Team:Paris Bettencourt/Project/Infiltrate
From 2013.igem.org
Marguerite (Talk | contribs) |
|||
| (49 intermediate revisions not shown) | |||
| Line 16: | Line 16: | ||
} | } | ||
</style> | </style> | ||
| + | <div id="page"> | ||
<div class="overbox"> | <div class="overbox"> | ||
<div class="bkgr"> | <div class="bkgr"> | ||
<h2>Background</h2> | <h2>Background</h2> | ||
| - | <p>Latent tuberculosis persists inside macrophages | + | <p>Latent tuberculosis persists inside macrophages, where it is partially protected from both the host immune system and conventional antibiotics.</p> |
</div> | </div> | ||
<div class="results"> | <div class="results"> | ||
<h2>Results</h2> | <h2>Results</h2> | ||
<ul> | <ul> | ||
| - | + | <li>We expressed the enzyme Trehalose Dimycolate Hydrolase (TDMH) in <i>E. coli</i> and showed that it is highly toxic to mycobacteria in culture.</li> | |
| - | + | <li>We expressed the listeriolyin O (LLO) gene in <i>E. coli</i> and showed that it is capable of entering the macrophage cytosol.</li> | |
| - | + | <li>We co-infected macrophages with both mycobacteria and our engineered <i>E. coli</i> to characterize the resulting phagocytosis and killing.</li> | |
| + | <li> We measured the effect of a range of pH (from 3 to 8,8) on <i>M. smegmatis</i> and the TDMH enzyme activity.</li> | ||
</ul> | </ul> | ||
| - | |||
</div> | </div> | ||
<div class="biocriks"> | <div class="biocriks"> | ||
| Line 63: | Line 64: | ||
<div style="clear: both;"></div> | <div style="clear: both;"></div> | ||
</div> | </div> | ||
| - | |||
<div id="Introduction"></div> | <div id="Introduction"></div> | ||
<h2>Introduction</h2> | <h2>Introduction</h2> | ||
| Line 69: | Line 69: | ||
<p> | <p> | ||
<i>Mycobacterium tuberculosis</i> (Mtb), the bacterium responsible for tuberculosis (TB), spreads by aerosol and infects its host through the airways. The bacterium is phagocytosed by macrophages in the lung, yet often evades death in the lysosome. Mtb can persist for years or even decades inside macrophages by inhibiting phagosome/lysosome fusion and supressing the normal acidification of the lysosome.</p> | <i>Mycobacterium tuberculosis</i> (Mtb), the bacterium responsible for tuberculosis (TB), spreads by aerosol and infects its host through the airways. The bacterium is phagocytosed by macrophages in the lung, yet often evades death in the lysosome. Mtb can persist for years or even decades inside macrophages by inhibiting phagosome/lysosome fusion and supressing the normal acidification of the lysosome.</p> | ||
| - | <p>An efficient treatment for persistent TB must enter infected macrophages and kill the pathogen there. In our system, E. coli is both the vector and the therapeutic agent agent by expressing the gene LLO to enter macrophages and TDMH to kill mycobacteria. | + | <p>An efficient treatment for persistent TB must enter infected macrophages and kill the pathogen there. In our system, <i>E. coli</i> is both the vector and the therapeutic agent agent by expressing the listeriolyin O gene LLO to enter macrophages and TDMH to kill mycobacteria. Additionally we thought about how we could deliver the E. coli to the lungs and decided that the most effective way would be through an inhaler. |
</p> | </p> | ||
</div> | </div> | ||
| Line 84: | Line 84: | ||
</div> | </div> | ||
<div style="clear: both;"></div> | <div style="clear: both;"></div> | ||
| + | <center><a href="https://static.igem.org/mediawiki/2013/1/1d/PB_inhalorinfiltrate.png" target="_blank"><img src="https://static.igem.org/mediawiki/2013/1/1d/PB_inhalorinfiltrate.png" width="1100"/></a></center> | ||
| + | <br><br> | ||
<div id="Design"></div> | <div id="Design"></div> | ||
| Line 90: | Line 92: | ||
<p> Mycobacterium species share a characteristic cell wall: thick, waxy, hydrophobic, and rich in mycolic acids. The low permeability of the envelope to hydrophilic solutes contributes to the intrinsic drug tolerance in mycobacteria.</p> | <p> Mycobacterium species share a characteristic cell wall: thick, waxy, hydrophobic, and rich in mycolic acids. The low permeability of the envelope to hydrophilic solutes contributes to the intrinsic drug tolerance in mycobacteria.</p> | ||
<p> | <p> | ||
| - | Trehalose Dimycolate Hydrolase (TDMH) is a cutinase-like serine esterase that triggers rapid lysis of the mycobacterial cell wall by degrading the mycolate layer. The enzyme was first isolated from Mycobacterium smegmatis and subsequently shown to hydrolyze purified TDM from various mycobacterial species. Exposure to TDMH triggers an immediate release of free mycolic acids, ultimately leading to lysis of many mycobacteria including Mtb (Yang et al. 2012). | + | Trehalose Dimycolate Hydrolase (TDMH) is a cutinase-like serine esterase that triggers rapid lysis of the mycobacterial cell wall by degrading the mycolate layer. The enzyme was first isolated from <i>Mycobacterium smegmatis</i> and subsequently shown to hydrolyze purified TDM from various mycobacterial species. Exposure to TDMH triggers an immediate release of free mycolic acids, ultimately leading to lysis of many mycobacteria including Mtb (Yang et al. 2012). |
</p> | </p> | ||
</div> | </div> | ||
<div class="rightparagraph"> | <div class="rightparagraph"> | ||
| - | <p> We have used <i>M. smegmatis</i> as a model system because Mtb are highly pathogenic and difficult to culture in the lab. M. smegmatis is a close relative of Mtb, shares many of its membrane properties, and is commonly used as a stand-in for Mtb physiology in the lab.</p> | + | <p> We have used <i>M. smegmatis</i> as a model system because Mtb are highly pathogenic and difficult to culture in the lab. <i>M. smegmatis</i> is a close relative of Mtb, shares many of its membrane properties, and is commonly used as a stand-in for Mtb physiology in the lab.</p> |
<p> In our system, described in the methods below, we used <i>E. coli</i> BL21 (DE3) as a chassis to express TDMH from an IPTG-inducible strong T7 promoter.</p> | <p> In our system, described in the methods below, we used <i>E. coli</i> BL21 (DE3) as a chassis to express TDMH from an IPTG-inducible strong T7 promoter.</p> | ||
</div> | </div> | ||
| Line 102: | Line 104: | ||
<h2>Killing mycobacteria with TDMH</h2> | <h2>Killing mycobacteria with TDMH</h2> | ||
<div class="leftparagraph"> | <div class="leftparagraph"> | ||
| - | + | <p> | |
| + | Figure 1 describes how we performed the setup of our experience by making a coculture of <i>M.smegmatis</i> and <i>E.coli</i>.</p> | ||
| + | <p>Figure 2(below) shows the killing of <i>M. smegmatis</i> with TDMH expressed by <i>E. coli</i>. We mixed liquid cultures of <i>E. coli</i> and <i>M. smegmatis</i> at equal cell densities as determined by plating assays. When expression of TMDH was induced with IPTG, nearly 99% of the <i>M. smegmatis</i> were killed within six hours. We saw no change in viability in cultures of <i>M. smegmatis</i> alone or when mixed with uninduced <i>E. coli</i>.</p> | ||
<p> | <p> | ||
| - | We next sought to quantify the effectiveness of TDMH killing. We mixed induced<i> E. coli</i> and mycobacteria in different ratios and used plating assays to measure viability. As shown in | + | We next sought to quantify the effectiveness of TDMH killing. We mixed induced<i> E. coli</i> and mycobacteria in different ratios and used plating assays to measure viability. As shown in figure 3 mycobacterial killing displayed dose dependence on the <i>E. coli</i> cell density. Small numbers of <i>E. coli</i> could kill many mycobacteria. For example, in mixed populations with 100 mycobactera for each <i>E. coli</i>, we still observed >50% mycobacterial killing after 2 hours. This indicates that, on average, each <i>E. coli</i> produced enough TDMH to kill 50 mycobacteria. We reason that this killing may be even more effective inside macrophages, where constrained volumes will increase the effective TDMH concentration.</p> |
</div> | </div> | ||
<div class="rightparagraph"> | <div class="rightparagraph"> | ||
<center><a href="https://static.igem.org/mediawiki/2013/d/d9/PB_FigIva.png" target="_blank"><img src="https://static.igem.org/mediawiki/2013/d/d9/PB_FigIva.png" width="70%" style="margin-top:-50px"/></a></center> | <center><a href="https://static.igem.org/mediawiki/2013/d/d9/PB_FigIva.png" target="_blank"><img src="https://static.igem.org/mediawiki/2013/d/d9/PB_FigIva.png" width="70%" style="margin-top:-50px"/></a></center> | ||
| - | <p><b>Figure | + | <p><b>Figure 1</b> Experimental setup of the <i>M. smegmatis</i> killing assay.<div style="font-size:90%"> <i>M. smegmatis</i> and <i>E.coli</i> cells (Uninduced and induced with various concentrations of IPTG) were cultured up to OD 1,then serially diluted. Equal volumes of <i>E.coli</i> and <i>M. smegmatis</i> cultures of the same dilutions were mixed, incubated for 6 hours, and plated at 0,3 and 6 hours. </div></p> |
</div> | </div> | ||
<div style="clear: both;"></div> | <div style="clear: both;"></div> | ||
<div class="leftparagraph"> | <div class="leftparagraph"> | ||
<a href="https://static.igem.org/mediawiki/2013/a/a7/PB_Fig1_Myco_Kill_Curve.png" target="_blank"><img src="https://static.igem.org/mediawiki/2013/a/a7/PB_Fig1_Myco_Kill_Curve.png" width="100%"/></a> | <a href="https://static.igem.org/mediawiki/2013/a/a7/PB_Fig1_Myco_Kill_Curve.png" target="_blank"><img src="https://static.igem.org/mediawiki/2013/a/a7/PB_Fig1_Myco_Kill_Curve.png" width="100%"/></a> | ||
| - | <p><b>Figure | + | <p><b>Figure 2</b> TDMH-expressing <i>E.coli</i> kill mycobacteria in culture.<div style="font-size:90%"> We mixed TDMH-expressing <i>E.coli</i> and WT <i>M. smegmatis</i> in LB media at an initial cell density of 107 cells/ml each. Plating assays were used to count specifically <i>M. smegmatis</i> after the indicated times. When TDMH-expression was fully induced with 1 mM IPTG, more than 98% of mycobacteria were killed after 6 hours (red line). Populations of mycobacteria alone (black line) and mycobacteria mixed with uninduced <i>E.coli</i> (blue line) were stable.</div></p> |
</div> | </div> | ||
<div class="rightparagraph"> | <div class="rightparagraph"> | ||
<center><a href="https://static.igem.org/mediawiki/2013/e/e0/PB_Fig2_Coli_Kill_Ratios.png" target="_blank"><img src="https://static.igem.org/mediawiki/2013/e/e0/PB_Fig2_Coli_Kill_Ratios.png" width="100%"/></a></center> | <center><a href="https://static.igem.org/mediawiki/2013/e/e0/PB_Fig2_Coli_Kill_Ratios.png" target="_blank"><img src="https://static.igem.org/mediawiki/2013/e/e0/PB_Fig2_Coli_Kill_Ratios.png" width="100%"/></a></center> | ||
| - | <p><b>Figure | + | <p><b>Figure 3</b> Dose-dependence in killing of mycobacteria by <i>E. coli.</i><div style="font-size:90%">We mixed <i>M. smegmatis</i> at a density of 10<sup>7</sup> cells/ml with <i>E. coli</i> at densities of 10<sup>5</sup> cells/ml (blue line) 10<sup>6</sup> cells/ml (green line) or 10<sup>7</sup> cells/ml to produce the ratios above. Even low densities of <i>E. coli</i> produced significant killing. Uninduced <i>E. coli</i> produced no significant killing.</div></p> |
</div> | </div> | ||
<div style="clear: both;"></div> | <div style="clear: both;"></div> | ||
| Line 123: | Line 127: | ||
<h2></h2> | <h2></h2> | ||
<div class="leftparagraph"> | <div class="leftparagraph"> | ||
| - | <p> We investigated further the mechanism of TDMH-mediated cell killing. The TDMH | + | <p> We investigated further the mechanism of TDMH-mediated cell killing. The TDMH enzyme in our system does not carry a secretion tag. Therefore, lysis of the <i>E. coli</i> membrane is probably required for the protein to reach (and lyse) <i>M. smegmatis</i>. We used plating assays to investigate the effect of TDMH induction on <i>E. coli</i> viability and Bradford assays to measure released protein. The results are presented in figure 4.</p> |
| - | <p>Figure | + | <p>Figure 4A shows that inducing TDMH-expressing <i>E. coli</i> with IPTG rapidly kills the <i>E. coli.</i> In figure 4B, we show that inducing TDMH expression increases the concentration of free protein in the media. In our model, TDMH induction leads to spontaneous lysis of TDMH-expressing <i>E. coli</i>. This could be simply due to the extremely high protein expression levels driven by the T7 promoter, or it may be partially due to the esterase activities of the TDMH enzyme. Once the TDMH is released from the <i>E. coli</i> it is free to act on mycobacterial membranes, causing lysis and the release of additional proteins. |
</p> | </p> | ||
</br> | </br> | ||
| - | <p><b>Figure | + | <p><b>Figure 4</b> Induction of TDMH kills <i>E. coli</i> and releases protein to the media.<div style="font-size:90%"><b>A)</b>Plating assays were used to determine <i>E. coli</i> viability in LB media after the indicated times. TDMH-induced <i>E. coli</i> rapidly lost viability (red line) Uninduced <i>E. coli</i> grew normally following a lag phase (black line).<b>B)</b>Bradford assays were used to measure free protein concentrations in mixed and induced bacterial cultures. When <i>E. coli</i> and mycobacteria are mixed and TDMH-expression is induced, free protein concentrations increase. This is consistent with a model in which TDMH induction leads to the lysis of the <i>E. coli</i> membrane. Action of TDMH on mycobacteria leads to further lysis and protein release.</div></p> |
</div> | </div> | ||
<div class="rightparagraph"> | <div class="rightparagraph"> | ||
| Line 138: | Line 142: | ||
<h2>Programmed drug delivery with LLO</h2> | <h2>Programmed drug delivery with LLO</h2> | ||
<div class="leftparagraph"> | <div class="leftparagraph"> | ||
| - | <p> <i>Listeria monocytogenes</i> is a bacterial pathogen that replicates in the cytosol of mammalian cells. The pathogen is initially internalized in host phagosomes, then lyses them to obtain access to the cytosol. A single gene, LLO, is responsible for this activity and is sufficient to convey the phenotype to <i>E. coli | + | <p> <i>Listeria monocytogenes</i> is a bacterial pathogen that replicates in the cytosol of mammalian cells. The pathogen is initially internalized in host phagosomes, then lyses them to obtain access to the cytosol. A single gene, LLO, is responsible for this activity and is sufficient to convey the phenotype to <i>E. coli</i>. |
</p> | </p> | ||
<p>The LLO protein is activated by the low pH of the lysosme. By forming large pores in the lysosomal membrane, it allows protein delivery to the cytosol of macrophages (Higgins et al. 1999). Using <i>E. coli</i> as a protein vector means we don't need to isolate and purify the TDMH enzyme. </p> | <p>The LLO protein is activated by the low pH of the lysosme. By forming large pores in the lysosomal membrane, it allows protein delivery to the cytosol of macrophages (Higgins et al. 1999). Using <i>E. coli</i> as a protein vector means we don't need to isolate and purify the TDMH enzyme. </p> | ||
| Line 146: | Line 150: | ||
<div class="rightparagraph"> | <div class="rightparagraph"> | ||
<center><img src="https://static.igem.org/mediawiki/2013/b/b7/PB_Killing_assay_macrophages1.png" width="80%"/></center> | <center><img src="https://static.igem.org/mediawiki/2013/b/b7/PB_Killing_assay_macrophages1.png" width="80%"/></center> | ||
| - | <p><b>Figure | + | <p><b>Figure 5</b> Experimental design of the killing assay inside the macrophages. We infected them with <i>M. smegmatis</i> and washed after 1 hour. Then, we infected them with <i>E. coli</i> and washed after 1 hour. The end of this final infection step refers to the time 0 on our graphs. At this time we started the observation under the microscope</p> |
</div> | </div> | ||
<div style="clear: both;"></div> | <div style="clear: both;"></div> | ||
| Line 152: | Line 156: | ||
<h2><i>E. coli</i> and <i>M. smegmatis</i> coinfections</h2> | <h2><i>E. coli</i> and <i>M. smegmatis</i> coinfections</h2> | ||
<div class="leftparagraph"> | <div class="leftparagraph"> | ||
| - | <p> Figure | + | <p> Figure 6 displays J774 macrophages one hour after coinfection with <i>E. coli</i> and <i>M. smegmatis</i>. As described in the methods, the <i>E. coli</i> express TDMH and LLO and fluorescent mRFP as a label. The <i>M. smegmatis</i> express GFP as a label, but are otherwise wild type.</p> |
| - | <p>The two movies displayed in figure | + | <p>The two movies displayed in figure 7 are time-lapse microscopy of live coinfected macrophages collected over a period of 6 hours. Fluorescence patterns revealing the localization of <i>E. coli</i> and <i>M. smegmatis</i> within the macrophages are relatively stable over this time period.</p> |
</div> | </div> | ||
<div class="rightparagraph"> | <div class="rightparagraph"> | ||
| Line 163: | Line 167: | ||
</br> | </br> | ||
<a href="https://static.igem.org/mediawiki/2013/5/51/PB_Fig4_Fresh_coinfection_zoom1.png" target="_blank"><img src="https://static.igem.org/mediawiki/2013/5/51/PB_Fig4_Fresh_coinfection_zoom1.png" width="48%"/></a> | <a href="https://static.igem.org/mediawiki/2013/5/51/PB_Fig4_Fresh_coinfection_zoom1.png" target="_blank"><img src="https://static.igem.org/mediawiki/2013/5/51/PB_Fig4_Fresh_coinfection_zoom1.png" width="48%"/></a> | ||
| - | <a href="https://static.igem.org/mediawiki/2013/ | + | <a href="https://static.igem.org/mediawiki/2013/c/c1/PB_Fig4_Fresh_coinfection_zoom2.png" target="_blank"><img src="https://static.igem.org/mediawiki/2013/c/c1/PB_Fig4_Fresh_coinfection_zoom2.png" width="48%" style="margin-left:3%"/></a> |
| - | <p><b>Figure | + | <p><b>Figure 6</b> Coinfection of J774 macrophages with mycobacteria and <i>E. coli</i> expressing TDMH and LLO<div style="font-size:90%"> <i>E. coli</i> (red) express mRFP as a marker and <i>M. smegmatis</i> (green) express GFP as a marker. Macrophages are observed to take up Mycobateria, <i>E. coli</i>, neither or both. This image was collected one hour after introducing the bacteria to the macrophage culture and following extensive washing to remove free bacteria.</div></p> |
</div> | </div> | ||
<div class="rightparagraph"> | <div class="rightparagraph"> | ||
<iframe frameborder="0" style="margin-bottom:20px;" width="535" height="300" src="http://www.dailymotion.com/embed/video/x15jnu6"></iframe> | <iframe frameborder="0" style="margin-bottom:20px;" width="535" height="300" src="http://www.dailymotion.com/embed/video/x15jnu6"></iframe> | ||
<iframe frameborder="0" width="535" height="300" src="http://www.dailymotion.com/embed/video/x15jnv8"></iframe> | <iframe frameborder="0" width="535" height="300" src="http://www.dailymotion.com/embed/video/x15jnv8"></iframe> | ||
| - | <p><b>Figure | + | <p><b>Figure 7</b> Temporal dynamics of macrophages coinfected with <i>E. coli</i> and <i>M. smegmatis.</i><div style="font-size:90%"><i>E. coli</i> and <i>M. smegmatis</i>, expressing mRFP and GFP respectively, were introduced to J774 macrophages. Fluorescent and bright field images were collected every 15 minutes for 6 hours. In these movies, the <i>E. coli</i> also express TDMH and LLO. Macrophages are observed to uptake <i>E. coli</i>, <i>M. smegmatis</i>, neither or both. The fluorescence patterns appear stable for these conditions and time period.</div></p> |
</div> | </div> | ||
| + | |||
| + | |||
| + | <h2>pH influence </h2> | ||
| + | <div class="leftparagraph"> | ||
| + | <p> We wanted to study the pH influence on <i>M. smegmatis</i> growth and on TDMH enzyme activity. We know that the phagosomal pH is usually acid (around 6) but can reach 4,5. We wondered if TDMH enzyme was working at this range of pH.</p> | ||
| + | <p>We studied 7 different buffers : 3 ; 4,5 ; 5,1 ; 6,4 ; 7,35 ; 8,2 and 8,8. </p> | ||
| + | |||
| + | <p>Figure 8 shows the activity of TDMH enzyme at various pH levels.</p> | ||
| + | |||
| + | <p>Mycobacteria was unable to grow at pH 3 with or without the presence of TDMH. Therefore we are unable to make any conclusions regarding the activity of the enzyme at very low pH values.</p> | ||
| + | |||
| + | <p>TDMH activity was active across a large range of pH from 4.5 to 8.8, though there was a small decrease in efficiency observed at pH 8.2. This is a promising result due to the difference in pH between the phagosome and the cytosol. Additionally, the high activity of TDMH even at low pH is promising as it will be required to function at low pH inside the phagosome. | ||
| + | </p> | ||
| + | |||
| + | |||
| + | </div> | ||
| + | <div class="rightparagraph"> | ||
| + | |||
| + | <center> <img src="https://static.igem.org/mediawiki/2013/e/e4/PB_TBC_PH_indepedence.png" width="60%"/></center> <p><b>Figure 8</b> pH influence on TDMH enzyme activity <div style="font-size:90%"> We add to <i>M.smegmatis</i> the proteins of an induced control, the uninduced strain of Bl21 and the induced strain producing TDMH.</div></p> | ||
| + | </div> | ||
| + | |||
| + | |||
<div style="clear: both;"></div> | <div style="clear: both;"></div> | ||
| Line 178: | Line 204: | ||
<div class="leftparagraph"> | <div class="leftparagraph"> | ||
<p> We have shown that <i>E. coli</i> can effectively kill mycobacteria in culture by expressing the TDMH enzyme, now a BioBrick. We have further shown that <i>E. coli</i> and mycobateria may co-localize within macrophages, where LLO-based systems can effectively deliver protein payloads.</p> | <p> We have shown that <i>E. coli</i> can effectively kill mycobacteria in culture by expressing the TDMH enzyme, now a BioBrick. We have further shown that <i>E. coli</i> and mycobateria may co-localize within macrophages, where LLO-based systems can effectively deliver protein payloads.</p> | ||
| - | <p>In the coming weeks, we will continue to refine our cell culture techniques to precisely characterize the effectiveness of our system in situ, within the living macrophage.</p> | + | <p>In the coming weeks, we will continue to refine our cell culture techniques to precisely characterize the effectiveness of our system <i>in situ</i>, within the living macrophage.</p> |
| + | <p><b>Figure 9</b> The effect of TDMH-induced <i>E. coli</i> on macrophage infection rates after 3 hours.<div style="font-size:90%">We scored fluorescence microscopy images of macrophages exposed to both <i>E. coli</i> and <i>M. smegmatis</i> expressing fluorescent protein markers. The <i>E. coli</i> expressed LLO and were induced or uninduced for TDMH expression. For both treatments, we observed an increase in the infection rates of both bacteria after 3 hours. We attribute this to combination of the media with free bacteria, which macrophages continue to consume over the course of the experiment. TDMH induction slowed the increase in <i>E. coli</i> infection rates, which is likely due to the toxic effects of TDMH expression on <i>E. coli</i>, as shown in figure 4.</div></p> | ||
</div> | </div> | ||
<div class="rightparagraph"> | <div class="rightparagraph"> | ||
| - | <center | + | <center><img src="https://static.igem.org/mediawiki/2013/1/15/PB_Fig6_Image_Counts.png" width="80%"/></center> |
| - | + | ||
</div> | </div> | ||
| - | <div style="clear: both;" | + | <div style="clear: both;"> |
| - | < | + | <h2>Bibliography</h2> |
<ul> | <ul> | ||
<li>Yang Y, Bhatti A, Ke D, Gonzalez-Juarrero M, Lenaerts A, Kremer L, Guerardel Y, Zhang P, Ojha AK (2012) : Exposure to a cutinase-like serine esterase triggers rapid lysis of multiple mycobacterial species. J Biol Chem. 2013 Jan 4;288(1):382-92.</li> | <li>Yang Y, Bhatti A, Ke D, Gonzalez-Juarrero M, Lenaerts A, Kremer L, Guerardel Y, Zhang P, Ojha AK (2012) : Exposure to a cutinase-like serine esterase triggers rapid lysis of multiple mycobacterial species. J Biol Chem. 2013 Jan 4;288(1):382-92.</li> | ||
| Line 193: | Line 219: | ||
</div> | </div> | ||
| + | <div style="clear: both;"></div> | ||
| + | <h2>Attributions</h2> | ||
| + | |||
| + | <p>pET21 1519 plasmid was kindly provided by Anil Ojha of the University of Pittsburgh.</p> | ||
| + | <p> Murine cell line macrophages J774 were a gift from Nicole Guiso and Nicolas Hegerle from Pasteur institute (Unit of Molecular Prevention and Therapy of Human Diseases. </p> | ||
| + | <p> <i>Mycobacterium smegmatis</i> was a gift from Brigitte Gicquel from Pasteur institute (Unit Mycobacterial genetics). </p> | ||
| + | <p> <i>Mycobacterium smegmatis</i> expressing GFP was kindly provided by Stephane Canaan from CNRS (Laboratory of Enzymology at Interfaces and Physiology of Lipolysis). </p> | ||
| + | <p>The project itself was designed and accomplished by Camélia Bencherif and Iva Atanaskovic with consultation with Edwin Wintermute, Mathias Toulouze and Ariel Lindner.</p> | ||
| + | <p> We also discussed the project with Christopher Anderson from Berkeley University (Synthetic biology Anderson lab) about the feasibility, good points and critical points of the project. </p> | ||
| + | <p> We want to thank all these people that made Infiltrate project happen !</p> | ||
| + | |||
| + | |||
| + | |||
| + | </div> | ||
</html> | </html> | ||
{{:Team:Paris_Bettencourt/footer}} | {{:Team:Paris_Bettencourt/footer}} | ||
Latest revision as of 03:44, 29 October 2013


Background
Latent tuberculosis persists inside macrophages, where it is partially protected from both the host immune system and conventional antibiotics.
Results
- We expressed the enzyme Trehalose Dimycolate Hydrolase (TDMH) in E. coli and showed that it is highly toxic to mycobacteria in culture.
- We expressed the listeriolyin O (LLO) gene in E. coli and showed that it is capable of entering the macrophage cytosol.
- We co-infected macrophages with both mycobacteria and our engineered E. coli to characterize the resulting phagocytosis and killing.
- We measured the effect of a range of pH (from 3 to 8,8) on M. smegmatis and the TDMH enzyme activity.
BioBricks
Aim
To create an E. coli strain capable of entering the macrophage cytosol and delivering a lytic enzyme to kill mycobacteria.
Skip to Introduction
Skip to Design
Skip to Killing Assay
Skip to Drug Delivery
Introduction
Mycobacterium tuberculosis (Mtb), the bacterium responsible for tuberculosis (TB), spreads by aerosol and infects its host through the airways. The bacterium is phagocytosed by macrophages in the lung, yet often evades death in the lysosome. Mtb can persist for years or even decades inside macrophages by inhibiting phagosome/lysosome fusion and supressing the normal acidification of the lysosome.
An efficient treatment for persistent TB must enter infected macrophages and kill the pathogen there. In our system, E. coli is both the vector and the therapeutic agent agent by expressing the listeriolyin O gene LLO to enter macrophages and TDMH to kill mycobacteria. Additionally we thought about how we could deliver the E. coli to the lungs and decided that the most effective way would be through an inhaler.
BOX 1: Why is it difficult to treat tuberculosis?
- Mtb can survive and replicate for years inside the macrophages by supressing the normal process of phagocytosis.
- The cell wall of M. tuberculosis is thick, waxy and rich in mycolic acids. It it difficult to penetrate with a small-molecule drug.
- The pathogen grows very slowly inside macrophages. Most conventional antibiotics target processes like DNA replication that are specific to growing cells.

TDMH and the mycobacterial cell wall
Mycobacterium species share a characteristic cell wall: thick, waxy, hydrophobic, and rich in mycolic acids. The low permeability of the envelope to hydrophilic solutes contributes to the intrinsic drug tolerance in mycobacteria.
Trehalose Dimycolate Hydrolase (TDMH) is a cutinase-like serine esterase that triggers rapid lysis of the mycobacterial cell wall by degrading the mycolate layer. The enzyme was first isolated from Mycobacterium smegmatis and subsequently shown to hydrolyze purified TDM from various mycobacterial species. Exposure to TDMH triggers an immediate release of free mycolic acids, ultimately leading to lysis of many mycobacteria including Mtb (Yang et al. 2012).
We have used M. smegmatis as a model system because Mtb are highly pathogenic and difficult to culture in the lab. M. smegmatis is a close relative of Mtb, shares many of its membrane properties, and is commonly used as a stand-in for Mtb physiology in the lab.
In our system, described in the methods below, we used E. coli BL21 (DE3) as a chassis to express TDMH from an IPTG-inducible strong T7 promoter.
Killing mycobacteria with TDMH
Figure 1 describes how we performed the setup of our experience by making a coculture of M.smegmatis and E.coli.
Figure 2(below) shows the killing of M. smegmatis with TDMH expressed by E. coli. We mixed liquid cultures of E. coli and M. smegmatis at equal cell densities as determined by plating assays. When expression of TMDH was induced with IPTG, nearly 99% of the M. smegmatis were killed within six hours. We saw no change in viability in cultures of M. smegmatis alone or when mixed with uninduced E. coli.
We next sought to quantify the effectiveness of TDMH killing. We mixed induced E. coli and mycobacteria in different ratios and used plating assays to measure viability. As shown in figure 3 mycobacterial killing displayed dose dependence on the E. coli cell density. Small numbers of E. coli could kill many mycobacteria. For example, in mixed populations with 100 mycobactera for each E. coli, we still observed >50% mycobacterial killing after 2 hours. This indicates that, on average, each E. coli produced enough TDMH to kill 50 mycobacteria. We reason that this killing may be even more effective inside macrophages, where constrained volumes will increase the effective TDMH concentration.

Figure 1 Experimental setup of the M. smegmatis killing assay.

Figure 2 TDMH-expressing E.coli kill mycobacteria in culture.

Figure 3 Dose-dependence in killing of mycobacteria by E. coli.
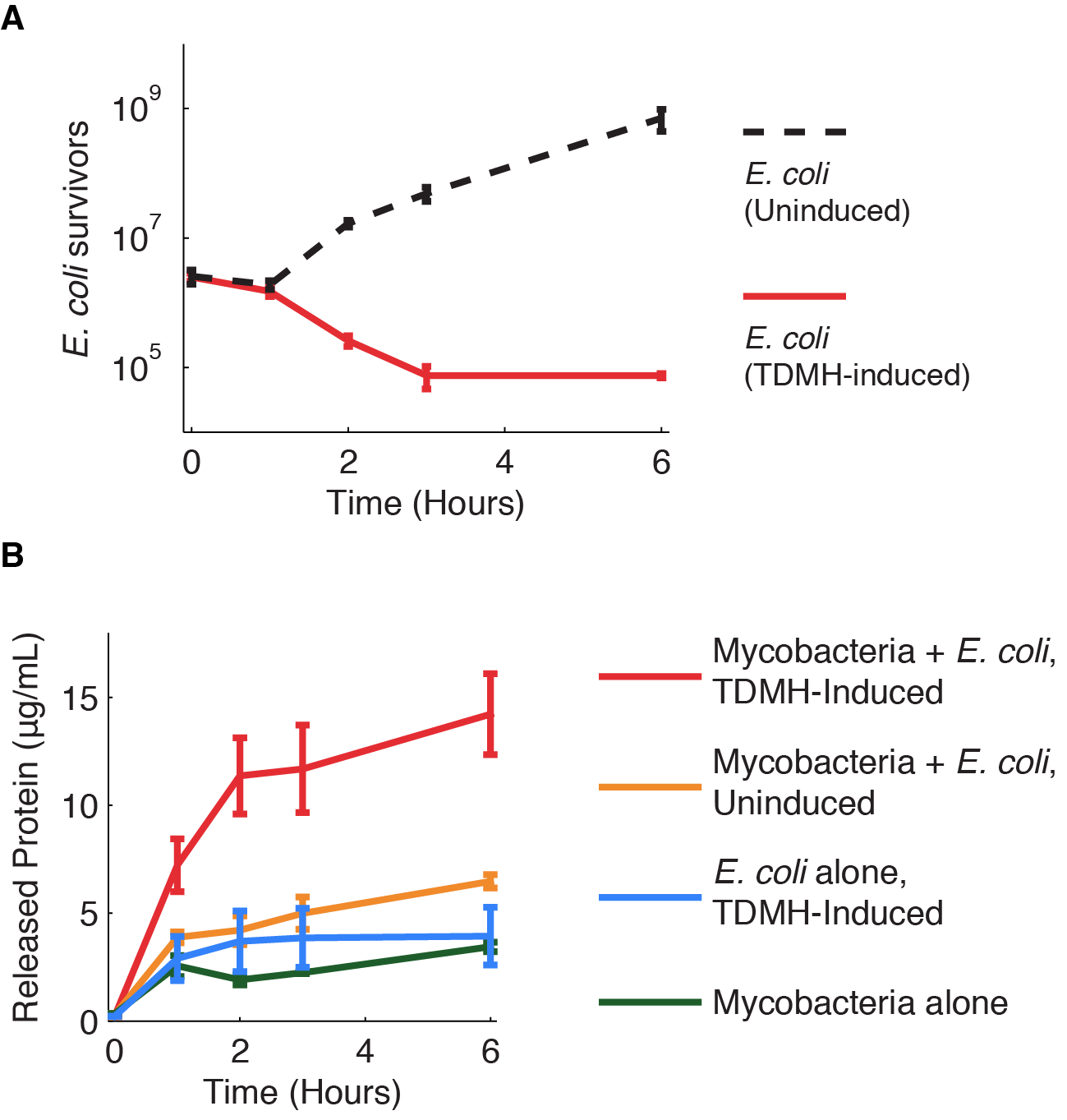
We investigated further the mechanism of TDMH-mediated cell killing. The TDMH enzyme in our system does not carry a secretion tag. Therefore, lysis of the E. coli membrane is probably required for the protein to reach (and lyse) M. smegmatis. We used plating assays to investigate the effect of TDMH induction on E. coli viability and Bradford assays to measure released protein. The results are presented in figure 4.
Figure 4A shows that inducing TDMH-expressing E. coli with IPTG rapidly kills the E. coli. In figure 4B, we show that inducing TDMH expression increases the concentration of free protein in the media. In our model, TDMH induction leads to spontaneous lysis of TDMH-expressing E. coli. This could be simply due to the extremely high protein expression levels driven by the T7 promoter, or it may be partially due to the esterase activities of the TDMH enzyme. Once the TDMH is released from the E. coli it is free to act on mycobacterial membranes, causing lysis and the release of additional proteins.
Figure 4 Induction of TDMH kills E. coli and releases protein to the media.
Programmed drug delivery with LLO
Listeria monocytogenes is a bacterial pathogen that replicates in the cytosol of mammalian cells. The pathogen is initially internalized in host phagosomes, then lyses them to obtain access to the cytosol. A single gene, LLO, is responsible for this activity and is sufficient to convey the phenotype to E. coli.
The LLO protein is activated by the low pH of the lysosme. By forming large pores in the lysosomal membrane, it allows protein delivery to the cytosol of macrophages (Higgins et al. 1999). Using E. coli as a protein vector means we don't need to isolate and purify the TDMH enzyme.
We hypothesized that a bacterial delivery system could be efficient and specific in targeting mycobacterial-infected macrophages. In effect, we might take advantage of the natural tendency of macrophages to phagocytose E. coli and their enzymatic payload.
Because LLO expression is known to be associated with Listeria pathogenicity, we took extra precautions in our cloning and experiments. These measures are detailed on our team safety page. Briefly, we worked in a Biosafety Level 2 lab under the supervision of two full-time lab managers. We researched and complied with French national and insitutional biosafety rules as they apply to this gene.

Figure 5 Experimental design of the killing assay inside the macrophages. We infected them with M. smegmatis and washed after 1 hour. Then, we infected them with E. coli and washed after 1 hour. The end of this final infection step refers to the time 0 on our graphs. At this time we started the observation under the microscope
E. coli and M. smegmatis coinfections
Figure 6 displays J774 macrophages one hour after coinfection with E. coli and M. smegmatis. As described in the methods, the E. coli express TDMH and LLO and fluorescent mRFP as a label. The M. smegmatis express GFP as a label, but are otherwise wild type.
The two movies displayed in figure 7 are time-lapse microscopy of live coinfected macrophages collected over a period of 6 hours. Fluorescence patterns revealing the localization of E. coli and M. smegmatis within the macrophages are relatively stable over this time period.
We next applied quantitative microscopy to characterize the behavior of our system inside live macrophages. We imaged macrophage populations under several conditions and time points and scored their rates of infection with E. coli, M. smegmatis or both. In all we scored over 13,000 individual cells collected from more than 100 separate images.
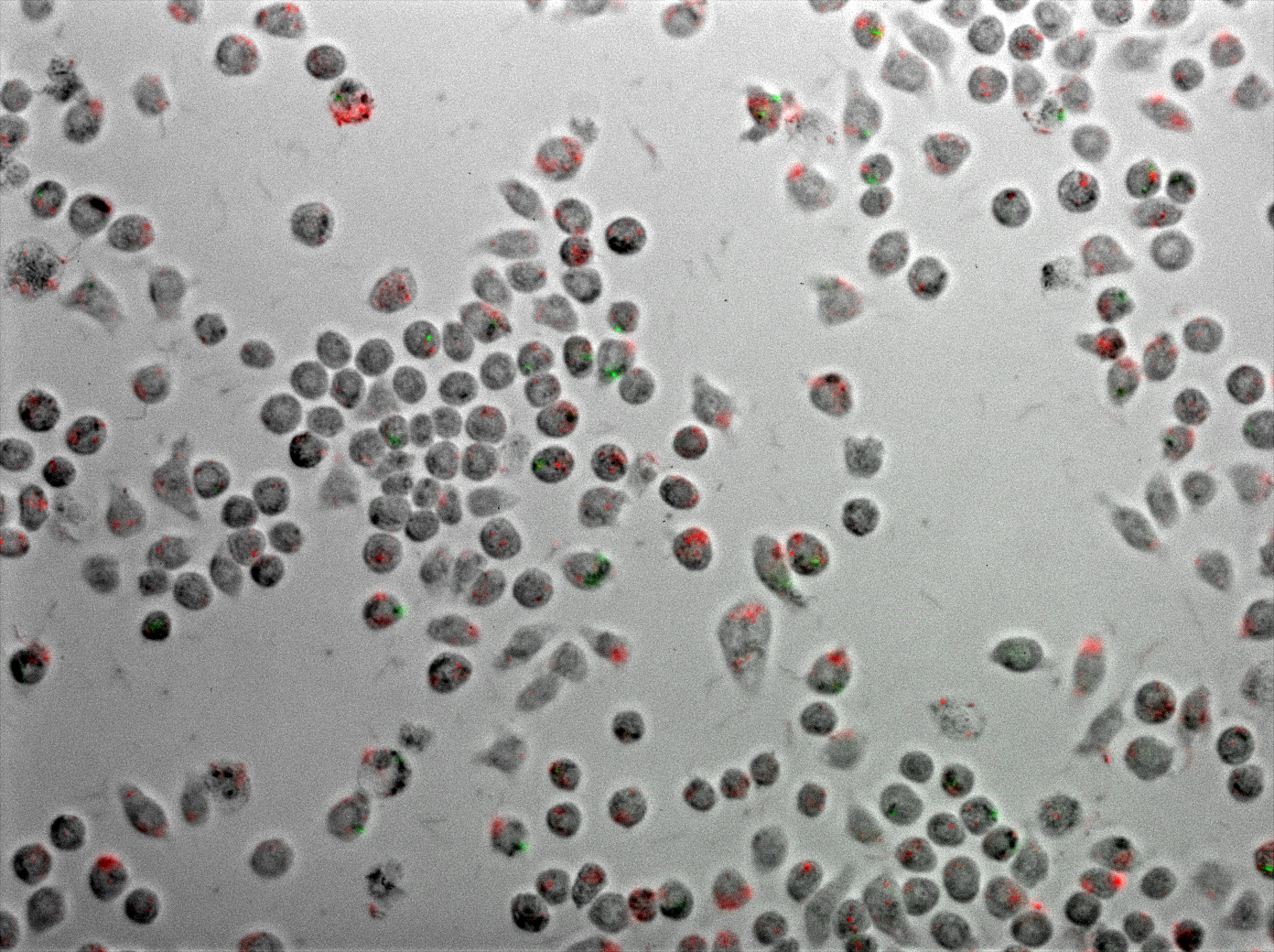


Figure 6 Coinfection of J774 macrophages with mycobacteria and E. coli expressing TDMH and LLO
Figure 7 Temporal dynamics of macrophages coinfected with E. coli and M. smegmatis.
pH influence
We wanted to study the pH influence on M. smegmatis growth and on TDMH enzyme activity. We know that the phagosomal pH is usually acid (around 6) but can reach 4,5. We wondered if TDMH enzyme was working at this range of pH.
We studied 7 different buffers : 3 ; 4,5 ; 5,1 ; 6,4 ; 7,35 ; 8,2 and 8,8.
Figure 8 shows the activity of TDMH enzyme at various pH levels.
Mycobacteria was unable to grow at pH 3 with or without the presence of TDMH. Therefore we are unable to make any conclusions regarding the activity of the enzyme at very low pH values.
TDMH activity was active across a large range of pH from 4.5 to 8.8, though there was a small decrease in efficiency observed at pH 8.2. This is a promising result due to the difference in pH between the phagosome and the cytosol. Additionally, the high activity of TDMH even at low pH is promising as it will be required to function at low pH inside the phagosome.

Figure 8 pH influence on TDMH enzyme activity
Conclusions and continuing work
We have shown that E. coli can effectively kill mycobacteria in culture by expressing the TDMH enzyme, now a BioBrick. We have further shown that E. coli and mycobateria may co-localize within macrophages, where LLO-based systems can effectively deliver protein payloads.
In the coming weeks, we will continue to refine our cell culture techniques to precisely characterize the effectiveness of our system in situ, within the living macrophage.
Figure 9 The effect of TDMH-induced E. coli on macrophage infection rates after 3 hours.

Bibliography
- Yang Y, Bhatti A, Ke D, Gonzalez-Juarrero M, Lenaerts A, Kremer L, Guerardel Y, Zhang P, Ojha AK (2012) : Exposure to a cutinase-like serine esterase triggers rapid lysis of multiple mycobacterial species. J Biol Chem. 2013 Jan 4;288(1):382-92.
- Rajesh Jayachandran, Varadharajan Sundaramurthy, Benoit Combaluzier , Philipp Mueller, Hannelie Korf, Kris Huygen, Toru Miyazaki, Imke Albrecht, Jan Massner, Jean Pieters (2007) : Survival of Mycobacteria in Macrophages Is Mediated by Coronin 1-Dependent Activation of Calcineurin. Cell, Volume 130, Issue 1, 13 July 2007, Pages 12-14
Attributions
pET21 1519 plasmid was kindly provided by Anil Ojha of the University of Pittsburgh.
Murine cell line macrophages J774 were a gift from Nicole Guiso and Nicolas Hegerle from Pasteur institute (Unit of Molecular Prevention and Therapy of Human Diseases.
Mycobacterium smegmatis was a gift from Brigitte Gicquel from Pasteur institute (Unit Mycobacterial genetics).
Mycobacterium smegmatis expressing GFP was kindly provided by Stephane Canaan from CNRS (Laboratory of Enzymology at Interfaces and Physiology of Lipolysis).
The project itself was designed and accomplished by Camélia Bencherif and Iva Atanaskovic with consultation with Edwin Wintermute, Mathias Toulouze and Ariel Lindner.
We also discussed the project with Christopher Anderson from Berkeley University (Synthetic biology Anderson lab) about the feasibility, good points and critical points of the project.
We want to thank all these people that made Infiltrate project happen !
 "
"







 +33 1 44 41 25 22/25
+33 1 44 41 25 22/25


