 µl | 1:10 dilution of O106 |
| 1.0 µl | 1:10 dilution of O107 |
| 18.0 µl | ddH2O |
| 1 µl | dNTP mix 50 mM |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
Reaction batch 2
| volume | reagent |
|---|---|
| 12.5 µl | Q5 Polymerase Mastermix |
| 1 µl | Plasmid P444 template |
| 1.25 µl | 1:10 dilution of O106 |
| 1.25 µl | 1:10 dilution of O107 |
| 8.0 µl | ddH2O |
| 1 µl | dNTP mix 50 mM |
PCR cycling parameters for batch 1
| Cycles | Temperature | Time |
|---|---|---|
| 1 | 95 °C | 30 sec |
| 20 | 95°C | 30 sec |
| 52°C | 1 min | |
| 68°C | 5 min | |
| 4°C | HOLD |
PCR cycling parameters for batch 2
| Cycles | Temperature | Time |
|---|---|---|
| 1 | 98 °C | 30 sec |
| 20 | 98°C | 10 sec |
| 55°C | 30 sec | |
| 72°C | 4 min | |
| 4°C | HOLD |
- Digestion of the parental DNA with DpnI: Addition of 1 µl DpnI to the PCR batch and incubate for 1 h at 37 °C.
The successful amplification was verified on an agerose gel and the PCR-product was transformed (by Jeff)
Ligation of F150 + F166 (IgKappa-SigP_NanoLuc in pSB1C3 + SpyTag) and negative control F150
Investigator: Jeff, Katrin
Aim of the experiment: Ligation of F150 + F166 (IgKappa-SigP_NanoLuc in pSB1C3 + SpyTag) and negative control F150
Procedure:
- Ligation batch for F150 + F166 (IgKappa-SigP_NanoLuc in pSB1C3 + SpyTag), vector:insert = 1:6:
| volume | reagent |
|---|---|
| 0.59 µl | F150 (169.3 ng/µl, 2660 bp) |
| 2.67 µl | F166 (3.3 ng/µl, 39 bp) |
| 13.79 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- Ligation was performed at 22 °C for 1h
- negative control was prepaed with water instead of insert F166
Test digestion of pAct in pSB1C3 (p435 to p448)
Investigator: Jeff
- App. 500 ng of DNA from 13 different clones were digeted with EcoRI and PstI
- Mastermix consisting of 3 µl of each enzyme 30 µl Cutsmart buffer and 120 µl water
- p444 and p447 were sent for sequencing
Transformation of E. coli XL1-blue with ligation product F150+F166 (IgKappa-SigP_NanoLuc in pSB1C3 + SpyTag), F142+F147 (Ig-Kappa-SigP_SpyCatcher + Nanoluc), F139+F141 (Ig-Kappa-SigP_Nanoluc + SpyCatcher) and negative controls
Investigator: Katrin
Aim of the experiment: Transformation of E. coli XL1-blue with ligation product F150+F166 (IgKappa-SigP_NanoLuc in pSB1C3 + SpyTag), F142+F147 (Ig-Kappa-SigP_SpyCatcher + Nanoluc), F139+F141 (Ig-Kappa-SigP_Nanoluc + SpyCatcher) and negative controls
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 8 µl of DNA were added to the tubes containing 150 µl of competent cells and gently mixed
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- The cell suspension were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated on a new chlorampenicol plate.
Preparative digestion of P434, P412, P462, P453
Investigator: Katrin
Aim of the experiment: Preparative digestion of P434, P412, P462, P453
- Batch for preparative digestion of P434
| volume | reagent |
|---|---|
| 20 µl | P434 |
| 4 µl | CutSmart Buffer (10x) |
| 1 µl | NgoMIV |
| 1 µl | PstI-HF |
| 14 µl | ddH2O |
| =40 µl | TOTAL |
- Batch for preparative digestion of P412
| volume | reagent |
|---|---|
| 20 µl | P412 |
| 4 µl | CutSmart Buffer (10x) |
| 1 µl | XbaI |
| 1 µl | PstI-HF |
| 14 µl | ddH2O |
| =40 µl | TOTAL |
- Batch for preparative digestion of P462
| volume | reagent |
|---|---|
| 20 µl | P462 |
| 4 µl | CutSmart Buffer (10x) |
| 1 µl | PstI-HF |
| 1 µl | SpeI |
| 14 µl | ddH2O |
| =40 µl | TOTAL |
- Batch for preparative digestion of P453
| volume | reagent |
|---|---|
| 20 µl | P453 |
| 4 µl | CutSmart Buffer (10x) |
| 1 µl | PstI-HF |
| 1 µl | SpeI |
| 14 µl | ddH2O |
| =40 µl | TOTAL |
- Incubation at 37 °C; 3 h, then put into freezer
Ligation of F142+F147 (IgK-SigP_Spycatcher + NanoLuc) and F141+F139 (SpyCatcher + IgK-SigP_NanoLuc)
Investigator: Louise
Aim of the experiment: Ligation of F142+F147 and F139+F141
Procedure:
- Ligation batch for F142+F147:
| volume | reagent |
|---|---|
| 0.81 µl | F142 (123.4 ng/µl, 2489 bp) |
| 3.51 µl | F147 (17.5 ng/µl, 510 bp) |
| 12.68 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- Ligation batch for F141+F139
| volume | reagent |
|---|---|
| 1.83 µl | F141 (54.6 ng/µl, 2425 bp) |
| 3.43 µl | F139 (21.0 ng/µl, 583 bp) |
| 11.74 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- Ligation was performed at 22 °C for 1h
- negative control was prepaed with water instead of inserts F147 and F139
Friday, July 5th
Ligation of F122+F169 (SERK-SigP_SERK-TMD + (GGGGS)x5-TEV-site-linker_NLS_NucA), F170+F168 (SERK-SigP_SpyCatcher_Strep-TagII-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1_IRES + Ig-Kappa-SigP_SpyTag_Nanoluc) and negative controls
Investigator: Katrin
Aim of the experiment: Ligation of F122+F169 (SERK-SigP_SERK-TMD + (GGGGS)x5-TEV-site-linker_NLS_NucA), F170+F168 (SERK-SigP_SpyCatcher_Strep-TagII-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1_IRES + Ig-Kappa-SigP_SpyTag_Nanoluc) and negative controls
Procedure:
- Ligation batch for F122+F169
| volume | reagent |
|---|---|
| 1.99 µl | F122 (50.3 ng/µl, 2369 bp) |
| 4.32 µl | F169 (18.1 ng/µl, 618 bp) |
| 10.69 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- Ligation batch for F170+F168
| volume | reagent |
|---|---|
| 2.01 µl | F170 (49.7 ng/µl, 4168 bp) |
| 4.11 µl | F168 (11.0 ng/µl, 628 bp) |
| 10.88 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- Ligation was performed at 22 °C for 1h
- negative control was prepaed with water instead of inserts F169 and F168
Transformation of E. coli XL1-blue with ligation product F122+F169 (SERK-SigP_SERK-TMD + (GGGGS)x5-TEV-site-linker_NLS_NucA), F170+F168 (SERK-SigP_SpyCatcher_Strep-TagII-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1_IRES + Ig-Kappa-SigP_SpyTag_Nanoluc) and negative controls
Investigator: Katrin
Aim of the experiment: TTransformation of E. coli XL1-blue with ligation product F122+F169 (SERK-SigP_SERK-TMD + (GGGGS)x5-TEV-site-linker_NLS_NucA), F170+F168 (SERK-SigP_SpyCatcher_Strep-TagII-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1_IRES + Ig-Kappa-SigP_SpyTag_Nanoluc) and negative controls
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 8 µl of DNA were added to the tubes containing 150 µl of competent cells and gently mixed
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- The cell suspension were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated on a new chlorampenicol plate.
Picking of E. coli XL1 blue transformed with ligation product F150+F166 (IgKappa-SigP_NanoLuc in pSB1C3 + SpyTag), F142+F147 (Ig-Kappa-SigP_SpyCatcher + Nanoluc), F139+F141 (Ig-Kappa-SigP_Nanoluc + SpyCatcher)
Investigator: Katrin
Aim of the experiment: Picking of E. coli XL1 blue transformed with ligation product F150+F166 (IgKappa-SigP_NanoLuc in pSB1C3 + SpyTag), F142+F147 (Ig-Kappa-SigP_SpyCatcher + Nanoluc), F139+F141 (Ig-Kappa-SigP_Nanoluc + SpyCatcher)
Procedure:
- Picked colonies were transferred into cell-culture tubes with air-permeable, sterile cover. Each tube contains 4 mL of LB-medium + 4 µL chloramphenicol(1000x).
- 2 colonies were picked for F150+F166 and F139+F141, 8 colonies were picked for F142+F147 (many colonies on negatibe controls)
- These tubes were transferred in a cell culture shaker at 37 °C and were incubated at 180 rpm overday.
Sequencing of P3 (pTEF2 in pTUM100), P341 (QCI of AlcR), P453 (SERK-SigP_SpyCatcher_Strep-tag-II-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1 + IRES in pSB1C3), P462 (SERK-SigP_SpyTag_Strep-tag-II-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1 + IRES in pSB1C3), P300 (Phytotchrome B)
Investigator: Rosario
Aim of the experiment: Sequencing of P3 (pTEF2 in pTUM100), P341 (QCI of AlcR), P453 (SERK-SigP_SpyCatcher_Strep-tag-II-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1 + IRES in pSB1C3), P462 (SERK-SigP_SpyTag_Strep-tag-II-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1 + IRES in pSB1C3), P300 (Phytotchrome B).
Procedure: The DNA was prepared for sequencing according to the Eurofins SmartSeq Kit protocol (15 µl of plasmid DNA (50 - 100 ng) and 2 µl sequencing primer).
The different genes we sequenced received the following barcodes:
| Label | Barcode1 | Barcode2 | Barcode3 | Barcode4 |
|---|---|---|---|---|
| P3 | FR02233147 (O5) | |||
| P341 | FR02233146 (O3) | FR02233145 (O4) | FR02233144 (O100) | FR02233143 (O101) |
| P453 | FR02233142 (O4) | |||
| P462 | FR02233141 (O4) | |||
| P300 | FR02233140 (O3) | FR02233139 (O4) | FR02233138 (O110) | FR02233137 (O111) |
Quikchange mutagenesis of P447 (Actin promoter) to remove forbidden EcoRI restriction site
Investigator: Jeff
Aim of the experiment: Quikchange mutagenesis of P447 (Actin promoter) to remove forbidden EcoRI restriction site.
Procedure:
Reaction batch 1:
| volume | reagent |
|---|---|
| 2.5 µl | 10x Pfu Ultra II buffer |
| 1 µl | Plasmid P447 template (1:100) |
| 1.0 µl | 1:10 dilution of O106 (=10 µM) |
| 1.0 µl | 1:10 dilution of O107 (=10 µM) |
| 18.0 µl | ddH2O |
| 1 µl | dNTP mix 50 mM |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U/µl) |
Reaction batch 2:
| volume | reagent |
|---|---|
| 25 µl | Q5 Polymerase Mastermix |
| 1 µl | Plasmid P447 template (1:100) |
| 2.5 µl | 1:10 dilution of O106 (=10 µM) |
| 2.5 µl | 1:10 dilution of O107 (=10 µM) |
| 8.0 µl | ddH2O |
| 1 µl | dNTP mix 50 mM |
PCR cycling parameters for batch 1
| Cycles | Temperature | Time |
|---|---|---|
| 1 | 95 °C | 30 sec |
| 20 | 95°C | 30 sec |
| 52°C | 1 min | |
| 68°C | 4 min | |
| 1 | 4°C | HOLD |
PCR cycling parameters for batch 2
| Cycles | Temperature | Time |
|---|---|---|
| 1 | 98 °C | 30 sec |
| 20 | 98°C | 10 sec |
| 55°C | 30 sec | |
| 72°C | 2 min | |
| 3 | 4°C | HOLD |
- Digestion of the parental DNA with DpnI: Addition of 1 µl DpnI to the PCR batch and incubate for 1 h at 37 °C.
QuikChange of pYes (P3)
Investigator: Rosario
Aim of the experiment: QuikChange of pYes (P3)
Reaction batch
| volume | reagent | Additional information |
|---|---|---|
| 2.5 µl | 10x Pfu Ultra II buffer | |
| 1 µl | Plasmid template (P3), (1:10 diluted) | need to get between 2.5 ng and 25 ng for a reaction batch of 25 µl |
| 1 µl | 1:10 dilution of O5 (=10 µM) | |
| 19 µl | ddH2O | |
| 1 µl | dNTP mix 50 mM | |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U/µl) |
PCR cycling parameters
| Segment | Cycles | Temperature | Time |
|---|---|---|---|
| 1 | 1 | 95 °C | 30 s |
| 2 | 20 | 95 °C | 30 s |
| 52 °C | 1 min | ||
| 68 °C | 4 min | ||
| 3 | 4 °C | hold |
- Digestion of the parental DNA with DpnI: Addition of 1 µl DpnI to the PCR batch and incubate for 1 h at 37 °C.
Analytical gelelectrophoresis of the DpnI-digested QuikChange products of P447 (pActin in pSB1C3) and P3 (pTEF2 in pTUM100)
Investigator: Jeff
Procedure: Analytical gelelectrophoresis of the DpnI-digested QuikChange products of P447 (pActin in pSB1C3) and P3 (pTEF2 in pTUM100) to check whether QuikChange might be worked.
Procedure:
- 4.5 µl of the QuikChange products of P447 (PfuUltraII Polymerase batch and Q5 Polymerase batch (see above)) and P3 (pTEF2 in pTUM100) were mixed together 0.5 µl of DNA loading buffer (10x).
- Analytical gelelectrophoresis was performed on a 1% agarose gel at 90 V for 40 min.
| Lane: | 2 log DNA ladder | QC of P3 with O44 & O45 (PfuUltraII) | QC of P447 with O106 & O107 (PfuUltraII) | QC of P447 with O106 & O107 (Q5) |
|---|---|---|---|---|
| Result: | as expected | no product visible | no product visible |
Transformation of E. coli XL1-blue with QuikChange products of P447 (PfuUltraII and Q5) and of P3 (pTEF2 in pTUM100)
Investigator: Rosario
Aim of the experiment: Transformation of E. coli XL1-blue with QuikChange products of P447 (PfuUltraII and Q5) and of P3 (pTEF2 in pTUM100) (see above).
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 8 µl of DNA were added to the tubes containing 150 µl of competent cells and gently mixed
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- The cell suspension were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated on a new chlorampenicol plate.
Saturday, July 6th
Quikchange mutagenesis of P447 (Actin promoter) to remove forbidden EcoRI restriction site
Investigator: Le swaffeleur
Aim of the experiment: Quikchange mutagenesis of P447 (Actin promoter) to remove forbidden EcoRI restriction site.
Procedure:
Reaction batch 1:
| volume | reagent |
|---|---|
| 2.5 µl | 10x Pfu Ultra II buffer |
| 4 µl | Plasmid P447 template (1:100) |
| 1.0 µl | 1:10 dilution of O106 (=10 µM) |
| 16.0 µl | ddH2O |
| 1 µl | dNTP mix 50 mM |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U/µl) |
| =20 µl | TOTAL |
Reaction batch 2:
| volume | reagent |
|---|---|
| 2.5 µl | 10x Pfu Ultra II buffer |
| 4 µl | Plasmid P447 template (1:100) |
| 1.0 µl | 1:10 dilution of O107 (=10 µM) |
| 16.0 µl | ddH2O |
| 1 µl | dNTP mix 50 mM |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U/µl) |
| =20 µl | TOTAL |
- The two batches were cycled seperately for the first 10 rounds.
PCR cycling parameters
| Cycles | Temperature | Time |
|---|---|---|
| 1 | 95 °C | 30 sec |
| 10 | 95°C | 30 sec |
| 54°C | 1 min | |
| 68°C | 4 min | |
| 1 | 4°C | HOLD |
- After the ten cycles the two reaction batches were pooled together and were additionally cycled for 20 rounds with the following conditions:
PCR cycling parameters after poooling the reaction batches together:
| Cycles | Temperature | Time |
|---|---|---|
| 1 | 98 °C | 30 sec |
| 20 | 98°C | 10 sec |
| 54°C | 30 sec | |
| 72°C | 2 min | |
| 3 | 4°C | HOLD |
- Digestion of the parental DNA with DpnI: Addition of 1 µl DpnI to the PCR batch and incubate for 1 h at 37 °C.
Analytical gelelectrophoresis of QuikChange products of P447 (pActin in pSB1C3)
Investigator: Jeff
Aim of the experiment: Analytical gelelectrophoresis of QuikChange products of P447 (pActin in pSB1C3).
Procedure:
- 9 µl of the QuikChange products were mixed with 1 µl of DNA loading buffer (10x).
- Analytical gelelectrophoresis was performed at 90 V for 30 min on a 1% agarose gel.
| Lane: | 2 log DNA ladder | QC of P447 with O106 & O107 (PfuUltraII, without DMSO) | QC of P447 with O106 & O107 (PfuUltraII, with DMSO) |
|---|---|---|---|
| Result: | as expected | as expected |
Transformation of E. coli XL1-blue with QuikChange products of P447 (pActin in pSB1C3)
Investigator: Jeff, Le Swaffeleur
Aim of the experiment: Transformation of E. coli XL1-blue with QuikChange products of P447 (pActin in pSB1C3).
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 8 µl of DNA were added to the tubes containing 150 µl of competent cells and gently mixed
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- The cell suspension were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated on a new chlorampenicol plate.
Picking of E. coli XL1-blue transformed with ligation product F122+F169 (SERK-SigP_SERK-TMD + (GGGGS)x5-TEV-site-linker_NLS_NucA) and F170+F168 (SERK-SigP_SpyCatcher_Strep-TagII-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1_IRES + Ig-Kappa-SigP_SpyTag_NanoLuc)
Investigator: Jeff
Aim of the experiment: Picking of E. coli XL1-blue transformed with ligation product F122+F169 (SERK-SigP_SERK-TMD + (GGGGS)x5-TEV-site-linker_NLS_NucA) and F170+F168 (SERK-SigP_SpyCatcher_Strep-TagII-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1_IRES + Ig-Kappa-SigP_SpyTag_NanoLuc).
Procedure:
- Picked colonies were transferred into cell-culture tubes with air-permeable, sterile cover. Each tube contains 4 mL of LB-medium + 4 µL chloramphenicol(1000x).
- 2 colonies were picked for F122+F169 and F170+F168.
- These tubes were transferred in a cell culture shaker at 37 °C and were incubated at 180 rpm overnight.
Sunday, July 7th
Miniprep of E. coli XL1 blue transformed with F170+F168(SERK-SigP_SpyCatcher_Strep-TagII-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1_IRES + Ig-Kappa-SigP_SpyTag_NanoLuc)
Investigator: Jeff, Katrin
Aim of the experiment: Miniprep of E. coli XL1 blue transformed with F170+F168(SERK-SigP_SpyCatcher_Strep-TagII-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1_IRES + Ig-Kappa-SigP_SpyTag_NanoLuc).
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN).
- The resulting tubes were labeled as P475 and P476.
Analytical digestion and gelelectrophoresis of P464 – P476 and P402 (control)
Investigator: Jeff, Katrin
Aim of the experiment: Analytical digestion and gelelectrophoresis of P464 – P476 and P402 (control)
- 16x Mastermix (EcoRI-HF & PstI-HF) for analytical digestion:
| volume | reagent |
|---|---|
| 17.5 µl | Mastermix for EcoRI & PstI |
| 2.5 µl | Plasmid DNA (P464 – P476, P402) |
| =20 µl | TOTAL |
- Analytical digestion was performed at 37 °C for 1.5 h.
- After incubation 2 µl of DNA loading buffer (10x) were added to the reaction batches.
- Analytical gelelectrophoresis was performed at 90 V for 1 h.
| Lane: | 1000 bp DNA ladder | Digestion of P464 with EcoRI & PstI | Digestion of P465 with EcoRI & PstI | Digestion of P466 with EcoRI & PstI | Digestion of P467 with EcoRI & PstI | Digestion of P402 with EcoRI & PstI (control) | Digestion of P475 with EcoRI & PstI | Digestion of P476 with EcoRI & PstI |
|---|---|---|---|---|---|---|---|---|
| Result: | as expected | as expected | as expected | as expected | as expected | as expected | as expected |
| Lane: | 1000 bp DNA ladder | Digestion of P468 with EcoRI & PstI | Digestion of P469 with EcoRI & PstI | Digestion of P470 with EcoRI & PstI | Digestion of P471 with EcoRI & PstI | Digestion of P472 with EcoRI & PstI | Digestion of P473 with EcoRI & PstI | Digestion of P474 with EcoRI & PstI |
|---|---|---|---|---|---|---|---|---|
| Result: | corrupt | corrupt | corrupt | corrupt | corrupt | corrupt | corrupt |
PCR of P378 (Proteinphosphotase 1)
Investigator: Katrin, Jeff
Aim of the experiment: PCR of P378 (Proteinphosphotase 1).
Procedure:
Reaction batch for PCR with Q5 Mastermix for P378 (Proteinphosphotase 1):
| volume | reagent |
|---|---|
| 25 µl | 2x Q5 Master Mix |
| 1 µl | dNTP mix (normally not necessary!) |
| 2.5 µl | 10 µM Forward Primer O112 (PP1 for) |
| 2.5 µl | 10 µM Reverse Primer O113 (PP1 rev) |
| 1 µl | Plasmid DNA (P378) - 1:1000 dilution (desired c between 1 pg and 1 ng) |
| 18 µL | ddH2O Water |
| =50 µL | TOTAL |
- The content has been mixed with a pipette
- The PCR program was performed after following scheme for P378(PP1 amplification):
Cycling parameters:
| Initial denaturation | 98 °C | 30 s |
|---|---|---|
| 30 cycles | 98 °C | 10 s |
| 62 °C | 30 s | |
| 72 °C | 30 s | |
| Final extension | 72 °C | 2 min |
| Hold | 4 °C | infinite |
- After PCR, the product was purified with QIAquick PCR Purification Kit, Qiagen.
- The resulting product was labeled as F174.
Analytical gelelectrophoresis of F174
Investigator: Jeff
Aim of the experiment: Analytical gelelectrophoresis of F174.
Procedure:
- 9 µl of the PCR product F174 (=PCR of P378 with O112/O113) products were mixed with 1 µl of DNA loading buffer (10x).
- Analytical gelelectrophoresis was performed at 90 V for 60 min on a 1% agarose gel.
| Lane: | 2 log DNA ladder | F174 |
|---|---|---|
| Result: | as expected |
Preparative digestion and gelelectrophoresis of P422 (FluA triple mutant), P464 (IgKappa-SigP_NanoLuc_SpyCatcher) and P466 (IgKappa-SigP_NanoLuc_SpyTag)
Investigator: Katrin, Jeff
Aim of the experiment: Preparative digestion and gelelectrophoresis of P422 (FluA triple mutant), P464 (IgKappa-SigP_NanoLuc_SpyCatcher) and P466 (IgKappa-SigP_NanoLuc_SpyTag).
Procedure:
Reaction batch for P422 (FluA triple mutant):
| volume | reagent |
|---|---|
| 20 µl | P422 |
| 4 µl | CutSmart Buffer (10x) |
| 1 µl | NgoMIV |
| 1 µl | SpeI |
| 14 µl | ddH2O |
| =40 µl | TOTAL |
Reaction batch for P464 (IgKappa-SigP_NanoLuc_SpyCatcher):
| volume | reagent |
|---|---|
| 20 µl | P464 |
| 4 µl | CutSmart Buffer (10x) |
| 1 µl | SpeI |
| 1 µl | PstI-HF |
| 14 µl | ddH2O |
| =40 µl | TOTAL |
Reaction batch for P466 (IgKappa-SigP_NanoLuc_SpyTag):
| volume | reagent |
|---|---|
| 20 µl | P466 |
| 4 µl | CutSmart Buffer (10x) |
| 1 µl | SpeI |
| 1 µl | PstI-HF |
| 14 µl | ddH2O |
| =40 µl | TOTAL |
- Reaction batches were incubated at 37 °C for 2.5 h.
- Preparative gelelectrophoresis was performed at 90 V for 1 h in an 1% agarose gel.
| Lane | Digestion of P422 with NgoMIV & SpeI | 2 log DNA ladder | Digestion of P464 with XbaI & PstI | Digestion of P466 with XbaI & PstI |
|---|---|---|---|---|
| Results | as expected; lower band was cut out | as expected; lower band was cut out | as expected; lower band was cut out |
- DNA bands were extracted with QIAquick Gel Extraction Kit, QIAGEN.
Preparative digestion of P379 (IgKappa-SigP_SpyCatcher) and F174 (Proteinphosphotase 1)
Investigator: Katrin, Jeff
Aim of the experiment: Preparative digestion of P379 (IgKappa-SigP_SpyCatcher) and F174 (Proteinphosphotase 1).
Procedure:
Reaction batch for P379 (IgKappa-SigP_SpyCatcher):
| volume | reagent |
|---|---|
| 20 µl | P379 |
| 4 µl | CutSmart Buffer (10x) |
| 1 µl | AgeI-HF |
| 1 µl | PstI-HF |
| 14 µl | ddH2O |
| =40 µl | TOTAL |
Reaction batch for F174 (Proteinphosphotase 1):
| volume | reagent |
|---|---|
| 25 µl | F174 |
| 5 µl | CutSmart Buffer (10x) |
| 1 µl | AgeI-HF |
| 1 µl | PstI-HF |
| 18 µl | ddH2O |
| =50 µl | TOTAL |
- Reaction batches were incubated at 37 °C for 4 h.
- After digestion, the product was purified with QIAquick PCR Purification Kit, Qiagen.
Ligation of F170+F179 (SERK-SigP_SpyCatcher_StrepTagII-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1_IRES in pSB1C3 + IgKappa-SigP_NanoLuc_SpyTag), F171+F178 (SERK-SigP_SpyTag_StrepTagII-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1_IRES in pSB1C3 + IgKappa-SigP_NanoLuc_SpyCatcher), F176+F147 (IgKappa-SigP_SpyCatcher in pSB1C3 + NanoLuc), F109+F177 (SERK-SigP in pSB1C3 + FluA triple mutant) and F123+F175 (pSB1C3 only + PP1)
Investigator: Jeff
Aim of the experiment: Ligation of F170+F179 (SERK-SigP_SpyCatcher_StrepTagII-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1_IRES in pSB1C3 + IgKappa-SigP_NanoLuc_SpyTag), F171+F178 (SERK-SigP_SpyTag_StrepTagII-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1_IRES in pSB1C3 + IgKappa-SigP_NanoLuc_SpyCatcher), F176+F147 (IgKappa-SigP_SpyCatcher in pSB1C3 + NanoLuc), F109+F177 (SERK-SigP in pSB1C3 + FluA triple mutant) and F123+F175 (pSB1C3 only + PP1).
Procedure:
Ligation batch for F170+F179 (SERK-SigP_SpyCatcher_StrepTagII-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1_IRES
in pSB1C3 + IgKappa-SigP_NanoLuc_SpyTag), vector:insert = 1:3:
| volume | reagent |
|---|---|
| 2.01 µl | F170 (49.7 ng/µl, 4168 bp) |
| 3.50 µl | F179 (12.9 ng/µl, 628 bp) |
| 11.49 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
Ligation batch for F171+F178 (SERK-SigP_SpyTag_StrepTagII-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1_IRES in pSB1C3 + IgKappa-SigP_NanoLuc_SpyCatcher), vector:insert = 1:3:
| volume | reagent |
|---|---|
| 1.65 µl | F171 (60.05 ng/µl, 3868 bp) |
| 5.80 µl | F178 (12.4 ng/µl, 928 bp) |
| 9.55 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
Ligation batch for F176+F147 (IgKappa-SigP_SpyCatcher in pSB1C3 + NanoLuc), vector:insert = 1:3:
| volume | reagent |
|---|---|
| 0.61 µl | F176 (163.9 ng/µl, 2489 bp) |
| 3.51 µl | F147 (17.5 ng/µl, 510 bp) |
| 12.88 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
Ligation batch for F109+F177 (SERK-SigP in pSB1C3 + FluA triple mutant), vector:insert = 1:3:
| volume | reagent |
|---|---|
| 0.92 µl | F109 (108.2 ng/µl, 2117 bp) |
| 2.90 µl | F177 (25.5 ng/µl, 522 bp) |
| 13.18 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
Ligation batch for F123+F175 (pSB1C3 only + PP1), vector:insert = 1:3:
| volume | reagent |
|---|---|
| 0.92 µl | F123 (49.3 ng/µl, 2086 bp) |
| 2.90 µl | F175 (12.3 ng/µl, 972 bp) |
| 3.59 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- Ligation was performed at 22 °C for 1 h
- Negative controls were also prepared with water instead of insert.
Transformation of E. coli XL1-blue with ligation products F170+F179 (SERK-SigP_SpyCatcher_StrepTagII-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1_IRES in pSB1C3 + IgKappa-SigP_NanoLuc_SpyTag), F171+F178 (SERK-SigP_SpyTag_StrepTagII-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1_IRES in pSB1C3 + IgKappa-SigP_NanoLuc_SpyCatcher), F176+F147 (IgKappa-SigP_SpyCatcher in pSB1C3 + NanoLuc), F109+F177 (SERK-SigP in pSB1C3 + FluA triple mutant) and F123+F175 (pSB1C3 only + PP1)
Investigator: Jeff
Aim of the experiment: Transformation of E. coli XL1-blue with ligation products F170+F179 (SERK-SigP_SpyCatcher_StrepTagII-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1_IRES in pSB1C3 + IgKappa-SigP_NanoLuc_SpyTag), F171+F178 (SERK-SigP_SpyTag_StrepTagII-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1_IRES in pSB1C3 + IgKappa-SigP_NanoLuc_SpyCatcher), F176+F147 (IgKappa-SigP_SpyCatcher in pSB1C3 + NanoLuc), F109+F177 (SERK-SigP in pSB1C3 + FluA triple mutant) and F123+F175 (pSB1C3 only + PP1).
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 10 µl of DNA were added to the tubes containing 150 µl of competent cells and gently mixed
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- The cell suspension were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated on new chlorampenicol plates.
Picking of E. coli XL1 blue transformed with QuikChange products of P447 (pActin) and ligation product F123+F152 ((GGGGS)x5-TEV-cleavage-site-linker)
Investigator: Jeff
Aim of the experiment: Picking of E. coli XL1 blue transformed with QuikChange products of P447 (pActin) and ligation product F123+F152 ((GGGGS)x5-TEV-cleavage-site-linker).
Procedure:
- Picked colonies were transferred into cell-culture tubes with air-permeable, sterile cover. Each tube contains 4 mL of LB-medium + 4 µL chloramphenicol (1000x).
- 8 colonies were picked for QuikChange products of P447 (pActin)and F139+F141 and 4 colonies were picked for ligation product F123+F152 ((GGGGS)x5-TEV-cleavage-site-linker).
- These tubes were transferred in a cell culture shaker at 37 °C and were incubated at 180 rpm overnight.
Week 12
Monday, July 8th
Analytical digestion and gelelectrophoresis of P477-P484 (pActin)
Investigator: Rosario, Ingmar
Aim of the experiment: Analytical digestion and gelelectrophoresis of P477-P484 (pActin in pSB1C3) after QC of forbidden EcoRI Restriction site.
- 10x Mastermix (EcoRI-HF & PstI-HF) for analytical digestion:
| volume | reagent |
|---|---|
| 15 µl | Cut Smart buffer |
| 2 µl | EcoRI |
| 2 µl | PstI |
| 30 µl | ddH2O |
| =49 µl | TOTAL |
- The volume of plasmid DNA added was calculated in order to have equal amounts of DNA of approximately 550 ng
- Analytical digestion was performed at 37 °C for 1.5 h.
- After incubation 2 µl of DNA loading buffer (10x) were added to the reaction batches.
- Analytical gelelectrophoresis was performed at 90 V for 1 h.
| Lane: | 1000 bp DNA ladder | Digestion of P477 with EcoRI & PstI | Digestion of P478 with EcoRI & PstI | Digestion of P479 with EcoRI & PstI | Digestion of P480 with EcoRI & PstI | Digestion of P481 with EcoRI & PstI (control) | Digestion of P482 with EcoRI & PstI | Digestion of P483 with EcoRI & PstI | Digestion of P484 with EcoRI & PstI |
|---|---|---|---|---|---|---|---|---|---|
| Result: | as expected | as expected | as expected | as expected | as expected | as expected | as expected | as expected |
- all quickchanges were successfull.
Quikchange mutagenesis of P477 (Actin promoter) to insert 50 bp deletion
Investigator: Rosario, Ingmar
Aim of the experiment: Quikchange mutagenesis of P477 (Actin promoter) to insert 50 bp deletion.
Procedure:
Reaction batch 1:
| volume | reagent |
|---|---|
| 2.5 µl | 10x Pfu Ultra II buffer |
| 2,67 µl | Plasmid P477 template (1:100) |
| 1.0 µl | 1:10 dilution of O108 (=10 µM) |
| 17.330 µl | ddH2O |
| 1 µl | dNTP mix 50 mM |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U/µl) |
| =25 µl | TOTAL |
Reaction batch 2:
| volume | reagent |
|---|---|
| 2.5 µl | 10x Pfu Ultra II buffer |
| 2.67 µl | Plasmid P477 template (1:100) |
| 1.0 µl | 1:10 dilution of O109 (=10 µM) |
| 17.33 µl | ddH2O |
| 1 µl | dNTP mix 50 mM |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U/µl) |
| =20 µl | TOTAL |
- The two batches were cycled seperately for the first 10 rounds.
PCR cycling parameters
| Cycles | Temperature | Time |
|---|---|---|
| 1 | 95 °C | 30 sec |
| 10 | 95°C | 30 sec |
| 54°C | 1 min | |
| 72°C | 4.5 min | |
| 1 | 4°C | HOLD |
- After the ten cycles the two reaction batches were pooled together and were additionally cycled for 20 rounds with the following conditions:
PCR cycling parameters after poooling the reaction batches together:
| Cycles | Temperature | Time |
|---|---|---|
| 1 | 98 °C | 30 sec |
| 20 | 98°C | 10 sec |
| 54°C | 30 sec | |
| 72°C | 2 min | |
| 3 | 4°C | HOLD |
- Digestion of the parental DNA with DpnI: Addition of 1 µl DpnI to the PCR batch and incubate for 1 h at 37 °C.
Sequencing of P477, P486, P487, P464, P466, P476, P435
Investigator: Rosario
Aim of the experiment: Sequencing of P477 (Actin promoter), P486, P487, P464, P466, P476, P435.
Procedure: The DNA was prepared for sequencing according to the Eurofins SmartSeq Kit protocol (15 µl of plasmid DNA (50 - 100 ng) and 2 µl sequencing primer).
The different genes we sequenced received the following barcodes:
| Label | Barcode1 | Barcode2 |
|---|---|---|
| P477 | FR02233136 (O3) | FR02233135 (O4) |
| P486 | FR02233134 (O3) | |
| P487 | FR02233133 (O3) | |
| P464 | FR02233132 (O4) | |
| P466 | FR02233131 (O4) | |
| P476 | FR02233130 (O3) | FR02233129 (O4) |
| P435 | FR02233128 (O3) |
Preparative digestion and gelelectrophoresis of P187 (LMU GFP)
Investigator: Rosario
Aim of the experiment: Preparative digestion and gelelectrophoresis of P187 (LMU GFP).
Procedure:
Reaction batch for P187 :
| volume | reagent |
|---|---|
| 20 µl | P187 |
| 4 µl | CutSmart Buffer (10x) |
| 1 µl | NgoMIV |
| 1 µl | SpeI |
| 14 µl | ddH2O |
| =40 µl | TOTAL |
- Reaction batch was incubated at 37 °C for 2.5 h.
- Preparative gelelectrophoresis was performed at 90 V for 1 h in an 1% agarose gel.
| Lane | Digestion of P187 with NgoMIV & SpeI | 2 log DNA ladder |
|---|---|---|
| Results | as expected; lower band was cut out |
- DNA bands were extracted with QIAquick Gel Extraction Kit, QIAGEN.
Ligation of F109+F180 (SERK-SigP in pSB1C3 + LMU GFP) and F92+F180 (IgKappa in pSB1C3 + LMU GFP)
Investigator: Rosario
Aim of the experiment: Ligation of F109+F180 (SERK-SigP in pSB1C3 + LMU GFP) and F92+F180 (IgKappa in pSB1C3 + LMU GFP).
Procedure:
Ligation batch for F109+F180 (SERK-SigP in pSB1C3 + LMU GFP), vector:insert = 1:3:
| volume | reagent |
|---|---|
| 4.1 µl | F180 (726 bp) |
| 0.9 µl | F179 (2177 bp) |
| 12 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
F92+F180 (IgKappa in pSB1C3 + LMU GFP), vector:insert = 1:3:
| volume | reagent |
|---|---|
| 4.15 µl | F180 (726 bp) |
| 0.85 µl | F92 (2144 bp) |
| 12 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- Ligation was performed at 22 °C for 1 h
- Negative controls were also prepared with water instead of insert.
Picking of E. coli XL1-blue transformed with ligation products F170+F179 (SERK-SigP_SpyCatcher_StrepTagII-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1_IRES in pSB1C3 + IgKappa-SigP_NanoLuc_SpyTag), F171+F178 (SERK-SigP_SpyTag_StrepTagII-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1_IRES in pSB1C3 + IgKappa-SigP_NanoLuc_SpyCatcher), F176+F147 (IgKappa-SigP_SpyCatcher in pSB1C3 + NanoLuc), F109+F177 (SERK-SigP in pSB1C3 + FluA triple mutant) and F123+F175 (pSB1C3 only + PP1))
Investigator: Rosario
Aim of the experiment: Picking of E. coli XL1-blue transformed with ligation products F170+F179 (SERK-SigP_SpyCatcher_StrepTagII-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1_IRES in pSB1C3 + IgKappa-SigP_NanoLuc_SpyTag), F171+F178 (SERK-SigP_SpyTag_StrepTagII-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1_IRES in pSB1C3 + IgKappa-SigP_NanoLuc_SpyCatcher), F176+F147 (IgKappa-SigP_SpyCatcher in pSB1C3 + NanoLuc), F109+F177 (SERK-SigP in pSB1C3 + FluA triple mutant) and F123+F175 (pSB1C3 only + PP1)).
Procedure:
- Picked colonies were transferred into cell-culture tubes with air-permeable, sterile cover. Each tube contains 4 mL of LB-medium + 4 µL chloramphenicol(1000x).
- 2 colonies were picked for all ligation products except F176+F147 for which 4 colonies were picked.
- These tubes were transferred in a cell culture shaker at 37 °C and were incubated at 180 rpm overnight.
Tuesday, July 9th
Miniprep of E. coli XL1 blue transformed with F176+F147 (IgKappa-SigP_SpyCatcher in pSB1C3 + NanoLuc), F123+F175 (pSB1C3 only + PP1), F109+F177 (SERK-SigP in pSB1C3 + FluA triple mutant), F171+F178 (SERK-SigP_SpyTag_StrepTagII-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1_IRES in pSB1C3 + IgKappa-SigP_NanoLuc_SpyCatcher) and F170+F179 (SERK-SigP_SpyCatcher_StrepTagII-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1_IRES in pSB1C3 + IgKappa-SigP_NanoLuc_SpyTag)
Investigator: Jeff, Florian
Aim of the experiment: Miniprep of E. coli XL1 blue transformed with F176+F147 (IgKappa-SigP_SpyCatcher in pSB1C3 + NanoLuc), F123+F175 (pSB1C3 only + PP1), F109+F177 (SERK-SigP in pSB1C3 + FluA triple mutant), F171+F178 (SERK-SigP_SpyTag_StrepTagII-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1_IRES in pSB1C3 + IgKappa-SigP_NanoLuc_SpyCatcher) and F170+F179 (SERK-SigP_SpyCatcher_StrepTagII-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1_IRES in pSB1C3 + IgKappa-SigP_NanoLuc_SpyTag).
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN).
- The resulting tubes were labeled as P488 to P499.
Analytical digestion and gelelectrophoresis of P488-P499 (several ligations)
Investigator: Florian
Aim of the experiment: Analytical digestion and gelelectrophoresis of P488-P499 (several ligations).
- 15x Mastermix (EcoRI-HF & PstI-HF) for analytical digestion:
| volume | reagent |
|---|---|
| 30 µl | Cut Smart buffer |
| 3.75 µl | EcoRI |
| 3.75 µl | PstI |
| 225 µl | ddH2O |
| =262,5 µl | TOTAL |
- 17.5 µl master mix and 2.5 µl plasmid DNA were mixed.
- Analytical digestion was performed at 37 °C for 1 h.
- After incubation 2 µl of DNA loading buffer (10x) were added to the reaction batches.
- Analytical gelelectrophoresis was performed at 90 V for 1 h.
| Lane: | 1 kb DNA ladder | Digestion of P488 F176+F147 (IgKappa-SigP_SpyCatcher in pSB1C3 + NanoLuc) with EcoRI & PstI | Digestion of P489 F176+F147 (IgKappa-SigP_SpyCatcher in pSB1C3 + NanoLuc) with EcoRI & PstI | Digestion of P490 F176+F147 (IgKappa-SigP_SpyCatcher in pSB1C3 + NanoLuc) with EcoRI & PstI | Digestion of P491 F176+F147 (IgKappa-SigP_SpyCatcher in pSB1C3 + NanoLuc) with EcoRI & PstI | Digestion of P492 F123+F175 (pSB1C3 only + PP1) with EcoRI & PstI | Digestion of P493 F123+F175 (pSB1C3 only + PP1) with EcoRI & PstI |
|---|---|---|---|---|---|---|---|
| Result: | as expected (950 bp) | as expected (950 bp) | as expected (950 bp) | as expected (950 bp) | as expected (972 bp) | as expected (972 bp) |
| Lane: | 1 kb DNA ladder | Digestion of P422 F123+F161 (FluA triple mutant + pSB1C3) with EcoRI & PstI (control) | Digestion of P494 F109+F177 (SERK-SigP in pSB1C3 + FluA triple mutant) with EcoRI & PstI | Digestion of P495 F109+F177 (SERK-SigP in pSB1C3 + FluA triple mutant) with EcoRI & PstI | Digestion of P496 F171+F178 (SERK-SigP_SpyTag_StrepTagII-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1_IRES in pSB1C3 + IgKappa-SigP_NanoLuc_SpyCatcher) with EcoRI & PstI | Digestion of P497 F171+F178 (SERK-SigP_SpyTag_StrepTagII-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1_IRES in pSB1C3 + IgKappa-SigP_NanoLuc_SpyCatcher) with EcoRI & PstI | Digestion of P498 F170+F179 (SERK-SigP_SpyCatcher_StrepTagII-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1_IRES in pSB1C3 + IgKappa-SigP_NanoLuc_SpyTag) with EcoRI & PstI | Digestion of P499 F170+F179 (SERK-SigP_SpyCatcher_StrepTagII-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1_IRES in pSB1C3 + IgKappa-SigP_NanoLuc_SpyTag) with EcoRI & PstI |
|---|---|---|---|---|---|---|---|---|
| Result: | as expected (522 bp) | strange | as expected (622 bp) | roughly as expected (2768 bp) | roughly as expected (2768 bp) | roughly as expected (2768 bp) | roughly as expected (2768 bp) |
Analytical gelelectrophoresis of QuikChange products of P477 (Actin promoter) to insert 50 bp deletion
Investigator: Florian
Aim of the experiment: Analytical gelelectrophoresis of QuikChange products of P477 (Actin promoter) to insert 50 bp deletion.
Procedure:
- 9 µl of the QuikChange products were mixed with 1 µl of DNA loading buffer (10x).
- Analytical gelelectrophoresis was performed at 90 V for 60 min on a 1% agarose gel.
| Lane: | 1 kb DNA ladder | QC of P477 |
|---|---|---|
| Result: | no product |
Quikchange mutagenesis of P477 (Actin promoter) to insert 60 bp deletion, second attempt
Investigator: Jeff
Aim of the experiment: Quikchange mutagenesis of P477 (Actin promoter) to insert 60 bp deletion.
Procedure:
Reaction batch 1:
| volume | reagent |
|---|---|
| 2.5 µl | 10x Pfu Ultra II buffer |
| 3 µl | Plasmid P477 template (1:100) |
| 1.0 µl | 1:10 dilution of O108 (=10 µM) |
| 17.0 µl | ddH2O |
| 1 µl | dNTP mix 50 mM |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U/µl) |
| =25 µl | TOTAL |
Reaction batch 2:
| volume | reagent |
|---|---|
| 2.5 µl | 10x Pfu Ultra II buffer |
| 3.0 µl | Plasmid P477 template (1:100) |
| 1.0 µl | 1:10 dilution of O109 (=10 µM) |
| 17.0 µl | ddH2O |
| 1 µl | dNTP mix 50 mM |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U/µl) |
| =20 µl | TOTAL |
- The two batches were cycled seperately for the first 25 rounds.
PCR cycling parameters
| Cycles | Temperature | Time |
|---|---|---|
| 1 | 95 °C | 30 sec |
| 10 | 95°C | 30 sec |
| 52°C | 1 min | |
| 72°C | 4.5 min | |
| 1 | 4°C | HOLD |
- After the ten cycles the two reaction batches were pooled together, 0.5 µl of polymerase and 1 µl of dNTP-mix was added and the reaction was cycled for 25 rounds with the following conditions:
PCR cycling parameters after poooling the reaction batches together:
| Cycles | Temperature | Time |
|---|---|---|
| 1 | 98 °C | 30 sec |
| 20 | 98°C | 10 sec |
| 52°C | 30 sec | |
| 72°C | 2 min | |
| 3 | 4°C | HOLD |
- Digestion of the parental DNA with DpnI: Addition of 1 µl DpnI to the PCR batch and incubate for 1 h at 37 °C.
Wednesday, July 10th
Preparative digestion of P495 (SERK-SigP_FluA)
Investigator: Jeff
Aim of the experiment: Preparative digestion of P495 (SERK-SigP_FluA).
Procedure:
Reaction batch for P495 (SERK-SigP_FluA):
| volume | reagent |
|---|---|
| 20 µl | P495 |
| 4 µl | CutSmart Buffer (10x) |
| 1 µl | AgeI-HF (20 U/µl) |
| 1 µl | PstI-HF (20 U/µl) |
| 14 µl | ddH2O |
| =40 µl | TOTAL |
- After digestion, linearized DNA was purified with QIAquick PCR purification kit, QIAGEN.
Preparative digestion and gelelectrophoresis of P486 ((GGGGS)x5-TEV-site-linker), P488 (IgKappa-SigP_SpyCatcher_NanoLuc) and P492 (Proteinphosphotase 1)
Investigator: Jeff
Aim of the experiment: Preparative digestion and gelelectrophoresis of P486 ((GGGGS)x5-TEV-site-linker), P488 (IgKappa-SigP_SpyCatcher_NanoLuc) and P492 (Proteinphosphotase 1).
Procedure:
Reaction batch for P486 ((GGGGS)x5-TEV-site-linker):
| volume | reagent |
|---|---|
| 20 µl | P486 |
| 4 µl | CutSmart Buffer (10x) |
| 1 µl | XbaI (20 U/µl) |
| 1 µl | AgeI-HF (20 U/µl) |
| 14 µl | ddH2O |
| =40 µl | TOTAL |
Reaction batch for P488 (IgKappa-SigP_SpyCatcher_NanoLuc):
| volume | reagent |
|---|---|
| 20 µl | P488 |
| 4 µl | CutSmart Buffer (10x) |
| 1 µl | XbaI (20 U/µl) |
| 1 µl | PstI-HF (20 U/µl) |
| 14 µl | ddH2O |
| =40 µl | TOTAL |
Reaction batch for P492 (Proteinphosphotase 1)):
| volume | reagent |
|---|---|
| 20 µl | P492 |
| 4 µl | CutSmart Buffer (10x) |
| 2 µl | NgoMIV (10 U/µl) |
| 2 µl | SpeI (10 U/µl) |
| 12 µl | ddH2O |
| =40 µl | TOTAL |
- Reaction batches were incubated at 37 °C for 2.5 h.
- Preparative gelelectrophoresis was performed at 90 V for 1 h in an 1% agarose gel.
| Lane | Digestion of P486 with XbaI & AgeI | 2 log DNA ladder | Digestion of P488 with XbaI & PstI | Digestion of P492 with NgoMIV & SpeI |
|---|---|---|---|---|
| Results | as expected; lower band was cut out | as expected; lower band was cut out | as expected; lower band was cut out |
- DNA bands were extracted with QIAquick Gel Extraction Kit, QIAGEN.
Ligation of F156+F182 (NLS_NucA in pSB1C3 + (GGGGS)x5-TEV-site-linker), F181+F135 (SERK-SigP_FluA in pSB1C3 + Strep-tag-II-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1), F171+F183 (SERK-SigP_SpyTag_Strep-tag-II-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1_IRES in pSB1C3 + IgKappa-SigP_SpyCatcher_NanoLuc), F109+F184 (SERK-SigP in pSB1C3 + Proteinphosphotase 1)
Investigator: Jeff
Aim of the experiment: Ligation of F156+F182 (NLS_NucA in pSB1C3 + (GGGGS)x5-TEV-site-linker), F181+F135 (SERK-SigP_FluA in pSB1C3 + Strep-tag-II-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1), F171+F183 (SERK-SigP_SpyTag_Strep-tag-II-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1_IRES in pSB1C3 + IgKappa-SigP_SpyCatcher_NanoLuc), F109+F184 (SERK-SigP in pSB1C3 + Proteinphosphotase 1).
Procedure:
Ligation batch for F156+F182 (NLS_NucA in pSB1C3 + (GGGGS)x5-TEV-site-linker), vector:insert = 1:3:
| volume | reagent |
|---|---|
| 0.63 µl | F156 (158.3 ng/µl, 2596 bp) |
| 2.88 µl | F182 (4.1 ng/µl, 102 bp) |
| 13.49 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
Ligation batch for F181+F135 (SERK-SigP_FluA in pSB1C3 + Strep-tag-II-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1), vector:insert = 1:3:
| volume | reagent |
|---|---|
| 0.54 µl | F181 (185.7 ng/µl, 2705 bp) |
| 6.21 µl | F135 (17.8 ng/µl, 996 bp) |
| 10.25 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
F171+F183 (SERK-SigP_SpyTag_Strep-tag-II-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1_IRES in pSB1C3 + IgKappa-SigP_SpyCatcher_NanoLuc), vector:insert = 1:3:
| volume | reagent |
|---|---|
| 1.65 µl | F171 (60.5 ng/µl, 3859 bp) |
| 1.22 µl | F183 (59 ng/µl, 928 bp) |
| 14.13 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
Ligation batch for F109+F184 (SERK-SigP in pSB1C3 + Proteinphosphotase 1), vector:insert = 1:3:
| volume | reagent |
|---|---|
| 0.92 µl | F109 (108.2 ng/µl, 2117 bp) |
| 2.44 µl | F184 (54.9 ng/µl, 972 bp) |
| 13.64 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- Ligation was performed at 16 °C overnight.
- Negative controls were also prepared with water instead of insert.
Thursday, July 11th
Transformation of E. coli XL1-Blue with ligation products F156+F182 (NLS_NucA in pSB1C3 + (GGGGS)x5-TEV-site-linker), F181+F135 (SERK-SigP_FluA in pSB1C3 + Strep-tag-II-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1), F171+F183 (SERK-SigP_SpyTag_Strep-tag-II-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1_IRES in pSB1C3 + IgKappa-SigP_SpyCatcher_NanoLuc), F109+F184 (SERK-SigP in pSB1C3 + Proteinphosphotase 1)
Investigator: Jeff
Aim of the experiment: Transformation of E. coli XL1-Blue with ligation products F156+F182 (NLS_NucA in pSB1C3 + (GGGGS)x5-TEV-site-linker), F181+F135 (SERK-SigP_FluA in pSB1C3 + Strep-tag-II-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1), F171+F183 (SERK-SigP_SpyTag_Strep-tag-II-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1_IRES in pSB1C3 + IgKappa-SigP_SpyCatcher_NanoLuc), F109+F184 (SERK-SigP in pSB1C3 + Proteinphosphotase 1).
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 10 µl of DNA were added to the tubes containing 100 µl of competent cells and gently mixed
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- The cell suspension were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated on new chlorampenicol plates.
Sequencing of P294, P296, P342, P488, P492, P495, P496 and P498
Investigator: Jeff
Aim of the experiment: Sequencing of P294, P296, P342, P488, P492, P495, P496 and P498.
Procedure:
- The DNA was prepared for sequencing according to the Eurofins SmartSeq Kit protocol (15 µl of plasmid DNA (50 - 100 ng) and 2 µl sequencing primer).
The different genes we sequenced received the following barcodes:
| Label | Barcode1 | Barcode2 | Barcode3 |
|---|---|---|---|
| P294 | FR02233072 (O100) | ||
| P296 | FR02233073 (O3) | FR02233074 (O4) | FR02233075 (O100) |
| P342 | FR02233076 (O3) | FR02233077 (O4) | FR02233078 (O100) |
| P488 | FR02233079 (O4) | ||
| P492 | FR02233080 (O3) | FR02233081 (O4) | |
| P495 | FR02233082 (O3) | ||
| P496 | FR02233083 (O3) | FR02233084 (O4) | |
| P498 | FR02233085 (O3) | FR02233086 (O4) |
Friday, July 12th
QuikChange of P477 (pActin) to insert missing 60 bp
Investigator: Jeff
Aim of the experiment: QuikChange of P477 (pActin) to insert missing 60 bp.
Procedure:
Reaction batch #1, with 5% DMSO and O108 as oligonucleotide:
| volume | reagent |
|---|---|
| 2.5 µl | 10x Pfu Ultra II buffer |
| 4 µl | Plasmid P477 template (1:100) |
| 1.0 µl | 1:10 dilution of O108 (=10 µM) |
| 1 µl | dNTP mix 50 mM |
| 1.25 µl | DMSO |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U/µl) |
| 14.75 µl | ddH2O |
| =25 µl | TOTAL |
Reaction batch #2, with 5% DMSO and O109 as oligonucleotide:
| volume | reagent |
|---|---|
| 2.5 µl | 10x Pfu Ultra II buffer |
| 4 µl | Plasmid P477 template (1:100) |
| 1.0 µl | 1:10 dilution of O109 (=10 µM) |
| 1 µl | dNTP mix 50 mM |
| 1.25 µl | DMSO |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U/µl) |
| 14.75 µl | ddH2O |
| =25 µl | TOTAL |
Reaction batch #3, O108 as oligonucleotide:
| volume | reagent |
|---|---|
| 2.5 µl | 10x Pfu Ultra II buffer |
| 4 µl | Plasmid P477 template (1:100) |
| 1.0 µl | 1:10 dilution of O108 (=10 µM) |
| 1 µl | dNTP mix 50 mM |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U/µl) |
| 16 µl | ddH2O |
| =25 µl | TOTAL |
Reaction batch #4, O109 as oligonucleotide:
| volume | reagent |
|---|---|
| 2.5 µl | 10x Pfu Ultra II buffer |
| 4 µl | Plasmid P477 template (1:100) |
| 1.0 µl | 1:10 dilution of O109 (=10 µM) |
| 1 µl | dNTP mix 50 mM |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U/µl) |
| 16 µl | ddH2O |
| =25 µl | TOTAL |
- Every reaction batch type was created twice; one batch of each type was used for the program with 55 °C as annealing temperature and one was used with 65 °C as annealing temperature.
- The batches were cycled seperately for the first 10 rounds
PCR cycling parameters with 55 °C annealing temperature:
| Cycles | Temperature | Time |
|---|---|---|
| 1 | 95 °C | 30 sec |
| 10 | 95°C | 30 sec |
| 55°C | 1 min | |
| 72°C | 4.5 min | |
| 1 | 4°C | HOLD |
PCR cycling parameters with 65 °C annealing temperature:
| Cycles | Temperature | Time |
|---|---|---|
| 1 | 95 °C | 30 sec |
| 10 | 95°C | 30 sec |
| 65°C | 1 min | |
| 72°C | 4.5 min | |
| 1 | 4°C | HOLD |
- After the ten cycles the reaction batches (#1 with #2 and #3 with #4) were pooled together and the reaction was cycled for further 20 rounds with the following conditions:
PCR cycling parameters with 55 °C annealing temperature:
| Cycles | Temperature | Time |
|---|---|---|
| 1 | 95 °C | 30 sec |
| 20 | 95°C | 30 sec |
| 55°C | 1 min | |
| 72°C | 4.5 min | |
| 1 | 4°C | HOLD |
PCR cycling parameters with 65 °C annealing temperature:
| Cycles | Temperature | Time |
|---|---|---|
| 1 | 95 °C | 30 sec |
| 20 | 95°C | 30 sec |
| 65°C | 1 min | |
| 72°C | 4.5 min | |
| 1 | 4°C | HOLD |
- Digestion of the parental DNA with DpnI: Addition of 1 µl DpnI to the pooled reaction batches and incubate for 1 h at 37 °C.
Analytical gelelectrophoresis of QuikChange products of P477 (Actin promoter) to insert 60 bp deletion
Investigator: Jeff
Aim of the experiment: Analytical gelelectrophoresis of QuikChange products of P477 (Actin promoter) to insert 60 bp deletion.
Procedure:
- 9 µl of the QuikChange products were mixed with 1 µl of DNA loading buffer (10x).
- Analytical gelelectrophoresis was performed at 100 V for 40 min on a 1% agarose gel.
| Lane: | 2 log DNA ladder | QC of P477 with DMSO and annealing temperature at 55 °C | QC of P477 with DMSO and annealing temperature at 65 °C | QC of P477 with annealing temperature at 55 °C | QC of P477 with annealing temperature at 65 °C |
|---|---|---|---|---|---|
| Result: | no product | no product | no product | no product |
Picking of E. coli XL1-Blue transformed with ligation products F156+F182 (NLS_NucA in pSB1C3 + (GGGGS)x5-TEV-site-linker), F181+F135 (SERK-SigP_FluA in pSB1C3 + Strep-tag-II-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1), F171+F183 (SERK-SigP_SpyTag_Strep-tag-II-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1_IRES in pSB1C3 + IgKappa-SigP_SpyCatcher_NanoLuc), F109+F184 (SERK-SigP in pSB1C3 + Proteinphosphotase 1)
Investigator: Jeff
Aim of the experiment: Picking of E. coli XL1-Blue transformed with ligation products F156+F182 (NLS_NucA in pSB1C3 + (GGGGS)x5-TEV-site-linker), F181+F135 (SERK-SigP_FluA in pSB1C3 + Strep-tag-II-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1), F171+F183 (SERK-SigP_SpyTag_Strep-tag-II-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1_IRES in pSB1C3 + IgKappa-SigP_SpyCatcher_NanoLuc), F109+F184 (SERK-SigP in pSB1C3 + Proteinphosphotase 1).
Procedure:
- Picked colonies were transferred into cell-culture tubes with air-permeable, sterile cover. Each tube contains 4 mL of LB-medium + 4 µL chloramphenicol (1000x).
- 2 colonies were picked for F156+F182, F171+F182, F109+F184 and 6 colonies were picked for ligation product F181+F135.
- These tubes were transferred in a cell culture shaker at 37 °C and were incubated at 180 rpm overnight.
Saturday, July 13th
Miniprep of E. coli XL1-Blue transformed with ligation products F156+F182 (NLS_NucA in pSB1C3 + (GGGGS)x5-TEV-site-linker), F181+F135 (SERK-SigP_FluA in pSB1C3 + Strep-tag-II-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1), F171+F183 (SERK-SigP_SpyTag_Strep-tag-II-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1_IRES in pSB1C3 + IgKappa-SigP_SpyCatcher_NanoLuc), F109+F184 (SERK-SigP in pSB1C3 + Proteinphosphotase 1)
Investigator: Jeff
Aim of the experiment: Miniprep of E. coli XL1-Blue transformed with ligation products F156+F182 (NLS_NucA in pSB1C3 + (GGGGS)x5-TEV-site-linker), F181+F135 (SERK-SigP_FluA in pSB1C3 + Strep-tag-II-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1), F171+F183 (SERK-SigP_SpyTag_Strep-tag-II-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1_IRES in pSB1C3 + IgKappa-SigP_SpyCatcher_NanoLuc), F109+F184 (SERK-SigP in pSB1C3 + Proteinphosphotase 1).
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Spin Miniprep Kit, QIAGEN).
- The resulting tubes were labeled as P508 to P519.
Analytical digestion and gelelectrophoresis of P508 – P519
Investigator: Le Swaffeleur
Aim of the experiment: Analytical digestion and gelelectrophoresis of P508 – P519.
Procedure:
- 15x Mastermix (EcoRI-HF & PstI-HF) for analytical digestion:
| volume | reagent |
|---|---|
| 26 µl | Cut Smart buffer |
| 3.25 µl | EcoRI-HF (20 U/µl) |
| 3.25 µl | PstI-HF (20 U/µl) |
| 195 µl | ddH2O |
| =227.5 µl | TOTAL |
- 17.5 µl master mix and 2.5 µl plasmid DNA (P508 – P519) were mixed.
- Analytical digestion was performed at 37 °C for 1 h.
- After incubation 2 µl of DNA loading buffer (10x) were added to the reaction batches.
- Analytical gelelectrophoresis was performed at 90 V for 1 h.
| Lane: | 2 log DNA ladder | P508, digested with EcoRI & PstI | P509, digested with EcoRI & PstI | P510, digested with EcoRI & PstI | P511, digested with EcoRI & PstI | P512, digested with EcoRI & PstI | P513, digested with EcoRI & PstI | P514, digested with EcoRI & PstI | P515, digested with EcoRI & PstI | P516, digested with EcoRI & PstI | P517, digested with EcoRI & PstI | P518, digested with EcoRI & PstI | P519, digested with EcoRI & PstI | 2 log DNA ladder |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Result: | as expected | as expected | as expected | as expected | as expected | as expected | as expected | as expected | as expected | as expected | as expected | as expected |
Sunday, July 14th
Preparative digestion and gelelectrophoresis of P428 (mMCSII_t35S_npt-casette) and P477 (pAct)
Investigator: Jeff
Aim of the experiment: Preparative digestion and gelelectrophoresis of P428 (mMCSII_t35S_npt-casette) and P477 (pAct).
Procedure:
Reaction batch for P428 (MCS-t35S-npt):
| volume | reagent |
|---|---|
| 15 µl | P428 |
| 4 µl | CutSmart Buffer (10x) |
| 1 µl | EcoRI (20 U/µl) |
| 1 µl | XbaI (20 U/µl) |
| 19 µl | ddH2O |
| =40 µl | TOTAL |
Reaction batch for P477 (pAct):
| volume | reagent |
|---|---|
| 15 µl | P477 |
| 4 µl | CutSmart Buffer (10x) |
| 1 µl | EcoRI (20 U/µl) |
| 2 µl | SpeI (10 U/µl) |
| 18 µl | ddH2O |
| =40 µl | TOTAL |
- Reaction batches were incubated at 37 °C for 4 h.
- Preparative gelelectrophoresis was performed at 90 V for 2 h in an 1% agarose gel.
| Lane | Digestion of P486 with XbaI & AgeI | 2 log DNA ladder | Digestion of P488 with XbaI & PstI | 2 log DNA ladder |
|---|---|---|---|---|
| Results | as expected; intense band was cut out | as expected; lowest band was cut out |
- DNA bands were extracted with QIAquick Gel Extraction Kit, QIAGEN.
Ligation of F185+F186 (pAct + MCS_t35S_npt)
Investigator: Jeff
Procedure:
Ligation batch for F185+F186 (pAct + MCS_t35S_npt), vector:insert = 1:3:
| volume | reagent |
|---|---|
| 4 µl | F185 (5000 bp) |
| 4 µl | F186 (1200 bp) |
| 1 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =10 µl | TOTAL |
- Ligation was performed at 22 °C for 1 h
- Negative controls were also prepared with water instead of insert.
Preparative digestion and gelelectrophoresis of P430 (PhyB_36AAlinker_C-TEV_IRES_N-TEV_36AAlinker_PIF3_IRES), P431 (PhyB_36AAlinker_C-TEV_IRES_N-TEV_36AAlinker_PIF6_IRES), P509 ((GGGGS)x5-TEV-site-linker_NLS_NucA) and P510 (SERK-SigP_PP1)
Investigator: Jeff
Aim of the experiment: Preparative digestion and gelelectrophoresis of P430 (PhyB_36AAlinker_C-TEV_IRES_N-TEV_36AAlinker_PIF3_IRES), P431 (PhyB_36AAlinker_C-TEV_IRES_N-TEV_36AAlinker_PIF6_IRES), P509 ((GGGGS)x5-TEV-site-linker_NLS_NucA) and P510 (SERK-SigP_PP1).
Procedure:
Reaction batch for P430 (PhyB_36AAlinker_C-TEV_IRES_N-TEV_36AAlinker_PIF3_IRES):
| volume | reagent |
|---|---|
| 20 µl | P430 |
| 4 µl | CutSmart Buffer (10x) |
| 2 µl | SpeI (20 U/µl) |
| 1 µl | PstI-HF (20 U/µl) |
| 13 µl | ddH2O |
| =40 µl | TOTAL |
Reaction batch for P431 (PhyB_36AAlinker_C-TEV_IRES_N-TEV_36AAlinker_PIF6_IRES):
| volume | reagent |
|---|---|
| 20 µl | P431 |
| 4 µl | CutSmart Buffer (10x) |
| 2 µl | SpeI (10 U/µl) |
| 1 µl | PstI-HF (20 U/µl) |
| 13 µl | ddH2O |
| =40 µl | TOTAL |
Reaction batch for P509 ((GGGGS)x5-TEV-site-linker_NLS_NucA):
| volume | reagent |
|---|---|
| 20 µl | P509 |
| 4 µl | CutSmart Buffer (10x) |
| 2 µl | NgoMIV (10 U/µl) |
| 1 µl | PstI-HF (20 U/µl) |
| 13 µl | ddH2O |
| =40 µl | TOTAL |
Reaction batch for P510 (SERK-SigP_PP1)):
| volume | reagent |
|---|---|
| 20 µl | P510 |
| 4 µl | CutSmart Buffer (10x) |
| 1 µl | AgeI-HF (20 U/µl) |
| 1 µl | PstI-HF (20 U/µl) |
| 14 µl | ddH2O |
| =40 µl | TOTAL |
- Reaction batches were incubated at 37 °C for 2.5 h.
- Preparative gelelectrophoresis was performed at 90 V for 1 h in an 1% agarose gel.
| Lane | Digestion of P430 with XbaI & AgeI | 2 log DNA ladder | Digestion of P431 with XbaI & PstI | Digestion of P509 with NgoMIV & SpeI |
|---|---|---|---|---|
| Results | as expected | as expected | as expected; lower band was cut out |
| Lane | 2 log DNA ladder | Digestion of P510 with AgeI & PstI |
|---|---|---|
| Results | as expected |
- DNA bands were extracted with QIAquick Gel Extraction Kit, QIAGEN.
Ligation of F122+F189 (SERK-SigP_SERK-TMD + (GGGGS)x5-TEV-site-linker_NLS_NucA), F190+F135 (SERK-SigP_PP1 + Strep-tag-II-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1)
Investigator: Jeff
Aim of the experiment: Ligation of F122+F189 (SERK-SigP_SERK-TMD + (GGGGS)x5-TEV-site-linker_NLS_NucA), F190+F135 (SERK-SigP_PP1 + Strep-tag-II-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1).
Procedure:
Ligation batch for F122+F189 (SERK-SigP_SERK-TMD + (GGGGS)x5-TEV-site-linker_NLS_NucA), vector:insert = 1:3:
| volume | reagent |
|---|---|
| 1.99 µl | F122 (50.3 ng/µl, 2369 bp) |
| 0.98 µl | F189 (79.9 ng/µl, 618 bp) |
| 14.03 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
Ligation batch for F190+F135 (SERK-SigP_PP1 + Strep-tag-II-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1), vector:insert = 1:3:
| volume | reagent |
|---|---|
| 0.45 µl | F190 (224.7 ng/µl, 3155 bp) |
| 5.32 µl | F135 (17.8 ng/µl, 996 bp) |
| 11.23 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- Ligation was performed at 16 °C overnight.
- Negative controls were also prepared with water instead of insert.
Week 13
Monday, July 15th
Transformation of the genes to be expressend in moss (retransformation) and F122+F189 (SERK-SigP_SERK-TMD + (GGGGS)x5-TEV-site-linker_NLS_NucA), F190+F135 (SERK-SigP_PP1 + Strep-tag-II-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1)
Investigator: Jeff
Procedure for the retransformations:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 0.5 µl of DNA were added to the tubes containing 100 µl of competent cells and gently mixed
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- The cell suspension were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- 50 µl of this cell suspension was plated on chlorampenicol plates.
Tuesday, July 16th
Preparative digestion and gelelectrophoresis of all effectors
Investigator: Jeff
Procedure:
Mastermix:
| volume | reagent |
|---|---|
| 224 | ddH2O |
| 64 µl | CutSmart Buffer (10x) |
| 16 µl | EcoRI-HF (20 U/µl) |
| 16 µl | PstI-HF (20 U/µl) |
- 20 µl of DNA were mixed with 20 µl of Mastermix and were incubated at 37 °C for 4 h.
- Preparative gelelectrophoresis was performed at 90 V for 1 h in an 1% agarose gel.
| Lane | 2 log DNA ladder | Digestion of P20 (GFP) with EcoRI & PstI | 2 log DNA ladder | Digestion of P196 (XylE) with EcoRI & PstI | 2 log DNA ladder | Digestion of P245 (EreB) with EcoRI & PstI |
|---|---|---|---|---|---|---|
| Results | as expected | as expected | as expected | as expected | as expected | as expected |
| Lane | 2 log DNA ladder | Digestion of P246 (nLuc) with EcoRI & PstI | 2 log DNA ladder | Digestion of P330 (SERK-SigP_NLuc) with EcoRI & PstI | 2 log DNA ladder | Digestion of P346 (Dehydrochlorinase) with EcoRI & PstI |
|---|---|---|---|---|---|---|
| Results | as expected | as expected | as expected | as expected | as expected | as expected |
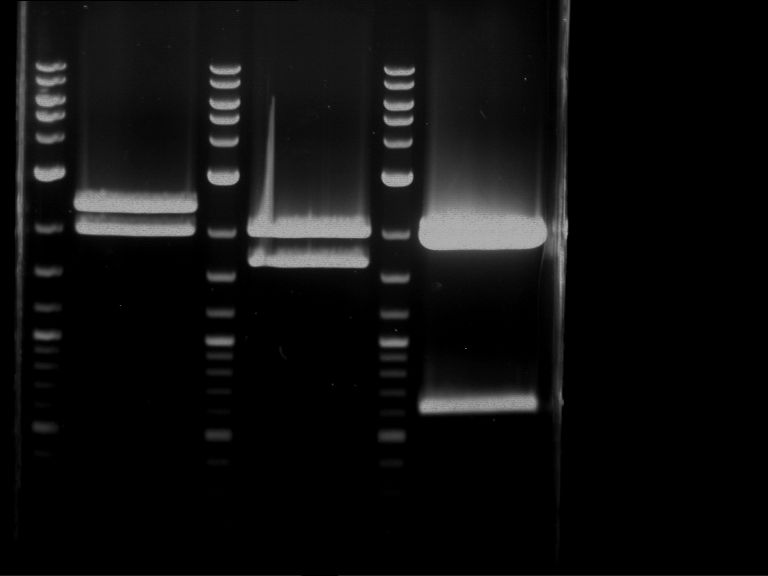
The digestion of P350 was separated for an additional hour because there might be contaminations of the vector that is smaller than the protein-coding BioBrick we want to express. The separated bands looked like that:
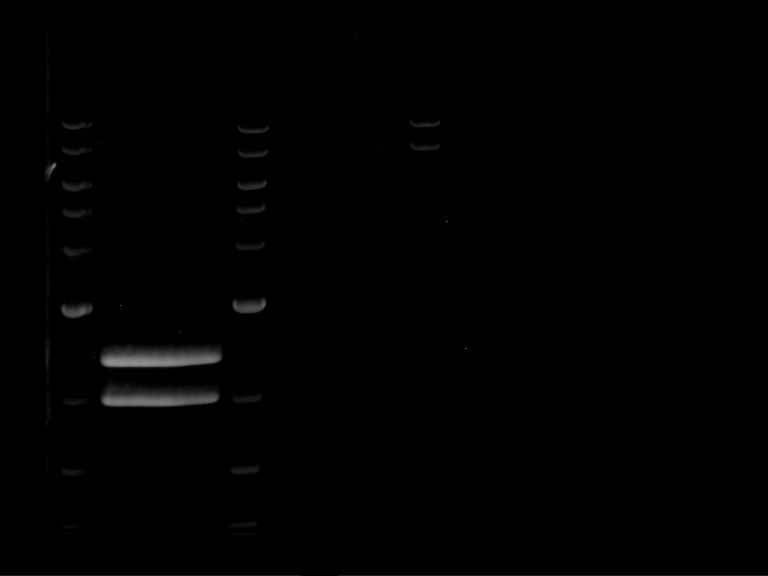
| Lane | 2 log DNA ladder | Digestion of P350 (EreB-Receptor) with EcoRI & PstI | 2 log DNA ladder | Digestion of P354 (nLuc-Receptor) with EcoRI & PstI | 2 log DNA ladder | Digestion of P402 (IgK-nLuc) with EcoRI & PstI |
|---|---|---|---|---|---|---|
| Results | as expected | as expected | as expected | as expected | as expected | as expected |
| Lane | 2 log DNA ladder | Digestion of P476 (SpyCatcher-Receptor_IRES_IgK_SpyTag-nLuc) with EcoRI & PstI | 2 log DNA ladder | Digestion of P496 (SpyTag-Receptor_IRES_nLuc_SpyCatcher with EcoRI & PstI | 2 log DNA ladder | Digestion of P498 (SpyCatcher-Receptor_IRES_IgK_nLuc-SpyTAG) with EcoRI & PstI |
|---|---|---|---|---|---|---|
| Results | as expected | as expected | as expected | as expected | as expected | as expected |
| Lane | 2 log DNA ladder | Digestion of P502 (IgK-GFP) with EcoRI & PstI | 2 log DNA ladder | Digestion of P512 (SpyTAG-Receptor-IRES_SpyCatcher-nLuc) with EcoRI & PstI | 2 log DNA ladder | Digestion of P516 (FluA-Recetpro) with EcoRI & PstI |
|---|---|---|---|---|---|---|
| Results | as expected | as expected | as expected | as expected | as expected | as expected |
- DNA bands were extracted with QIAquick Gel Extraction Kit, QIAGEN.
Miniprep of P520 to P525: P520 (Bba_KK509000,Cauliflower Mosaic Virus 35S promoter) P521-525 (pAct(with_del)-MCS-T35S-npt
Investigator: Jeff
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Spin Miniprep Kit, QIAGEN).
- The resulting tubes were labeled as P520 to P525.
Analytical digestion and gelelectrophoresis of P520 - P525
Investigator: Jeff
Procedure:
- 7x Mastermix (EcoRI-HF & PstI-HF) for analytical digestion:
| volume | reagent |
|---|---|
| 7 µl | Cut Smart buffer |
| 1.75 µl | EcoRI-HF (20 U/µl) |
| 1.75 µl | PstI-HF (20 U/µl) |
| 38.5 µl | ddH2O |
| =49 µl | TOTAL |
- 7 µl master mix and 3 µl plasmid DNA were mixed.
- Analytical digestion was performed at 37 °C for 1.5bsp;h.
- After incubation 2 µl of DNA loading buffer (10x) were added to the reaction batches.
- Analytical gelelectrophoresis was performed at 90 V for 1 h.
| Lane: | 2 log DNA ladder | P428, dsted with EcoRI & PstI | P522, digested with EcoRI & PstI | P522, digested with EcoRI & PstI | P523, digested with EcoRI & PstI | P524, digested with EcoRI & PstI | P525, digested with EcoRI & PstI |
|---|---|---|---|---|---|---|---|
| Result: | control | no insert ;( | no insert ;( | no insert ;( | no insert ;( | no insert ;( | no insert ;( |
Wednesday, July 17th
Thursday, July 18th
Analytical digestion and gelelectrophoresis of P537 – P540 and analytical gelelectrophoresis of F206 and F207
Investigator: Jeff
Aim of the experiment: Analytical digestion and gelelectrophoresis of P537 – P540 and analytical gelelectrophoresis of F206 and F207.
Procedure:
| volume | reagent |
|---|---|
| 2.5 µl | Plasmid DNA (P537 – P540) |
| 2 µl | Cut Smart buffer |
| 0.25 µl | EcoRI-HF |
| 0.25 µl | PstI-HF |
| 15 µl | ddH2O |
| =20 µl | TOTAL |
- Analytical digestion was performed at 37 °C for 1.5 h.
- After incubation 2 µl of DNA loading buffer (10x) were added to the reaction batches and were loaded on a 1% agarose gel.
- Analytical gelelectrophoresis was performed at 90 V for 1 h.
| Lane: | 2 log DNA ladder | Digestion of P537 F122+F189 (SERK-SigP_SERK-TMD in pSB1C3 + (GGGGS)x5-TEV-site-linker_NLS_NucA) with EcoRI & PstI | Digestion of P538 F122+F189 (SERK-SigP_SERK-TMD in pSB1C3 + (GGGGS)x5-TEV-site-linker_NLS_NucA) with EcoRI & PstI | Digestion of P539 F190+F135 (SERK-SigP_PP1 in pSB1C3 only + Strep-tag-II-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1) with EcoRI & PstI | Digestion of P540 F190+F135 (SERK-SigP_PP1 in pSB1C3 only + Strep-tag-II-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1) with EcoRI & PstI | F206 | F207 |
|---|---|---|---|---|---|---|---|
| Result: | as expected (916 bp) | as expected (916 bp) | as expected (2080 bp) | as expected (2080 bp) | as expected (3484 bp) | as expected (3484 bp) |
PCR to insert 60 missing base-pairs into pActin (new method)
Investigator: Jeff
Aim of the experiment: PCR to insert 60 missing base-pairs into pActin (new method)
Procedure:
Reaction batch for PCR with Q5 Mastermixes for P477 (pActin):
| volume | reagent |
|---|---|
| 25 µl | 2x Q5 Master Mix |
| 2.5 µl | 10 µM Forward Primer O114 (pAct_ins_fw) |
| 2.5 µl | 10 µM Reverse Primer O115 (pAct_ins_rv) |
| 1 µl | Plasmid DNA (P477) - 1:1000 dilution (desired c between 1 pg and 1 ng) |
| 19 µL | ddH2O Water |
| =50 µL | TOTAL |
| volume | reagent |
|---|---|
| 25 µl | 2x Q5 Master Mix |
| 2.5 µl | 10 µM Forward Primer O114 (pAct_ins_fw) |
| 2.5 µl | 10 µM Reverse Primer O115 (pAct_ins_rv) |
| 1 µl | Plasmid DNA (P477) - 1:100 dilution (desired c between 1 pg and 1 ng) |
| 19 µL | ddH2O Water |
| =50 µL | TOTAL |
- The content has been mixed with a pipette
- The PCR program was performed after following scheme:
Cycling parameters:
| Initial denaturation | 98 °C | 30 s |
|---|---|---|
| 30 cycles | 98 °C | 10 s |
| 72 °C | 30 s | |
| 72 °C | 2 min | |
| Final extension | 72 °C | 2 min |
| Hold | 4 °C | infinite |
- After PCR, the product was purified with QIAquick PCR Purification Kit, Qiagen.
- The resulting product was labeled as F206 for the 1:1000 batch and F207 for the 1:100 batch.
Friday, July 19th
Saturday, July 20th
Sunday, July 21st
Week 14
Monday, July 22nd
Arrival of the correct pActin 5 from Physcomitrella patens from Reski lab
This plasmid was named p541
Tuesday, July 23rd
Wednesday, July 24th
PCR of P541 (PppActin5)
Investigator: Jeff
Aim of the experiment: PCR of P541 (PppActin5).
Procedure:
Reaction batch for PCR with Q5 Mastermix for P541 (PppActin5):
| volume | reagent |
|---|---|
| 25 µl | 2x Q5 Master Mix |
| 1 µl | dNTP mix (normally not necessary!) |
| 2.5 µl | 10 µM Forward Primer O116 (PppAct_f) |
| 2.5 µl | 10 µM Reverse Primer O117 (PppAct_r) |
| 1 µl | Plasmid DNA (P541) - 1:1000 dilution (desired c between 1 pg and 1 ng) |
| 18 µL | ddH2O Water |
| =50 µL | TOTAL |
- The content has been mixed with a pipette
- The PCR program was performed after following scheme:
Cycling parameters:
| Initial denaturation | 98 °C | 30 s |
|---|---|---|
| 30 cycles | 98 °C | 10 s |
| 53 °C | 30 s | |
| 72 °C | 30 s | |
| Final extension | 72 °C | 2 min |
| Hold | 4 °C | infinite |
- After PCR, the product was purified with QIAquick PCR Purification Kit, Qiagen.
- The resulting product was labeled as F208.
Analytical gelelectrophoresis of F208
Investigator: Jeff
Aim of the experiment: Analytical gelelectrophoresis of F208.
Procedure:
- 5 µl of the PCR product F208 (=PCR of P541 with O116/O117) products were mixed with 2 µl of DNA loading buffer (10x).
- Analytical gelelectrophoresis was performed at 90 V for 60 min on a 1% agarose gel.
| Lane: | 2 log DNA ladder | F208 |
|---|---|---|
| Result: | as expected |
Preparative digestion of P208 (PCR of PppActin5)
Investigator: Jeff
Procedure:
Reaction batch for F208 (PCR of PppActin5):
| volume | reagent |
|---|---|
| 25 µl | F208 |
| 5 µl | CutSmart Buffer (10x) |
| 1 µl | EcoRI-HF |
| 1 µl | SpeI-HF |
| 18 µl | ddH2O |
| =45 µl | TOTAL |
- Reaction batch was incubated at 37 °C for 2 h.
- After digestion, the product was purified with QIAquick PCR Purification Kit, Qiagen.
Ligation and transformation of F209 + F41 (PppActin5 in pSB1C3)
Investigator: Jeff
Procedure:
Ligation batch for F209 (PppActin5), vector:insert = 1:3:
| volume | reagent |
|---|---|
| 7 µl | F209 (36.9 ng/µl, ??? bp) |
| 1 µl | F189 (89.9 ng/µl, ??? bp) |
| 1 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =10 µl | TOTAL |
- Ligation was performed at RT for 1 hour.
- Negative controls were also prepared with water instead of insert.
The ligation products were transformed into E. coli XL-1 Blue.
Thursday, July 25th
Picking of E. coli XL1 blue transformed with ligation product of F208 and F41 (PppActin5 in pSB1C3)
Investigator: Jeff
Procedure:
- Picked colonies were transferred into cell-culture tubes with air-permeable, sterile cover. Each tube contains 4 mL of LB-medium + 4 µL chloramphenicol (1000x).
- 6 colonies were picked for ligation of F208 and F41 (PppActin5 in pSB1C3).
- These tubes were transferred in a cell culture shaker at 37 °C and were incubated at 180 rpm overnight.
Friday, July 26th
Miniprep of P542 to P547 (pSB1C2_PppActin5)
Investigator: Jeff
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Spin Miniprep Kit, QIAGEN).
- The resulting tubes were labeled as P542 to P547.
Analytical digestion and gelelectrophoresis of P542 – P547
Investigator: Jeff
Procedure:
| volume | reagent |
|---|---|
| 3 µl | Plasmid DNA (P542 – P547) |
| 1 µl | Cut Smart buffer |
| 0.25 µl | EcoRI-HF |
| 0.25 µl | PstI-HF |
| 5.5 µl | ddH2O |
| =10 µl | TOTAL |
- Analytical digestion was performed at 37 °C for 1.5 h.
- After incubation 2 µl of DNA loading buffer (10x) were added to the reaction batches and were loaded on a 1% agarose gel.
- Analytical gelelectrophoresis was performed at 90 V for 1 h.
| Lane: | 2 log DNA ladder | Digestion of P542 (PppActin5) with EcoRI & PstI | Digestion of P543 (PppActin5) with EcoRI & PstI | Digestion of P544 (PppActin5) with EcoRI & PstI | Digestion of P545 (PppActin5) with EcoRI & PstI | Digestion of P546 (PppActin5) with EcoRI & PstI | Digestion of P547 (PppActin5) with EcoRI & PstI |
|---|---|---|---|---|---|---|---|
| Result: | result unclear because insert and backbone have the same length! | result unclear because insert and backbone have the same length! | result unclear because insert and backbone have the same length! | result unclear because insert and backbone have the same length! | result unclear because insert and backbone have the same length! | result unclear because insert and backbone have the same length! |
Unfortunately the correctness of the clones could not be seen in this experiment. Thus a second digestion was performed:
| volume | reagent |
|---|---|
| 3 µl | Plasmid DNA (P542 – P547) |
| 1 µl | Cut Smart buffer |
| 0.25 µl | EcoRI-HF |
| 0.25 µl | PstI-HF |
| 0.25 µl | NcoI (form Fermentas |
| 5.25 µl | ddH2O |
| =10 µl | TOTAL |
- Analytical digestion was performed at 37 °C for 1.5 h.
- After incubation 2 µl of DNA loading buffer (10x) were added to the reaction batches and were loaded on a 1% agarose gel.
- Analytical gelelectrophoresis was performed at 90 V for 1 h.
| Lane: | 2 log DNA ladder | Digestion of P542 (PppActin5) with EcoRI & PstI | Digestion of P543 (PppActin5) with EcoRI & PstI | Digestion of P544 (PppActin5) with EcoRI & PstI | Digestion of P545 (PppActin5) with EcoRI & PstI | Digestion of P546 (PppActin5) with EcoRI & PstI | Digestion of P547 (PppActin5) with EcoRI & PstI |
|---|---|---|---|---|---|---|---|
| Result: | Clone correct: backbone and PppActin5 | Clone wrong: only backbone, no insert | Clone correct: backbone and PppActin5 | Clone correct: backbone and PppActin5 | Clone wrong: only backbone, no insert | Clone correct: backbone and PppActin5 |
Sequencing of P544 and P545
Investigator: Ingmar
Procedure: The DNA was prepared for sequencing according to the Eurofins SmartSeq Kit protocol (15 µl of plasmid DNA (50 - 100 ng) and 2 µl sequencing primer).
The different genes we sequenced received the following barcodes:
| Label | Barcode1 | Barcode2 |
|---|---|---|
| P544 | FR02233095 | FR02233087 |
| P545 | FR02233093 | FR02233094 |
Saturday, July 27th
Sunday, July 28st
Week 15
Monday, July 29th
QuikChange of P544 (PppActin5)
Investigator: Jeff
Procedure:
| volume | reagent |
|---|---|
| 2.5 µl | 10x Pfu Ultra II buffer |
| 4 µl | Plasmid P544 template (1:100) |
| 1.0 µl | 1:10 dilution of O118 (=10 µM) |
| 1.0 µl | 1:10 dilution of O119 (=10 µM) |
| 1 µl | dNTP mix 50 mM |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U/µl) |
| 15 µl | ddH2O |
| =25 µl | TOTAL |
| volume | reagent |
|---|---|
| 2.5 µl | 10x Pfu Ultra II buffer |
| 4 µl | Plasmid P544 template (1:100) |
| 1.0 µl | 1:10 dilution of O120 (=10 µM) |
| 1.0 µl | 1:10 dilution of O121 (=10 µM) |
| 1 µl | dNTP mix 50 mM |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U/µl) |
| 15 µl | ddH2O |
| =25 µl | TOTAL |
PCR cycling parameters with 55 °C annealing temperature:
| Cycles | Temperature | Time |
|---|---|---|
| 1 | 95 °C | 30 sec |
| 10 | 95°C | 30 sec |
| 55°C | 1 min | |
| 72°C | 5 min | |
| 1 | 4°C | HOLD |
- Digestion of the parental DNA with DpnI: Addition of 1 µl DpnI to the pooled reaction batches and incubate for 1 h at 37 °C.
Tuesday, July 30th
Wednesday, July 31st
Preparative digestion and gelelectrophoresis of P537 (SERK-SigP_SERK-TMD_(GGGGS)x5-TEV-site-linker_NLS_NucA)
Investigator: Jeff
Aim of the experiment: Preparative digestion and gelelectrophoresis of P537 (SERK-SigP_SERK-TMD_(GGGGS)x5-TEV-site-linker_NLS_NucA).
Procedure:
Reaction batch for P537 (SERK-SigP_SERK-TMD_(GGGGS)-TEV-site-linker_NLS_NucA):
| volume | reagent |
|---|---|
| 20 µl | P537 |
| 4 µl | CutSmart Buffer (10x) |
| 2 µl | XbaI (20 U/µl) |
| 2 µl | PstI-HF (20 U/µl) |
| 12 µl | ddH2O |
| =40 µl | TOTAL |
- Reaction batches were incubated at 37 °C for 2 h.
- Preparative gelelectrophoresis was performed at 90 V for 1 h in an 1% agarose gel.
| Lane | 2 log DNA ladder | Digestion of P537 with XbaI & PstI |
|---|---|---|
| Results | as expected; lower band was cut out |
- DNA bands were extracted with QIAquick Gel Extraction Kit, QIAGEN.
- Resulting fragment was named as F210.
Ligation of F187+F210 (PhyB_36AAlinker_C-TEV_IRES_N-TEV_36AAlinker_PIF3_IRES in pSB1C3 + SERK-SigP_SERK-TMD_(GGGGS)x5-TEV-site-linker_NLS_NucA) and F188+F210 (PhyB_36AAlinker_C-TEV_IRES_N-TEV_36AAlinker_PIF6_IRES in pSB1C3 + SERK-SigP_SERK-TMD_(GGGGS)x5-TEV-site-linker_NLS_NucA)
Investigator: Jeff
Aim of the experiment: Ligation of F187+F210 (PhyB_36AAlinker_C-TEV_IRES_N-TEV_36AAlinker_PIF3_IRES in pSB1C3 + SERK-SigP_SERK-TMD_(GGGGS)x5-TEV-site-linker_NLS_NucA) and F188+F210 (PhyB_36AAlinker_C-TEV_IRES_N-TEV_36AAlinker_PIF6_IRES in pSB1C3 + SERK-SigP_SERK-TMD_(GGGGS)x5-TEV-site-linker_NLS_NucA).
Procedure:
Ligation batch for F187+F210 (PhyB_36AAlinker_C-TEV_IRES_N-TEV_36AAlinker_PIF3_IRES in pSB1C3 + SERK-SigP_SERK-TMD_(GGGGS)x5-TEV-site-linker_NLS_NucA, vector:insert = 1:3:
| volume | reagent |
|---|---|
| 0.53 µl | F187 (189.9 ng/µl, 7346 bp) |
| 0.68 µl | F210 (55.3 ng/µl, 916 bp) |
| 15.79 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
Ligation batch for F188+F210 (PhyB_36AAlinker_C-TEV_IRES_N-TEV_36AAlinker_PIF6_IRES in pSB1C3 + SERK-SigP_SERK-TMD_(GGGGS)x5-TEV-site-linker_NLS_NucA, vector:insert = 1:3:
| volume | reagent |
|---|---|
| 0.73 µl | F188 (136.7 ng/µl, 7346 bp) |
| 0.68 µl | F210 (55.3 ng/µl, 916 bp) |
| 15.59 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- Ligation was performed at 16 °C overnight.
- Negative controls were also prepared with water instead of insert.
Thursday, August 1st
Transformation of E. coli XL1-Blue with BBa_K864100 (SYFP2), BBa_E0020 (eCFP) and ligation products F187+F210 (PhyB_36AAlinker_C-TEV_IRES_N-TEV_36AAlinker_PIF3_IRES in pSB1C3 + SERK-SigP_SERK-TMD_(GGGGS)x5-TEV-site-linker_NLS_NucA) and F188+F210 (PhyB_36AAlinker_C-TEV_IRES_N-TEV_36AAlinker_PIF6_IRES in pSB1C3 + SERK-SigP_SERK-TMD_(GGGGS)x5-TEV-site-linker_NLS_NucA)
Investigator: Jeff
Aim of the experiment: Transformation of E. coli XL1-Blue with BBa_K864100 (SYFP2), BBa_E0020 (eCFP) and ligation products F187+F210 (PhyB_36AAlinker_C-TEV_IRES_N-TEV_36AAlinker_PIF3_IRES in pSB1C3 + SERK-SigP_SERK-TMD_(GGGGS)x5-TEV-site-linker_NLS_NucA) and F188+F210 (PhyB_36AAlinker_C-TEV_IRES_N-TEV_36AAlinker_PIF6_IRES in pSB1C3 + SERK-SigP_SERK-TMD_(GGGGS)x5-TEV-site-linker_NLS_NucA).
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 10 µl of DNA were added to the tubes containing 100 µl of competent cells and gently mixed
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- The cell suspension were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated on new chlorampenicol plates.
Friday, August 2nd
Picking of E. coli XL1-Blue transformed with BBa_K864100 (SYFP2), BBa_E0020 (eCFP) and ligation products F187+F210 (PhyB_36AAlinker_C-TEV_IRES_N-TEV_36AAlinker_PIF3_IRES in pSB1C3 + SERK-SigP_SERK-TMD_(GGGGS)x5-TEV-site-linker_NLS_NucA) and F188+F210 (PhyB_36AAlinker_C-TEV_IRES_N-TEV_36AAlinker_PIF6_IRES in pSB1C3 + SERK-SigP_SERK-TMD_(GGGGS)x5-TEV-site-linker_NLS_NucA)
Investigator: Jeff
Aim of the experiment: Picking of E. coli XL1-Blue transformed with BBa_K864100 (SYFP2), BBa_E0020 (eCFP) and ligation products F187+F210 (PhyB_36AAlinker_C-TEV_IRES_N-TEV_36AAlinker_PIF3_IRES in pSB1C3 + SERK-SigP_SERK-TMD_(GGGGS)x5-TEV-site-linker_NLS_NucA) and F188+F210 (PhyB_36AAlinker_C-TEV_IRES_N-TEV_36AAlinker_PIF6_IRES in pSB1C3 + SERK-SigP_SERK-TMD_(GGGGS)x5-TEV-site-linker_NLS_NucA).
Procedure:
- Picked colonies were transferred into cell-culture tubes with air-permeable, sterile cover. Each tube contains 4 mL of LB-medium + 4 µL chloramphenicol (1000x).
- 2 colonies were picked for BBa_K864100, BBa_E0020, F187+F210 and F188+F210 each.
- These tubes were transferred in a cell culture shaker at 37 °C and were incubated at 180 rpm overnight.
Saturday, August 3rd
Miniprep of E. coli Xl1-Blue transformed with BBa_K864100 (SYFP2), BBa_E0020 (eCFP) and ligation products F187+F210 (PhyB_36AAlinker_C-TEV_IRES_N-TEV_36AAlinker_PIF3_IRES in pSB1C3 + SERK-SigP_SERK-TMD_(GGGGS)x5-TEV-site-linker_NLS_NucA) and F188+F210 (PhyB_36AAlinker_C-TEV_IRES_N-TEV_36AAlinker_PIF6_IRES in pSB1C3 + SERK-SigP_SERK-TMD_(GGGGS)x5-TEV-site-linker_NLS_NucA)
Investigator: Jeff
Aim of the experiment: Miniprep of E. coli Xl1-Blue transformed with BBa_K864100 (SYFP2), BBa_E0020 (eCFP) and ligation products F187+F210 (PhyB_36AAlinker_C-TEV_IRES_N-TEV_36AAlinker_PIF3_IRES in pSB1C3 + SERK-SigP_SERK-TMD_(GGGGS)x5-TEV-site-linker_NLS_NucA) and F188+F210 (PhyB_36AAlinker_C-TEV_IRES_N-TEV_36AAlinker_PIF6_IRES in pSB1C3 + SERK-SigP_SERK-TMD_(GGGGS)x5-TEV-site-linker_NLS_NucA).
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Spin Miniprep Kit, QIAGEN).
- The resulting tubes were labeled as P558 to P565.
Analytical digestion and gelelectrophoresis of P558 – P565
Investigator: Jeff
Aim of the experiment: Analytical digestion and gelelectrophoresis of P558 – P565.
Procedure:
Mastermix for analytical digestion with EcoRI and PstI
| volume | reagent |
|---|---|
| 20 µl | Cut Smart buffer |
| 2.5 µl | EcoRI-HF |
| 2.5 µl | PstI-HF |
| 150 µl | ddH2O |
| =175 µl | TOTAL |
- 17.5 µl of the mastermix were mixed together with 2.5 µl of Plasmid DNA (P558 – P565)
- Analytical digestion was performed at 37 °C for 1.5 h.
- After incubation 2 µl of DNA loading buffer (10x) were added to the reaction batches and were loaded on a 1% agarose gel.
- Analytical gelelectrophoresis was performed at 90 V for 1 h.
| Lane: | Digestion of P558 (BBa_K864100) | Digestion of P559 (BBa_K864100) | 2 log DNA ladder | Digestion of P560 (BBa_E0020) | Digestion of P561 (BBa_E0020) |
|---|---|---|---|---|---|
| Result: | as expected (723 bp) | as expected (723 bp) | as expected (723 bp) | as expected (723 bp) |
| Lane: | Digestion of P562 (F187+F210) | Digestion of P563 (F187+F210) | 2 log DNA ladder | Digestion of P564 (F188+F210) | Digestion of P565 (F188+F210) |
|---|---|---|---|---|---|
| Result: | as expected (6191 bp) | as expected (6191 bp) | not as expected; corrupt (6191 bp) | as expected (6191 bp) |
Sunday, August 4th
Week 16
Monday, August 5th
Preparative digestion and gelelectrophoresis of P540 (SERK-SigP_PP1_Strep-tag-II-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1), P558 (SYFP2), P560 (eCFP), P562 (PhyB_36AAlinker_C-TEV_IRES_N-TEV_36AAlinker_PIF3_IRES_SERK-SigP_SERK-TMD_(GGGGS)x5-TEV-site-linker_NLS_NucA) and P565 (PhyB_36AAlinker_C-TEV_IRES_N-TEV_36AAlinker_PIF6_IRES_SERK-SigP_SERK-TMD_(GGGGS)x5-TEV-site-linker_NLS_NucA)
Investigator: Jeff
Aim of the experiment: Preparative digestion and gelelectrophoresis of P540 (SERK-SigP_PP1_Strep-tag-II-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1), P558 (SYFP2), P560 (eCFP), P562 (PhyB_36AAlinker_C-TEV_IRES_N-TEV_36AAlinker_PIF3_IRES_SERK-SigP_SERK-TMD_(GGGGS)x5-TEV-site-linker_NLS_NucA) and P565 (PhyB_36AAlinker_C-TEV_IRES_N-TEV_36AAlinker_PIF6_IRES_SERK-SigP_SERK-TMD_(GGGGS)x5-TEV-site-linker_NLS_NucA).
Procedure:
Reaction batch for P540:
| volume | reagent |
|---|---|
| 20 µl | P540 |
| 4 µl | CutSmart Buffer (10x) |
| 2 µl | EcoRI-HF (20 U/µl) |
| 2 µl | PstI-HF (20 U/µl) |
| 4 µl | XhoI (10 U/µl) |
| 8 µl | ddH2O |
| =40 µl | TOTAL |
Reaction batch for P558, P560, P562, P565:
| volume | reagent |
|---|---|
| 20 µl | P558, P560, P562, P565 |
| 4 µl | CutSmart Buffer (10x) |
| 2 µl | EcoRI-HF (20 U/µl) |
| 2 µl | PstI-HF (20 U/µl) |
| 12 µl | ddH2O |
| =40 µl | TOTAL |
- Reaction batches were incubated at 37 °C for 2 h.
- Preparative gelelectrophoresis was performed at 90 V for 1 h in an 1% agarose gel.
| Lane | Digestion of P540 with EcoRI & PstI & XhoI | 2 log DNA ladder | Digestion of P558 with EcoRI & PstI | Digestion of P560 with EcoRI & PstI |
|---|---|---|---|---|
| Results | as expected; upper band was cut out | as expected; lower band was cut out | as expected; lower band was cut out |
| Lane | Digestion of P562 with EcoRI & PstI | 2 log DNA ladder | Digestion of P565 with EcoRI & PstI |
|---|---|---|---|
| Results | as expected; upper band was cut out | as expected; upper band was cut out |
- DNA bands were extracted with QIAquick Gel Extraction Kit, QIAGEN.
- Resulting fragment was named as F214 – F218.
PCR of P558 (SYFP2) and P560 (eCFP) to introduce RFC25 pre- and suffixes
Investigator: Jeff
Aim of the experiment: PCR of P558 (SYFP2) and P560 (eCFP) to introduce RFC25 pre- and suffixes. Procedure:
Reaction batch for PCR with Q5 Mastermixes for P558 (SYFP2):
| volume | reagent |
|---|---|
| 25 µl | 2x Q5 Master Mix |
| 2.5 µl | 10 µM Forward Primer O122 (SYFP2_fw) |
| 2.5 µl | 10 µM Reverse Primer O123 (SYFP2_rv) |
| 1 µl | Plasmid DNA (P558) - 1:100 dilution (desired c between 1 pg and 1 ng) |
| 19 µL | ddH2O Water |
| =50 µL | TOTAL |
Reaction batch for PCR with Q5 Mastermixes for P560 (eCFP):
| volume | reagent |
|---|---|
| 25 µl | 2x Q5 Master Mix |
| 2.5 µl | 10 µM Forward Primer O124 (eCFP_fw) |
| 2.5 µl | 10 µM Reverse Primer O125 (eCFP_rv) |
| 1 µl | Plasmid DNA (P560) - 1:100 dilution (desired c between 1 pg and 1 ng) |
| 19 µL | ddH2O Water |
| =50 µL | TOTAL |
- The content has been created on ice and mixed with a pipette
- The gradient PCR program was performed after following scheme with following conditions (Tm=67 °C; ΔG=2 °C; P558 in row 1(=65.0 °C); P560 in row 12 (=69.2 °C)):
Cycling parameters:
| Initial denaturation | 98 °C | 30 s |
|---|---|---|
| 30 cycles | 98 °C | 10 s |
| Tm=67 °C; ΔG=2 °C | 30 s | |
| 72 °C | 22 s | |
| Final extension | 72 °C | 2 min |
| Hold | 4 °C | infinite |
- After PCR, the product was purified with QIAquick PCR Purification Kit, Qiagen.
- The resulting product was labeled as F212 for the the PCR products of P558 and F213 for the PCR product of P560.
Analytical gelelectrophoresis of F212 (SYFP2 RFC25) and F213 (eCFP RFC25)
Investigator: Jeff
Aim of the experiment: Analytical gelelectrophoresis of F212 (SYFP2 RFC25) and F213 (eCFP RFC25).
Procedure:
- 4.5 µl of the PCR product F212 and F213 products were mixed with 0.5 µl of DNA loading buffer (10x).
- Analytical gelelectrophoresis was performed at 90 V for 60 min on an 1% agarose gel.
| Lane: | F212 | 2 log DNA ladder | F213 |
|---|---|---|---|
| Result: | as expected | as expected |
Tuesday, August 6th
Oligohybridization of O128(G-TEV-site-G_fw) and O129 (G-TEV-site-G_rv)
Investigator: Jeff
Aim of the experiment: Oligohybridization of O128(G-TEV-site-G_fw) and O129 (G-TEV-site-G_rv).
Procedure:
- 25 µL of O128 (100 µM) and 25 µL of O129 (100 µM) were pooled together.
- Heating up to 95 °C for 5 min.
- Cooling at RT in a styropor box overnight.
Preparative digestion of F212 (SYFP2 RFC25) and F213 (eCFP RFC25)
Investigator: Jeff
Aim of the experiment: Preparative digestion of F212 (SYFP2 RFC25) and F213 (eCFP RFC25).
Procedure:
Reaction batch for F212:
| volume | reagent |
|---|---|
| 25 µl | F212 |
| 5 µl | CutSmart Buffer (10x) |
| 1 µl | XbaI (20 U/µl) |
| 1 µl | AgeI-HF (20 U/µl) |
| 18 µl | ddH2O |
| =50 µl | TOTAL |
Reaction batch for F213:
| volume | reagent |
|---|---|
| 25 µl | F213 |
| 5 µl | CutSmart Buffer (10x) |
| 1 µl | XbaI (20 U/µl) |
| 1 µl | AgeI-HF (20 U/µl) |
| 18 µl | ddH2O |
| =50 µl | TOTAL |
- Reaction batches were incubated at 37 °C for 2 h.
- Digested products were purified with QIAquick PCR Purification Kit, QIAGEN.
Ligation of F123+F219 (pSB1C3 + SYFP2) and F123+F220 (eCFP)
Investigator: Jeff
Aim of the experiment: Ligation of F123+F219 (pSB1C3 + SYFP2) and F123+F220 (eCFP).
Procedure:
Ligation batch for F123+F219 (pSB1C3 + SYFP2), vector:insert = 1:3:
| volume | reagent |
|---|---|
| 1 µl | F123 (49.3 ng/µl, 2086 bp) |
| 0.4 µl | F219 (114.9 ng/µl, 723 bp) |
| 15.6 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
Ligation batch for F123+F220 (pSB1C3 + eCFP), vector:insert = 1:3:
| volume | reagent |
|---|---|
| 1 µl | F123 (49.3 ng/µl, 2086 bp) |
| 1.2 µl | F220 (41.7 ng/µl, 723 bp) |
| 14.8 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- Ligation was performed at 16 °C overnight.
Analytical digestion and gelelectrophoresis of further clones
Investigator: Jeff
Procedure:
The plasmids were cut with 0.2 µl of EcoRI and SpeI in a volume of 10 µl.
- Analytical digestion was performed at 37 °C for 2 h.
- After incubation 3 µl of DNA loading buffer (10x) were added to the reaction batches and were loaded on a 1% agarose gel.
- Analytical gelelectrophoresis was performed at 90 V for 1 h.
| Lane: | 2log DNA ladder | clone 1 | clone 2 | clone 3 | clone 4 | clone 5 | clone 6 | clone 7 | clone 8 |
|---|---|---|---|---|---|---|---|---|---|
| Result: | wrong result | wrong result | wrong result | wrong result | wrong result | wrong result | wrong result | wrong result |
Preparative digestion of P428 (mMCS-T35S-npt) and P554 (PppAct5-QCII)
Investigator: Jeff
Procedure:
Reaction batch for P428:
| volume | reagent |
|---|---|
| 20 µl | P428 |
| 5 µl | CutSmart Buffer (10x) |
| 1 µl | EcoRI (20 U/µl) |
| 1 µl | XbaI (20 U/µl) |
| 23 µl | ddH2O |
| =50 µl | TOTAL |
Reaction batch for P554:
| volume | reagent |
|---|---|
| 20 µl | F213 |
| 5 µl | CutSmart Buffer (10x) |
| 1 µl | EcoRI (20 U/µl) |
| 2 µl | SpeI (10 U/µl) |
| 2 µl | NcoI (10 U/µl) |
| 18 µl | ddH2O |
| =50 µl | TOTAL |
- Reaction batches were incubated at 37 °C for 2 h.
| Lane | 2 log DNA ladder | Digestion of P428 with EcoRI & XbaI | Digestion of P554 with EcoRI & SpeI |
|---|---|---|---|
| Results | as expected; highest band was cut out | as expected; highest band was cut out |
The bands were extracted from the gel and termed F221 and F222. The concentrations were determined to 42 and 37 ng/µl.
Ligation of F221+F222 (PppAct5 + pSB1C3-mMCS-T35S-npt)
Investigator: Jeff
Procedure:
Ligation batch for F221+F222 (PppAct5 + pSB1C3-mMCS-T35S-npt), vector:insert = 1:3:
| volume | reagent |
|---|---|
| 5 µl | F221 (42 ng/µl, 3700 bp) |
| 11 µl | F222 (37 ng/µl, 2000 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 2 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- Ligation was performed at room temperature overnight.
Transformation of E. coli XL1-Blue with ligation F221+F222
Investigator: Jeff
Procedure:
- After 1 h of ligation 10 µl were used for transformation.
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 10 µl of DNA were added to the tubes containing 100 µl of competent cells and gently mixed
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- The cell suspension were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated on new chlorampenicol plates.
Wednesday, August 7th
Transformation of E. coli XL1-Blue with ligation F221+F222
Investigator: Jeff
Procedure:
- After 1 h of ligation 10 µl were used for transformation.
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 10 µl of DNA were added to the tubes containing 100 µl of competent cells and gently mixed
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- The cell suspension were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated on new chlorampenicol plates.
Picking of of E. coli XL1 blue with PppAct5-mMCS-T35S-npt from the ligation performed after 1 h
Investigator: Jeff
Procedure:
- PppAct5-mMCS-T35S-npt: Colonies were picked from chloramphenicol plates.
- Picked pipette tips was transferred into cell-culture tubes with air-permeable, sterile cover. Each tube contain 4 mL of LB-medium + 4 µL chloramphenicol(1000x).
- 12 colonies were picked.
- These tubes were transferred in a cell culture shaker at 37 °C and were incubated overnight
Thursday, August 8th
Analytical digestion and gelelectrophoresis of further clones
Investigator: Jeff
Procedure:
The plasmids were cut with 0.2 µl of EcoRI and SpeI in a volume of 10 µl.
- Analytical digestion was performed at 37 °C for 2 h.
- After incubation 3 µl of DNA loading buffer (10x) were added to the reaction batches and were loaded on a 1% agarose gel.
- Analytical gelelectrophoresis was performed at 90 V for 1 h.
| Lane: | 2log DNA ladder | clone 1 | clone 2 | clone 3 | clone 4 | clone 5 | clone 6 | clone 7 | clone 8 | clone 9 | clone 10 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Result: | wrong result | wrong result | wrong result | wrong result | wrong result | wrong result | wrong result | wrong result | wrong result | wrong result |
All these clones were discarded.
Additionally the fragments for the ligation were controlled.
| Lane: | 2log DNA ladder | F185 | F211 | F221 | F222 |
|---|---|---|---|---|---|
| Result: | correct | correct | correct | correct |
Picking of of E. coli XL1 blue with PppAct5-mMCS-T35S-npt from the ligation performed over night
Investigator: Jeff
Procedure:
- PppAct5-mMCS-T35S-npt: Colonies were picked from chloramphenicol plates.
- Picked pipette tips was transferred into cell-culture tubes with air-permeable, sterile cover. Each tube contain 4 mL of LB-medium + 4 µL chloramphenicol(1000x).
- 40 colonies were picked.
- These tubes were transferred in a cell culture shaker at 37 °C and were incubated overnight
Friday, August 9th
Miniprep of E. coli Xl1-Blue transformed with ligation of PppAct5 into mMCS-T35S-npt
Investigator: Jeff
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Spin Miniprep Kit, QIAGEN).
- The resulting tubes were labeled as P574 to P613.
Analytical digestion and gelelectrophoresis of P574 – P613
Investigator: Jeff
Aim of the experiment: Analytical digestion and gelelectrophoresis of P574 – P613.
Procedure:
Mastermix for analytical digestion with EcoRI
| volume | reagent |
|---|---|
| 45 µl | Cut Smart buffer |
| 8 µl | EcoRI-HF |
| 400 µl | ddH2O |
| =453 µl | TOTAL |
- 9 µl of the mastermix were mixed together with 1 µl of Plasmid DNA
- Analytical digestion was performed at 37 °C for 2 h.
- After incubation 3 µl of DNA loading buffer (10x) were added to the reaction batches and were loaded on a 1% agarose gel.
- Analytical gelelectrophoresis was performed at 90 V for 1 h.
| Lane: | 2log DNA ladder | clone 1 | clone 2 | clone 3 | clone 4 | clone 5 | clone 6 | clone 7 | clone 8 | clone 9 | clone 10 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Result: | wrong result | wrong result | wrong result | wrong result | wrong result | wrong result | wrong result | wrong result | wrong result | good clone => only faint additional band |
| Lane: | 2log DNA ladder | clone 1 | clone 2 | clone 3 | clone 4 | clone 5 | clone 6 | clone 7 | clone 8 | clone 9 | clone 10 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Result: | wrong result | wrong result | wrong result | wrong result | wrong result | wrong result | wrong result | best clone => no additional band (best clone) | wrong result | wrong result |
| Lane: | 2log DNA ladder | clone 1 | clone 2 | clone 3 | clone 4 | clone 5 | clone 6 | clone 7 | clone 8 | clone 9 | clone 10 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Result: | wrong result | wrong result | wrong result | wrong result | wrong result | wrong result | wrong result | wrong result | correct size, but additional bands | correct size, but additional bands |
| Lane: | 2log DNA ladder | clone 1 | clone 2 | clone 3 | clone 4 | clone 5 | clone 6 | clone 7 | clone 8 | clone 9 | clone 10 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Result: | wrong result | wrong result | correct size, but additional bands | wrong result | wrong result | correct size, but additional bands | wrong result | wrong result | wrong result | wrong result |
Saturday, August 10th
Miniprep of E. coli Xl1-Blue transformed with ligation products F123+F219 (SYFP RFC25) and F123+F220 (eCFP RFC25)
Investigator: Jeff
Aim of the experiment: Miniprep of E. coli Xl1-Blue transformed with ligation products F123+F219 (SYFP RFC25) and F123+F220 (eCFP RFC25).
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Spin Miniprep Kit, QIAGEN).
- The resulting tubes were labeled as P614 to P617.
Analytical digestion and gelelectrophoresis of P614 – P617
Investigator: Jeff
Aim of the experiment: Analytical digestion and gelelectrophoresis of P614 – P617.
Procedure:
Mastermix for analytical digestion with EcoRI and PstI
| volume | reagent |
|---|---|
| 2.5 µl | Plasmid DNA (P614 – P617) |
| 2 µl | Cut Smart buffer |
| 0.25 µl | EcoRI-HF |
| 0.25nbsp;µl | PstI-HF |
| 15 µl | ddH2O |
| =20 µl | TOTAL |
- Analytical digestion was performed at 37 °C for 1 h.
- After incubation 2 µl of DNA loading buffer (10x) were added to the reaction batches and were loaded on a 1% agarose gel.
- Analytical gelelectrophoresis was performed at 120 V for 30 min.
| Lane: | 2 log DNA ladder | Digestion of P614 (SYFP2 RFC25) | Digestion of P615 (SYFP2 RFC25) | Digestion of P616 (eCFP RFC25) | Digestion of P617 (eCFP RFC25) |
|---|---|---|---|---|---|
| Result: | as expected (723 bp) | as expected (723 bp) | as expected (723 bp) | as expected (723 bp) |
Sunday, August 11th
Preparative digestion of P615 (SYFP2 RFC25)
Investigator: Jeff
Aim of the experiment: Preparative digestion of P615 (SYFP2 RFC25)
Procedure:
Preparative digestion and gelelectrophoresis of P617 (eCFP RFC25)
Investigator: Jeff
Aim of the experiment: Preparative digestion of P617 (eCFP RFC25)
Procedure:
Ligation of F224+F223 (creation of Gly-TEV-site-Gly_SYFP2)
Investigator: Jeff
Aim of the experiment: Ligation of F224+F223 (creation of Gly-TEV-site-Gly_SYFP2).
Procedure:
Ligation batch for F224+F223 (SYFP2 + Gly-TEV-site-Gly), vector:insert = 1:3:
| volume | reagent |
|---|---|
| 0.66 µl | F224 (152.1 ng/µl, 2086 bp) |
| 0.34 µl | F223 (2506.9 ng/µl, 35 bp) |
| 16 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- Ligation was performed at 16 °C overnight.
Miniprep of Retrafos of P516, P330, P245, P496, P346, P196, P20, P476, P554, P498, P246, P354, P428, P512, P402, P197, P350, P502
Investigator: Louise, Christopher
Aim of the experiment: Miniprep of Retrafos of P516, P330, P245, P496, P346, P196, P20, P476, P554, P498, P246, P354, P428, P512, P402, P197, P350, P502.
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Spin Miniprep Kit, QIAGEN).
- The new plasmids were numbered P618 - P635 (see Inventory)
Week 17
Monday, August 12th
Sequencing of P509, P512, P516, P537, P540, P562, P565, P583, P591
Investigator: Louise
Aim of the experiment: Sequencing of P509, P512, P516, P537, P540, P562, P565, P583, P591
Procedure: The DNA was prepared for sequencing according to the Eurofins SmartSeq Kit protocol (15 µl of plasmid DNA (50 - 100 ng) and 2 µl sequencing primer).
The different genes we sequenced received the following barcodes:
| Label | Barcode1 (fw) | Barcode2 (rev) |
|---|---|---|
| P509 | FR02233103 | |
| P632 | FR02233102 | FR02233101 |
| P633 | FR02233100 | FR02233099 |
| P537 | FR02233098 | FR02233097 |
| P540 | FR02233096 | FR02233104 |
| P562 | FR02233105 | FR02233106 |
| P565 | FR02233107 | FR02233108 |
| P583 | FR02233109 | FR02233110 (Primer O131) |
| P591 | FR02233111 | FR02233088 (Primer O131) |
Transformation of E. coli XL1-Blue with P337,P540, P583 and P591
Investigator: Johanna
Aim of the experiment: Retransformation of E. coli XL1-Blue with P337,P540, P583 and P591
Procedure:
- 1,5 µl of dd H2O were added to the empty tubes
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 1,5 µl of DNA were added to the tubes containing 150 µl of competent cells and gently mixed
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- The cell suspension were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated on new chlorampenicol plates.
Transformation of E. coli XL1-Blue with ligation product F224+F223 (SYFP2 + G-TEV-site-G) and negative control of F224
Investigator: Louise
Aim of the experiment: Transformation of E. coli XL1-Blue with ligation product F224+F223 (SYFP2 + G-TEV-site-G) and negative control of F224
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 1,5 µl of DNA were added to the tubes containing 150 µl of competent cells and gently mixed
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- The cell suspension were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated on new chlorampenicol plates.
Tuesday, August 13th
Gelextraction and Ligation of digested P583 (PppActin5-MCS-T35S-npt) and P591 (PppActin5-MCS-T35S-npt)
Investigator: Louise
Aim of the experiment: Gelextraction and selfligation to verify correct cut of digested P583 (PppActin5-MCS-T35S-npt) and P591 (PppActin5-MCS-T35S-npt)
Procedure:
- After preparative digestion of P583 and P591 with SpfI and MfeI the gel extraction was performed after manufacturer`s protocol (QIAquick Gel Extraction Kit, QIAGEN)
- the extracted fragments were labelled F226 (digested P583) and F227 (digested P591)
- ligation was performed without extra inserts with just 100 ng of cut plasmid to test if it was cut as expected
- Ligation batch for self-ligation of F226
| volume | reagent |
|---|---|
| 0.6 µl | F226 (164.1 ng/µl) |
| 16.4 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- Ligation batch for self-ligation of F227
| volume | reagent |
|---|---|
| 0.4 µl | F227 (243.4 ng/µl) |
| 16.6 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
Transformation of E. coli XL1-Blue with ligation product of self-ligated F226 and F227 and with Biobrick BBa_J45014
Investigator: Louise
Aim of the experiment: Transformation of E. coli XL1-Blue with ligation product of self-ligated F226 and F227 and with Biobrick BBa_245014
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 1,5 µl of DNA were added to the tubes containing 150 µl of competent cells and gently mixed
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- The cell suspension were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated on new chlorampenicol plates.
Picking of Retrafos of P337, P540, P583 and P591 and of Ligation F224 + F223
Investigator: Jeff
Aim of the experiment: Picking of Retrafos of P337 (SERK-SigP_XylE), P540 (SERK-SigP_PP1_Strep-tag-II-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1), P583 (PppActin5-MCS-T35S-npt) and P591 (PppActin5-MCS-T35S-npt) and of Ligation F224 + F223 (SYFP2 RFC25 in pSB1C3 + Oligohybridization of G-TEV-site-G)
Procedure:
- Retrafos 1 clone each, Ligation 4 clones due to approx. 50 % positive neg. ctrl
- 4 ml LB medium w/ 4 µl chloramphenicol
- Incubation overnight at 37 °C in the 180 rpm
Wednesday, August 14th
Picking of of E. coli XL1 blue transformed with BBa_J45014 (banana odour)
Investigator: Louise
Aim of the experiment: Picking of of E. coli XL1 blue transformed with BBa_J45014 (banana odour)
Procedure:
- BBa_J45014 (banana odour): 4 Colonies were picked from chloramphenicol plates.
- Each colony was transferred into a tube containing 4 mL of LB-medium + 4 µL chloramphenicol(1000x).
- These tubes were transferred into a cell culture shaker at 37 °C and were incubated overnight
Analytical digestion and gelelectrophoresis of P640-P643 (G-TEV-site-G_SYFP2) and P615 with EcoRI and PstI and P637, P639 with EcoRI and MfeI (old and new)
Investigator: Louise, Johanna
Aim of the experiment: Analytical digestion and gelelectrophoresis of P640-P643 (G-TEV-site-G_SYFP2) and P615 (SYFP2, RFC25) with EcoRI and PstI and P637 (PppActin5-MCS-T35S-npt), P639 (PppActin5-MCS-T35S-npt) with EcoRI and MfeI (old and new)
Procedure:
- batch for digestion of P640-P643
| volume | reagent |
|---|---|
| 8.5 µl | Plasmid DNA |
| 2 µl | Cut Smart buffer |
| 0.25 µl | EcoRI-HF |
| 0.25 µl | PstI-HF |
| 9 µl | ddH2O |
| =20 µl | TOTAL |
- batch for digestion of P615
| volume | reagent |
|---|---|
| 2.5 µl | Plasmid DNA |
| 2 µl | Cut Smart buffer |
| 0.25 µl | EcoRI-HF |
| 0.25 µl | PstI-HF |
| 15 µl | ddH2O |
| =20 µl | TOTAL |
- batch for digestion of P637 and P639
| volume | reagent |
|---|---|
| 8.5 µl | Plasmid DNA |
| 2 µl | Cut Smart buffer |
| 0.25 µl | EcoRI-HF |
| 0.25 µl | MfeI (old or new) |
| 9 µl | ddH2O |
| =20 µl | TOTAL |
- Analytical digestion was performed at 37 °C for 1.5 h.
- After incubation 2 µl of DNA loading buffer (10x) were added to the reaction batches and were loaded on a 1% agarose gel.
- Analytical gelelectrophoresis was performed at 90 V for 1 h.
| Lane: | DNA ladder | Digestion of P615 (SYFP2 RFC25-reference) | Digestion of P637 (with old MfeI) | Digestion of P637 (with new MfeI) | Digestion of P639 (with old MfeI) | Digestion of P639 (with new MfeI) | Digestion of P640 | Digestion of P641 | Digestion of P642 | Digestion of P643 |
|---|---|---|---|---|---|---|---|---|---|---|
| Result: | as expected | corrupt MfeI | as expected | corrupt MfeI | as expected | as expected | as expected | as expected | as expected |
Sequencing of P614, P617, P637, P639 and P641
Investigator: Flo, Johanna
Aim of the experiment: Sequencing of P614, P617, P637, P639 and P641
Procedure: The DNA was prepared for sequencing according to the Eurofins SmartSeq Kit protocol (15 µl of plasmid DNA (50 - 100 ng) and 2 µl sequencing primer).
The different genes we sequenced received the following barcodes:
| Label | Barcode | Primer |
|---|---|---|
| P614 | FR02233112 | 03 |
| P617 | FR02233113 | 03 |
| P641 | FR02233114 | 03 |
| P637 | FR02233119 | 061 |
| P639 | FR02233115 | 061 |
Miniprep of Retrafos of P337, P583, P540, P591 and of ligation product F224+F223
Investigator: Johanna
Aim of the experiment: Miniprep of Retrafos of P337, P583, P540, P591 and of ligation product F224+F223
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Spin Miniprep Kit, QIAGEN).
- The new plasmids were numbered as followed:
| Plasmid | Miniprep-Plasmid |
|---|---|
| P337 | P636 |
| P583 | P637 |
| P540 | P638 |
| P591 | P639 |
| F224+223 (version 1) | P640 |
| F224+223 (version 2) | P641 |
| F224+223 (version 3) | P642 |
| F224+223 (version 4) | P643 |
Miniprep of Retrafos of banana odour, P583 and P591
Investigator: Florian
Aim of the experiment: Miniprep of Retrafos of banana odour, P583 and P591.
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Spin Miniprep Kit, QIAGEN).
- The new plasmids were numbered as followed:
| Plasmid | Miniprep-Plasmid |
|---|---|
| Banana 1 | P644 |
| Banana 2 | P645 |
| Banana 3 | P646 |
| Banana 4 | P647 |
| P583 | P648 |
| P591 | P649 |
Preparative digestion of P644 (banana odour), P648 & P649 (PppAct5-mMCS-T35S-npt)
Investigator: Florian
Procedure:
Reaction batch for P644:
| volume | reagent |
|---|---|
| 20 µl | P644 |
| 5 µl | CutSmart Buffer (10x) |
| 1 µl | EcoRI (20 U/µl) |
| 1 µl | PstI (20 U/µl) |
| 23 µl | ddH2O |
| =50 µl | TOTAL |
Reaction batch for P648:
| volume | reagent |
|---|---|
| 40 µl | P648 |
| 10 µl | CutSmart Buffer (10x) |
| 2 µl | MfeI (20 U/µl) |
| 0 µl was empty :'( | SbfI (20 U/µl) |
| 46 µl | ddH2O |
| =50 µl | TOTAL |
Reaction batch for P649:
| volume | reagent |
|---|---|
| 40 µl | P649 |
| 10 µl | CutSmart Buffer (10x) |
| 2 µl | MfeI (20 U/µl) |
| 2 µl | SbfI (20 U/µl) |
| 46 µl | ddH2O |
| =50 µl | TOTAL |
- Reaction batches were incubated at 37 °C for 3 h.
| Lane | 2 log DNA ladder | Digestion of P428 with EcoRI & XbaI | Digestion of P554 with EcoRI & SpeI |
|---|---|---|---|
| Results | as expected; highest band was cut out | as expected; highest band was cut out |
The bands were extracted from the gel and termed F221 and F222. The concentrations were determined to 42 and 37 ng/µl.
Thursday, August 15th
Preparative digestion of P414 and P640 + Preparative Gelelectrophoresis + Gelextraction
Investigator: Andi
Procedure:
Reaction batch for P414:
| volume | reagent |
|---|---|
| 20 µl | P414 |
| 4 µl | CutSmart Buffer (10x) |
| 1 µl | EcoRI (20 U/µl) |
| 1 µl | PstI (20 U/µl) |
| 14 µl | ddH2O |
| =40 µl | TOTAL |
Reaction batch for P640:
| volume | reagent |
|---|---|
| 20 µl | P640 |
| 4 µl | CutSmart Buffer (10x) |
| 1 µl | EcoRI (20 U/µl) |
| 2 µl | NgoMIV (20 U/µl) |
| 13 µl | ddH2O |
| =40 µl | TOTAL |
- Reaction batches were incubated at 37 °C for 2.5 h.
- Preparative gelelectrophoresis was performed at 90 V for 1 h in an 1% agarose gel.
| Lane | Digestion of P414 with EcoRI & PstI | 1kB DNA ladder | Digestion of P640 with EcoRI & NgoMIV |
|---|---|---|---|
| Results | as expected; upper band was cut out | as expected; lower band was cut out |
- Gelextraction was performed after preparative Gelelectrophoresis
- The resulting fragments were named F231 (Laccase) + F232
Ligation of F230 with F191, F192, F193, F194, F195, F196, F197, F198, F199, F200, F201, F202, F203, F204, F205, F214, F217 and F218
Investigator: Louise, Florian
Aim of the experiment: Ligation of F230 with F191, F192, F193, F194, F195, F196, F197, F198, F199, F200, F201, F202, F203, F204, F205, F214, F217 and F218
Procedure:
Ligation batch for F230+F191 (plasmid with mini MCS + GFP cytoplasmatic), vector:insert = 1:3
| volume | reagent |
|---|---|
| 0.41 µl | F230 (243.7 ng/µl, 5796 bp) |
| 18.7 µl | F191 (2 ng/µl, 723 bp) |
| 0.0 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
Ligation batch for F230+F192 (plasmid with mini MCS + catecholdioxygenase cytoplasmatic), vector:insert = 1:3
| volume | reagent |
|---|---|
| 0.41 µl | F230 (243.7 ng/µl, 5796 bp) |
| 5.94 µl | F192 (8 ng/µl, 918 bp) |
| 10.65 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
Ligation batch for F230+F193 (plasmid with mini MCS + EreB cytoplasmatic), vector:insert = 1:3
| volume | reagent |
|---|---|
| 0.41 µl | F230 (243.7 ng/µl, 5796 bp) |
| 1.85 µl | F193 (35 ng/µl, 1254 bp) |
| 14.74 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
Ligation batch for F230+F194 (plasmid with mini MCS + Nanoluciferase cytoplasmatic), vector:insert = 1:3
| volume | reagent |
|---|---|
| 0.41 µl | F230 (243.7 ng/µl, 5796 bp) |
| 8.80 µl | F194 (3 ng/µl, 510 bp) |
| 7.79 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
Ligation batch for F230+F195 (plasmid with mini MCS + nanoluciferase secretory_SERK), vector:insert = 1:3
| volume | reagent |
|---|---|
| 0.41 µl | F230 (243.7 ng/µl, 5796 bp) |
| 3.99 µl | F195 (8 ng/µl, 616 bp) |
| 12.60 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
Ligation batch for F230+F196 (plasmid with mini MCS + gutathiontransferase cytoplasmatic), vector:insert = 1:3
| volume | reagent |
|---|---|
| 0.41 µl | F230 (243.7 ng/µl, 5796 bp) |
| 1.17 µl | F196 (28 ng/µl, 633 bp) |
| 15.42 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
Ligation batch for F230+F197 (plasmid with mini MCS + EreB membrane associated), vector:insert = 1:3
| volume | reagent |
|---|---|
| 0.41 µl | F230 (243.7 ng/µl, 5796 bp) |
| 11.12 µl | F197 (11 ng/µl, 2362 bp) |
| 5.47 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
Ligation batch for F230+F198 (plasmid with mini MCS + Nanoluc membrane associated), vector:insert = 1:3
| volume | reagent |
|---|---|
| 0.41 µl | F230 (243.7 ng/µl, 5796 bp) |
| 6.44 µl | F198 (13 ng/µl, 1618 bp) |
| 10.15 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
Ligation batch for F230+F199 (plasmid with mini MCS + nanoluc secretory_Ig Kappa), vector:insert = 1:3
| volume | reagent |
|---|---|
| 0.41 µl | F230 (243.7 ng/µl, 5796 bp) |
| 1.16 µl | F199 (26 ng/µl, 583 bp) |
| 15.43 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
Ligation batch for F230+F200 (plasmid with mini MCS + Nanoluc+IRES+Spytag_Nanoluc), vector:insert = 1:3
| volume | reagent |
|---|---|
| 0.41 µl | F230 (243.7 ng/µl, 5796 bp) |
| 3.00 µl | F200 (47 ng/µl, 2725 bp) |
| 13.59 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
Ligation batch for F230+F201 (plasmid with mini MCS + Nanoluc+IRES+Nanoluc_Spycatcher), vector:insert = 1:3
| volume | reagent |
|---|---|
| 0.41 µl | F230 (243.7 ng/µl, 5796 bp) |
| 3.81 µl | F201 (37 ng/µl, 2725 bp) |
| 12.78 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
Ligation batch for F230+F202 (plasmid with mini MCS + Nanoluc+IRES+Nanoluc_Spytag), vector:insert = 1:3
| volume | reagent |
|---|---|
| 0.41 µl | F230 (243.7 ng/µl, 5796 bp) |
| 2.66 µl | F202 (53 ng/µl, 2725 bp) |
| 13.93 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
Ligation batch for F230+F203 (plasmid with mini MCS + IgKappa_GFP secretory), vector:insert = 1:3
| volume | reagent |
|---|---|
| 0.41 µl | F230 (243.7 ng/µl, 5796 bp) |
| 1.09 µl | F203 (38 ng/µl, 800 bp) |
| 15.50 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
Ligation batch for F230+F204 (plasmid with mini MCS + Nanoluc+IRES+Spycatcher_Nanoluc), vector:insert = 1:3
| volume | reagent |
|---|---|
| 0.41 µl | F230 (243.7 ng/µl, 5796 bp) |
| 2.17 µl | F204 (65 ng/µl, 2725 bp) |
| 14.42 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
Ligation batch for F230+F205 (plasmid with mini MCS + FluA membrane associated), vector:insert = 1:3
| volume | reagent |
|---|---|
| 0.41 µl | F230 (243.7 ng/µl, 5796 bp) |
| 2.11 µl | F205 (40 ng/µl, 1630 bp) |
| 14.48 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
Ligation batch for F230+F214 (plasmid with mini MCS + PP1 membrane associated), vector:insert = 1:3
| volume | reagent |
|---|---|
| 0.41 µl | F230 (243.7 ng/µl, 5796 bp) |
| 0.95 µl | F214 (113 ng/µl, 2080 bp) |
| 15.64 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
Ligation batch for F230+F217 (plasmid with mini MCS + PIF3 safety), vector:insert = 1:3
| volume | reagent |
|---|---|
| 0.41 µl | F230 (243.7 ng/µl, 5796 bp) |
| 2.76 µl | F217 (116 ng/µl, 6191 bp) |
| 13.83 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
Ligation batch for F230+F217 (plasmid with mini MCS + PIF3 safety), vector:insert = 3:1
| volume | reagent |
|---|---|
| 1.15 µl | F230 (243.7 ng/µl, 5796 bp) |
| 0.86 µl | F217 (116 ng/µl, 6191 bp) |
| 14.99 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
Ligation batch for F230+F218 (plasmid with mini MCS + PIF6 safety), vector:insert = 1:3
| volume | reagent |
|---|---|
| 0.41 µl | F230 (243.7 ng/µl, 5796 bp) |
| 2.50 µl | F218 (128 ng/µl, 6191 bp) |
| 14.09 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
Ligation batch for F230+F218 (plasmid with mini MCS + PIF3 safety), vector:insert = 3:1
| volume | reagent |
|---|---|
| 1.15 µl | F230 (243.7 ng/µl, 5796 bp) |
| 0.78 µl | F218 (116 ng/µl, 6191 bp) |
| 15.07 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- Ligation was performed at roomtemperature, 1 h
Ligation of F230 with F231
Investigator: Louise, Christopher
Aim of the experiment: Ligation of F230 with F231
Procedure:
Ligation batch for F230+F231 (plasmid with mini MCS + Laccase membrane-associated), vector:insert = 1:3
| volume | reagent |
|---|---|
| 0.41 µl | F230 (243.7 ng/µl, 5796 bp) |
| 14.33 µl | F231 (9.5 ng/µl, ~2630 bp) |
| 2.26 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- Ligation was performed at room temperature, 1 h
Transformation of E. coli XL1-Blue with ligation products F230+F191, F230+F192, F230+F193, F230+F194, F230+F195, F230+F196, F230+F197, F230+F198, F230+F199, F230+F200, F230+F201, F230+F202, F230+F203, F230+F204, F230+F205, F230+F214, F230+F217, F230+F218, F217+F230, F218+F230 and negative control of F230
Investigator: Louise, Christopher
Aim of the experiment: Transformation of E. coli XL1-Blue with ligation products F230+F191, F230+F192, F230+F193, F230+F194, F230+F195, F230+F196, F230+F197, F230+F198, F230+F199, F230+F200, F230+F201, F230+F202, F230+F203, F230+F204, F230+F205, F230+F214, F230+F217, F230+F218, F217+F230, F218+F230 and negative control of F230
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 1,5 µl of DNA were added to the tubes containing 150 µl of competent cells and gently mixed
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- The cell suspension were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated on new chlorampenicol plates.
Friday, August 16th
Sequencing of P640 (SYFP2 RFC25 in pSB1C3 + Oligohybridization of G-TEV-site-G)
Investigator: Rosario
Aim of the experiment: Sequencing of P640.
Procedure: The DNA was prepared for sequencing according to the Eurofins SmartSeq Kit protocol (15 µl of plasmid DNA (50 - 100 ng) and 2 µl sequencing primer).
The different genes we sequenced received the following barcodes:
| Label | Barcode | Primer |
|---|---|---|
| P640 | FR02233116 | 03 |
Preparative digestion of P640 (SYFP2 RFC25 in pSB1C3 + Oligohybridization of G-TEV-site-G) with EcoRI+NgoMIV
Investigators: Louise, Jeff
Aim of the experiment: Preparative digestion of P640 (SYFP2 RFC25 in pSB1C3 + Oligohybridization of G-TEV-site-G) with EcoRI+NgoMIV
Procedure:
| volume | reagent |
|---|---|
| 33 µl | P640 |
| 4 µl | CutSmart Buffer (10x) |
| 2 µl | NgoMIV (10 U/µl) |
| 1 µl | EcoRI Hf (20 U/µl) |
| =40 µl | TOTAL |
- Reaction batches were incubated at 37 °C for 3 h.
| Lane | 2 log DNA ladder | Digestion of P640 with EcoRI & NgoMIV |
|---|---|---|
| Results | as expected; band was cut out |
The band was extracted from the gel via QIAGEN Gel Extraction Kit and termed F233. The concentration was 31.1 ng/µl.
Transformation of E. coli XL1-Blue with ligation product F230+F231(plasmid with mMCS+membrane-bound Laccase) and negative control of F230, Retrafo of P541 (PppActin5) and P449(stress inducible Promoter_RFP +t35S-NPT-pSB1C3)
Investigator: Rosario
Aim of the experiment: Transformation of E. coli XL1-Blue with ligation product F230+F231(plasmid with mMCS+membrane-bound Laccase) and negative control of F230, Retrafo of P541 (PppActin5) and P449(stress inducible Promoter_RFP +t35S-NPT-pSB1C3).
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 1,5 µl of DNA were added to the tubes containing 150 µl of competent cells and gently mixed
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- The cell suspension were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated on new chlorampenicol plates.
Ligation of F233+F225 (creation of eCFP_Gly-TEV-site-Gly_SYFP2)
Investigator: Jeff
Aim of the experiment: Creation of eCFP_Gly-TEV-site-Gly_SYFP2
Procedure:
Ligation batch, vector:insert = 1:3
| volume | reagent |
|---|---|
| 3.22 µl | F233 (31.1 ng/µl, 2834 bp) |
| 1.93 µl | F225 (39.6 ng/µl, 723 bp) |
| 11.85 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- neg. ctrl. w/ water instead of insert
- Ligation was performed at room temperature, 1 h
Miniprep of E. coli XL1-Blue transformed with ligation products F230+F191, F230+F192, F230+F193, F230+F194, F230+F195, F230+F196, F230+F197, F230+F198, F230+F199, F230+F200, F230+F201, F230+F202, F230+F203, F230+F204, F230+F205, F230+F214, F230+F217, F217+F230, F230+F218, F218+F230, F230+F228
Investigator: Jeff, Rosario, Louise, Christopher
Aim of the experiment: Miniprep of E. coli XL1-Blue transformed with ligation products F230+F191, F230+F192, F230+F193, F230+F194, F230+F195, F230+F196, F230+F197, F230+F198, F230+F199, F230+F200, F230+F201, F230+F202, F230+F203, F230+F204, F230+F205, F230+F214, F230+F217, F217+F230, F230+F218, F218+F230, F230+F228
Procedure:
- Ligation products were labelled B to V (in the same order as in Aim of the experiment, above)
- Two clones for every ligation product except for H, L and O
- Miniprep was performed after manufacturer's protocol (QIAprep Spin Miniprep Kit, QIAGEN).
- The resulting tubes were labeled as P650 to P688.
Analytical digestion and gelelectrophoresis of P650-P688 with XbaI and PstI-HF
Investigator: Jeff, Rosario, Louise, Christopher
Aim of the experiment: Analytical digestion and gelelectrophoresis of P650-P688 with XbaI and PstI-HF
Procedure:
- batch for digestion of P650-P688
| volume | reagent |
|---|---|
| 5 µl | Plasmid DNA |
| 1 µl | Cut Smart buffer |
| 0.2 µl | XbaI |
| 0.2 µl | PstI-HF |
| 3.6 µl | ddH2O |
| =10 µl | TOTAL |
- Analytical digestion was performed at 37 °C for 1 h.
- After incubation 2 µl of DNA loading buffer (10x) were added to the reaction batches and were loaded on a 1% agarose gel.
- Analytical gelelectrophoresis was performed at 90 V for 1 h.
Saturday, August 17th
Picking of of E. coli XL1 blue transformed with P449, P541, F230+F231, 2x K-Platte, 1x L-Platte, 2x M-Platte, 1x O-Platte
Investigator: Andi, Johanna
Aim of the experiment: Picking of of E. coli XL1 blue transformed with P449, P541, F230+F231, 2x K-Platte, 1x L-Platte, 2x M-Platte, 1x O-Platte
Procedure:
- F230+F231: 8 Colonies were picked from chloramphenicol plates.
- P449: 4 Colonies were picked from chloramphenicol plates.
- P541: 4 Colonies were picked from ampicilin plates.
- K-Plate: 4 Colonies were picked from chloramphenicol plates.
- L-Plate: 4 Colonies were picked from chloramphenicol plates.
- M-Plate: 4 Colonies were picked from chloramphenicol plates.
- O-Plate: 4 Colonies were picked from chloramphenicol plates.
- Each colony was transferred into a tube containing a relation of 1:1000 of LB-medium : antibiotic
- These tubes were transferred into a cell culture shaker at 37 °C and were incubated 16 h
Preparative digestion of P625, P516, P644 + Preparative Gelelectrophoresis + Gelextraction
Investigator: Andi, Johanna
Aim of Experiment: Obtaining digested fragments of P625 (SERK-SigP_EreB_Strep-tag-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1), P516 (SERK-SigP_FluA_Strep-tag-II-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1)and P644 (Banana odour BBa_J45014)
Procedure:
Reaction batch for P625, P516 and P644:
| volume | reagent |
|---|---|
| 20 µl | PXXX |
| 4 µl | CutSmart Buffer (10x) |
| 2 µl | EcoRI (20 U/µl) |
| 2 µl | PstI (20 U/µl) |
| 12 µl | ddH2O |
| =40 µl | TOTAL |
- Reaction batches were incubated at 37 °C for 2.5 h.
- Preparative gelelectrophoresis was performed at 90 V for 1 h in an 1% agarose gel.
- Gelextraction was performed after preparative Gelelectrophoresis
- The resulting fragments were named F234 (digestion of P516), F235 (digestion of P625), F236 (digestion of P644).
Ligation of F230+ F234, F230+ F235, F230+F236
Investigator: Johanna, Andi
Aim of the experiment: Ligation of F230+ F234, F230+ F235, F230+F236
Procedure:
Ligation batch for F230+F234, vector:insert = 1:3
| volume | reagent |
|---|---|
| 6.3 µl | F234(228 ng/µl, 1600 bp) |
| 0.4 µl | F230 (100 ng/µl, bp) |
| 10.3 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
Ligation batch for F230+F235, vector:insert = 1:3
| volume | reagent |
|---|---|
| 9.0 µl | F235(214 ng/µl, 2300 bp) |
| 0.4 µl | F230 (100 ng/µl, bp) |
| 7.6 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
Ligation batch for F230+F236, vector:insert = 1:3
| volume | reagent |
|---|---|
| 16.6 µl | F236(330 ng/µl, 1500 bp) |
| 0.4 µl | F230 (100 ng/µl, bp) |
| 0.0 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- neg. ctrl. w/ water instead of insert
- Ligation was performed at room temperature, 1 h
Miniprep of F230+F231 (version 1-4), P541 (version 1-2), K3, K4, L2, L3, M3, M4, O2, O3, P3-P10 and P449 (version 1-2)
Investigator: Johanna, Andi
Aim of the experiment: Miniprep of F230+F231 (version 1-4), P541 (version 1-2), K3, K4, L2, L3, M3, M4, O2, O3, P3-P10 and P449 (version 1-2)
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Spin Miniprep Kit, QIAGEN).
- The new plasmids were numbered as followed:
| Plasmid | Miniprep-Plasmid |
|---|---|
| F230+F231 (version 1) | P689 |
| F230+F231 (version 2) | P690 |
| F230+F231 (version 3) | P691 |
| F230+231 (version 4) | P692 |
| P541 (version 1) | P693 |
| P541 (version 2) | P694 |
| K3 | P695 |
| K4 | P696 |
| L2 | P697 |
| L3 | P698 |
| M3 | P699 |
| M4 | P700 |
| O2 | P701 |
| O3 | P702 |
| P3 | P703 |
| P4 | P704 |
| P5 | P705 |
| P6 | P706 |
| P7 | P707 |
| P8 | P708 |
| P9 | P709 |
| P10 | P710 |
| P449 (version 1) | P711 |
| P449 (version 2) | P712 |
Analytical digestion and gelelectrophoresis of P689-P712 with XbaI and PstI
Investigator: Andi, Johanna
Aim of the experiment: Analytical digestion and gelelectrophoresis of P689-P712 with XbaI and PstI
Procedure:
- batch for digestion of P689-712
| volume | reagent |
|---|---|
| 5 µl | Plasmid DNA |
| 1 µl | Cut Smart buffer |
| 0.2 µl | XbaI-HF |
| 0.2 µl | PstI-HF |
| 3.6 µl | ddH2O |
| =10 µl | TOTAL |
- Analytical digestion was performed at 37 °C for 1.5 h.
- After incubation 2 µl of DNA loading buffer (10x) were added to the reaction batches and were loaded on a 1% agarose gel.
- Analytical gelelectrophoresis was performed at 90 V for 1 h.
| Lane | 1kB DNA ladder | Digestion of P689 | Digestion of P690 | Digestion of P691 | Digestion of P692 | Digestion of P693 | Digestion of P694 | Digestion of P695 | Digestion of P696 | Digestion of P697 | Digestion of P698 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Results | as expected | as expected | as expected | as expected | as expected | weird | weird | as expected | as expected | as expected | weird |
| Lane | 1kB DNA ladder | Digestion of P699 | Digestion of P700 | Digestion of P701 | Digestion of P702 | Digestion of P703 | Digestion of P704 | Digestion of P705 | Digestion of P706 | Digestion of P707 | Digestion of P708 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Results | as expected | as expected | as expected | weird | as expected | as expected | as expected | as expected | as expected | as expected | as expected |
| Lane | 1kB DNA ladder | Digestion of P709 | Digestion of P710 | Digestion of P711 | Digestion of P712 |
|---|---|---|---|---|---|
| Results | as expected | as expected | as expected | as expected | as expected |
Transformation of E. coli XL1-Blue with ligation product F230+F234, F230+F235, F230+F236
Investigator: Johanna, Andi
Aim of the experiment: Transformation of E. coli XL1-Blue with ligation product F230+F234, F230+F235, F230+F236.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 1,5 µl of DNA were added to the tubes containing 150 µl of competent cells and gently mixed
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- The cell suspension were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated on new chlorampenicol plates.
Sunday, August 18th
BITTE VERVOLLSTÄNDIGEN (P-Nummern) Midiprep of Safety_PIF3, PP1 membrane bound, Safety_PIF6, Safety_PIF3, Positve control mit PppActin5, EreB cyt., NanoLuc cyt., XylE cyt., NanoLuc sek. (IgKappa), NanoLuc sek. (SERK), GFP cyt., GFP sek. (IgKappa), FluA membrane bound, stress-inducible promoter, GST, NanoLuc membrane bound
Investigator: Andreas, Rosario
Aim of the experiment: Midiprep of RFC25 compatible RFP generator in (pSB1C3)
Procedure:
- Midiprep was performed after manufacturer's protocol (QIAprep Midiprep, QIAGEN)
Digestion of BITTE VERVOLLSTÄNDIGEN
Investigator: Ingmar
Aim of the experiment:
Procedure:
Week 18
Monday, August 19th
Analytical Gelelectrophoresis of digested Midi-Preps F243 - F260
Investigator: Ingmar
Aim of the experiment: Verification of linearisation of digest fragments F234-F260
Procedure:
- Concentrations of digestions were measured (blanked against EB-buffer), see Inventory List
- 5 µl of DNA loading buffer (10x) mixed with 1 µl Midiprep-DNA, on 1% agarose gel.
- 90 V 1 h.
| Lane | 1kB DNA ladder | F234 | F235 | F236 | F237 | F238 | F239 | F240 | F241 | F242 | F243 | F244 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Name | - | Nanoluc secretory | GFP cytoplasmatic | Nanoluc membrane bound | Nanoluc cytoplasmatic | Ere B cytoplasm | PIF6 | PIF3 clone1 | PIF3 clone2 | GFP_IgKappa | PP1 | C1 |
| Results |
| Lane | 1kB DNA ladder | F245 | F246 | F247 | F248 | F249 | F250 | F251 | F252 |
|---|---|---|---|---|---|---|---|---|---|
| Name | - | Safety_PIF3 | PP1 mebrane bound | Safety_PIF6 | Safety_PIF? | pos ctrl mit Ppp Actin5 | Ere B cytoplasm | Nanoluc cytoplasm | XylE cytoplasm |
| Results |
| Lane | 1kB DNA ladder | F253 | F254 | F255 | F256 | F257 | F258 | F259 | F260 |
|---|---|---|---|---|---|---|---|---|---|
| Name | - | Nanoluc secretory IgKappa | Nanoluc secretory SERK | GFP cytoplasm | GFP secretory IgKappa | FluA membrane bound | stress inducible promotor | GST | Nanoluc membrane bound |
| Results | - |
Preparation of the linearized DNA (F234-F260) for the Moss Transformation
Investigator: Ingmar
Aim of the experiment: Preparation of the linearized DNA (F234-F260) for the Moss Transfection
Procedure:
- The DNA pellets were solved in 500 µl ddH2O
- Linerization of the DNA with EcoRI: 56,6 µl CutSmart buffer and 10 µl EcoRI were added to the 500 µl DNA. Incubation over night at 37 °C.
- For purification the DNA was again precipitated with pure isopropanol: Addition of 500 µl isopropanol, mixing by inverting, centrifugation at 16,200 g for 40 min; supernatant was discarded
- Wash pellet with 200 µl 70 % ethanol, centrifugation at 16,200 g for 40 min, supernatant discarded. The pellet was air dried at 50 °C for 8 min.
- The DNA pellets were dissolved in the respectively calculated volumes of 0,1 M Ca(NO3)2 soultion to obtain final concentrations of 250 ng/µl.
| fragment | name | concentration | concentration w/ loss | volume 0,1 M Ca(NO3)2 |
|---|---|---|---|---|
| F234 | Nanoluc SERK-SigP | 440,8 | 396,72 | 872,78 |
| F235 | GFP cytoplasm | 241,8 | 217,62 | 478,76 |
| F236 | Nanoluc membrane | 339,9 | 305,91 | 673,00 |
| F237 | Nanoluc cytoplasm | 219,1 | 197,19 | 433,82 |
| F238 | EreB cytoplasm | 355,9 | 320,31 | 704,68 |
| F239 | PIF6 | 669,9 | 602,91 | 1326,40 |
| F240 | PIF3 (1) | 438,1 | 394,29 | 867,438 |
| F241 | PIF3 (2) | 642,9 | 578,61 | 1272,942 |
| F242 | GFP IgKappa | 339,4 | 305,46 | 672,012 |
| F243 | PP1 receptor | 333,5 | 300,15 | 660,33 |
| F244 | Catecholdioxygenase cytoplasm | 342,4 | 308,16 | 677,952 |
Tuesday, August 20th
Preparative digestion + Gelelectrophoreses + Gelextraction of P643/F261
Investigator: Andi, Rosario, Johanna
Aim of the experiment: Preparative digestion + Gelelectrophoreses + Gelextraction of P643/F261.
Procedure:
Reaction batch for P643:
| volume | reagent |
|---|---|
| 20 µl | P643 |
| 5 µl | CutSmart Buffer (10x) |
| 2 µl | EcoRI (20 U/µl) |
| 2 µl | NgoMIV (20 U/µl) |
| 12 µl | ddH2O |
| =40 µl | TOTAL |
Result of the Gelelectrophoresis:
- Reaction batch was incubated at 37 °C for 2 h.
- Digested product was purified with QIAquick PCR Purification Kit, QIAGEN.
- Gelelectrophoresis was performed for 1 h and 90V.
- Gelextraction was performed after manufacturers protocol and the resulting Fragment was called F261.
Sequencing of P643 (SYFP2+ G-TEV-site-G)
Investigator: Andi
Aim of the experiment: Sequencing of P643 (SYFP2+ G-TEV-site-G)
Procedure: The DNA was prepared for sequencing according to the Eurofins SmartSeq Kit protocol (15 µl of plasmid DNA (50 - 100 ng) and 2 µl sequencing primer).
The different genes we sequenced received the following barcodes:
| Label | Barcode | Primer |
|---|---|---|
| P643 | FR0223317 | 03 |
Ligation of F261+F225 (G-TEV-site-G_SYFP2 + eCFP)
Investigator: Rosario, Andi, Johanna
Aim of the experiment: Ligation of F261+F225 (G-TEV-site-G_SYFP2 + eCFP)
Procedure:
Ligation batch for F261+F225, vector:insert = 1:3
| volume | reagent |
|---|---|
| 1,5 µl | F225 |
| 1,2 µl | F261 |
| 14,3 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- neg. ctrl. w/ water instead of insert
- Ligation was performed at room temperature, 1 h
Transformation of E. coli XL1-Blue with ligation product F225+F261 and Retrafo of P643
Investigator: Rosario
Aim of the experiment: Transformation of E. coli XL1-Blue with ligation product F225+F261 and Retrafo of P643.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 5 µl of DNA (Ligation) and 1 µl of DNA (Retrafo) were added to the tubes containing 150 µl of competent cells and gently mixed
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- The cell suspensions were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated on new chlorampenicol plates.
Transformation of Physcomitrella patens with F234-F260
Investigator: Ingmar, Jeff
Aim of the experiment: Transformation of Physcomitrella patens with F234-F260
Procedure:
According to the Reski protocol [http://www.plant-biotech.net/]
- Prepare 4 % driselase solution in 0.5 M mannitol, vortex, keep on rotating platform for 45 min (protected from light) and centrifuge at 3500 rpm, 10 min; sterile filter
- Extraction of moss from bioreactor culture by filtering through protoplast sieve (mesh size 100 µm)
- Addition of 12 ml 0.5 M mannitol solution and 4 ml of driselase stock solution (final concentration: 1 % w/v) and Incubation for 2 h at room temperature on rotating platform (protected from light)
- Gently filter moss material through protoplast sieve (mesh size 100 µm), then filtrate through 50 µm sieve
- Centrifugation at 500 rpm for 10 min with slow acceleration rates in a glass tube and discard supernatant; wash protoplasts with mannitol solution, repeat.
- Resuspend pellet in total 10 ml mannitol
- Determine concentration of protoplasts/ ml
- 100 µl of 250 ng/µl DNA solution with 350 µl PEG 4000 solution (8 g PEG4000 ad 20 g 3M medium, sterile filtered), incubate 30 min while gently mixing every 5 min
- Dilution with 3M medium every 5 min: 1 ml, then 2 ml, then 3, then 4
- Centrifugation at 500 rpm for 10 min, discard supernatant, resuspend in 4 ml regeneration medium and divide into 2 6wells
- Incubation in the dark over night, then light/dark regime of 16/8 h
Wednesday, August 21st
Picking of of E. coli XL1 blue transformed with ligation product F225+F261 and Retrafo of P643
Investigator: Rosario
Aim of the experiment: Picking of of E. coli XL1 blue transformed with ligation product F225+F261 and Retrafo of P643.
Procedure:
- F225+F261: 3 Colonies were picked from chloramphenicol plates.
- P643: 1 Colony was picked from a chloramphenicol plate.
- Each colony was transferred into a tube containing a relation of 1000:1 of LB-medium : antibiotic
- These tubes were transferred into a cell culture shaker at 37 °C and were incubated 12 h
Transformation of E. coli XL1-Blue with ligation product F225+F261
Investigator: Rosario
Aim of the experiment: Transformation of E. coli XL1-Blue with ligation product F225+F261.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 5 µl of DNA were added to the tubes containing 150 µl of competent cells and gently mixed
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- The cell suspensions were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated on new chlorampenicol plates.
Thursday, August 22nd
Miniprep of E. coli Xl1-Blue transformed with ligation product F261 + F225 and Retrafo of P643
Investigator: Florian
Aim of the experiment: Miniprep of E. coli Xl1-Blue transformed with ligation product F261 + F225 and Retrafo of P643.
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Spin Miniprep Kit, QIAGEN).
- The resulting tubes were labeled as P714 to P716.
Analytical digestion and gelelectrophoresis of P714 & P715
Investigator: Florian
Aim of the experiment: Analytical digestion and gelelectrophoresis of P714 & P715.
Procedure:
Batch for digestion with EcoRI & PstI
| volume | reagent |
|---|---|
| 2.5 µl | Plasmid |
| 2 µl | Cut Smart buffer |
| 0.25 µl | EcoRI-HF |
| 0.25 µl | PstI-HF |
| 15 µl | ddH2O |
| =20 µl | TOTAL |
- Analytical digestion was performed at 37 °C for 1 h.
- After incubation 2 µl of DNA loading buffer (10x) were added to the reaction batches and were loaded on a 1% agarose gel.
- Analytical gelelectrophoresis was performed at 90 V for 1 h.
| Lane: | 2log DNA ladder | P714 | P715 |
|---|---|---|---|
| Result: | as expected | as expected |
ReTrafo of E. coli XL1-Blue with P650, P652,P654, P656, P658, P661, P663, P666, P672, P678, P679, P682, P685, P706, P709, P625, P414, P628, P629, P630, P632, P713
Investigator: Rosario
Aim of the experiment: ReTrafo of E. coli XL1-Blue with P650, P652,P654, P656, P658, P661, P663, P666, P672, P678, P679, P682, P685, P706, P709, P625, P414, P628, P629, P630, P632, P713.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 5 µl of DNA (Ligation) and 1 µl of DNA (Retrafo) were added to the tubes containing 150 µl of competent cells and gently mixed
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- The cell suspensions were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated on new chlorampenicol plates.
Preparative digestion of P625, P414 with EcoRI, PstI and MlyI, P628, P629, P630, P632 with EcoRI, PstI and SspI + Preparative Gelelectrophoresis + Gelextraction
Investigator: Johanna, Louise
Procedure:
Reaction batch for P625, P414:
| volume | reagent |
|---|---|
| 20 µl | Plasmid DNA |
| 4 µl | CutSmart Buffer (10x) |
| 1 µl | EcoRI (20 U/µl) |
| 1 µl | PstI (20 U/µl) |
| 2 µl | MlyI (20 U/µl) |
| 12 µl | ddH2O |
| =40 µl | TOTAL |
Reaction batch for P628,P629,P630, P632:
| volume | reagent |
|---|---|
| 20 µl | Plasmid DNA |
| 4 µl | CutSmart Buffer (10x) |
| 1 µl | EcoRI (20 U/µl) |
| 1 µl | PstI (20 U/µl) |
| 2 µl | SspI (20 U/µl) |
| 12 µl | ddH2O |
| =40 µl | TOTAL |
- Reaction batches were incubated at 37 °C for 2.5 h.
- Preparative gelelectrophoresis was performed at 90 V for 1 h in an 0,5% agarose gel.
- Gelextraction was performed after preparative Gelelectrophoresis
- The resulting fragments were named F262 (digestion of P625), F264 (digestion of P628), F263 (digestion of P629, F265 (digestion of P630), F266 (digestion of P632) and F267 (digestion of P414).
Ligation of F230+F262, F230+F263, F230+F264, F230+F265, F230+F266, F230+F267
Investigator: Flo, Rosario, Jeff
Aim of the experiment: Ligation of F230+F262 (SERK-SigP_EreB_Strep-tag-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1 in PppActin_mMCS(insert location)_t35S_npt-cassette_pSB1C3), F230+F263 (SERK-SigP_SpyTag_Strep-Tag-II-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1_IRES_IgKappa-SigP_NanoLuc_SpyCatcher in PppActin_mMCS(insert location)_t35S_npt-cassette_pSB1C3), F230+F264 (SERK-SigP_SpyCatcher_Strep-Tag-II-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1_IRES_IgKappa-SigP_SpyTag_NanoLuc in PppActin_mMCS(insert location)_t35S_npt-cassette_pSB1C3), F230+F265 (SERK-SigP_SpyCatcher_Strep-Tag-II-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1_IRES_IgKappa-SigP_NanoLuc_SpyTag in PppActin_mMCS(insert location)_t35S_npt-cassette_pSB1C3), F230+F266 (SERK-SigP_SpyTag_Strep-Tag-II-TEV-site-linker_SERK-TMD_8AAlinker_GFPmut1_IRES_IgKappa-SigP_SpyCatcher_NanoLuc in PppActin_mMCS(insert location)_t35S_npt-cassette_pSB1C3), F230+F267 (SERK-SigP_Laccase_Strep-tag-II-TEV-site-Linker_SERK-TMD_8AALinker_GFPmut1 in PppActin_mMCS(insert location)_t35S_npt-cassette_pSB1C3)
Procedure:
Friday, August 23rd
Preparative digestion of P715 with XbaI and PstI + Preparative Gelelectrophoresis + Gelextraction
Investigator: Louise, Flo
Procedure:
Reaction batch for P715:
| volume | reagent |
|---|---|
| 20 µl | Plasmid DNA |
| 4 µl | CutSmart Buffer (10x) |
| 1 µl | XbaI (20 U/µl) |
| 1 µl | PstI (20 U/µl) |
| 14 µl | ddH2O |
| =40 µl | TOTAL |
- Reaction batches were incubated at 37 °C for 2.5 h.
- Preparative gelelectrophoresis was performed at 90 V for 1 h in an 0,5% agarose gel.
- The lower band was cut out
- Gelextraction was performed after preparative Gelelectrophoresis
Transformation of E. coli XL1-Blue with P6 (pGAL in pTUM 100)
Investigator: Louise
Aim of the experiment: Transformation of E. coli XL1-Blue with P6 (pGAL in pTUM 100). Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 5 µl of DNA were added to the tubes containing 150 µl of competent cells and gently mixed
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- The cell suspensions were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated on new ampicillin plates.
Sequencing of P715 (sYFP_G_TEV-site_G_cYFP)
Investigator: Louise
Aim of the experiment: Sequencing of P715 (sYFP_G_TEV-site_G_cYFP)
Procedure: The DNA was prepared for sequencing according to the Eurofins SmartSeq Kit protocol (15 µl of plasmid DNA (50 - 100 ng) and 2 µl sequencing primer).
The gene we sequenced received the following barcode:
| Label | Barcode | Primer |
|---|---|---|
| P715 | FR02233118 | 03 |
Ligation of F187+F268 and F188+F268
Investigator: Flo
Aim of the experiment: Ligation of F187+F268 and F188+F268
Procedure:
- Ligation batch for F187+F268, vector:insert = 1:3
| volume | reagent |
|---|---|
| 0.527 µl | F187 |
| 1.406 µl | F268 |
| 15.067 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- Ligation batch for F188+F268, vector:insert = 1:3
| volume | reagent |
|---|---|
| 0.732 µl | F188 |
| 1.406 µl | F268 |
| 14.862 µl | ddH2O |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- neg. ctrl. water instead of insert
- Ligation was performed at room temperature for 1 h
Picking of transformed Plasmid E. coli XL1 blue with P650, P652,P654, P656, P658, P661, P663, P666, P672, P678, P679, P682, P685, P706, P709, P625, P414, P628, P629, P630, P632, P713
Investigator: Louise
Aim of the experiment: Picking of transformed Plasmid E. coli XL1 blue with P650, P652,P654, P656, P658, P661, P663, P666, P672, P678, P679, P682, P685, P706, P709, P625, P414, P628, P629, P630, P632, P713
Procedure:
- Picking and overnight culture after standard laboratory's protocol. (6 µl Cam, 6 ml LB-medium)
- 1 colony was picked for every transformation product.
Transformation of E. coli XL1-Blue with ligation product F225+F261
Investigator: Rosario
Aim of the experiment: Transformation of E. coli XL1-Blue with ligation product F225+F261.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 5 µl of DNA were added to the tubes containing 150 µl of competent cells and gently mixed
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- The cell suspensions were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated on new chlorampenicol plates.
 "
"