Team:Paris Bettencourt/Project/Infiltrate
From 2013.igem.org
| Line 38: | Line 38: | ||
<br> | <br> | ||
<h2>Killing mycobacteria with TDMH</h2> | <h2>Killing mycobacteria with TDMH</h2> | ||
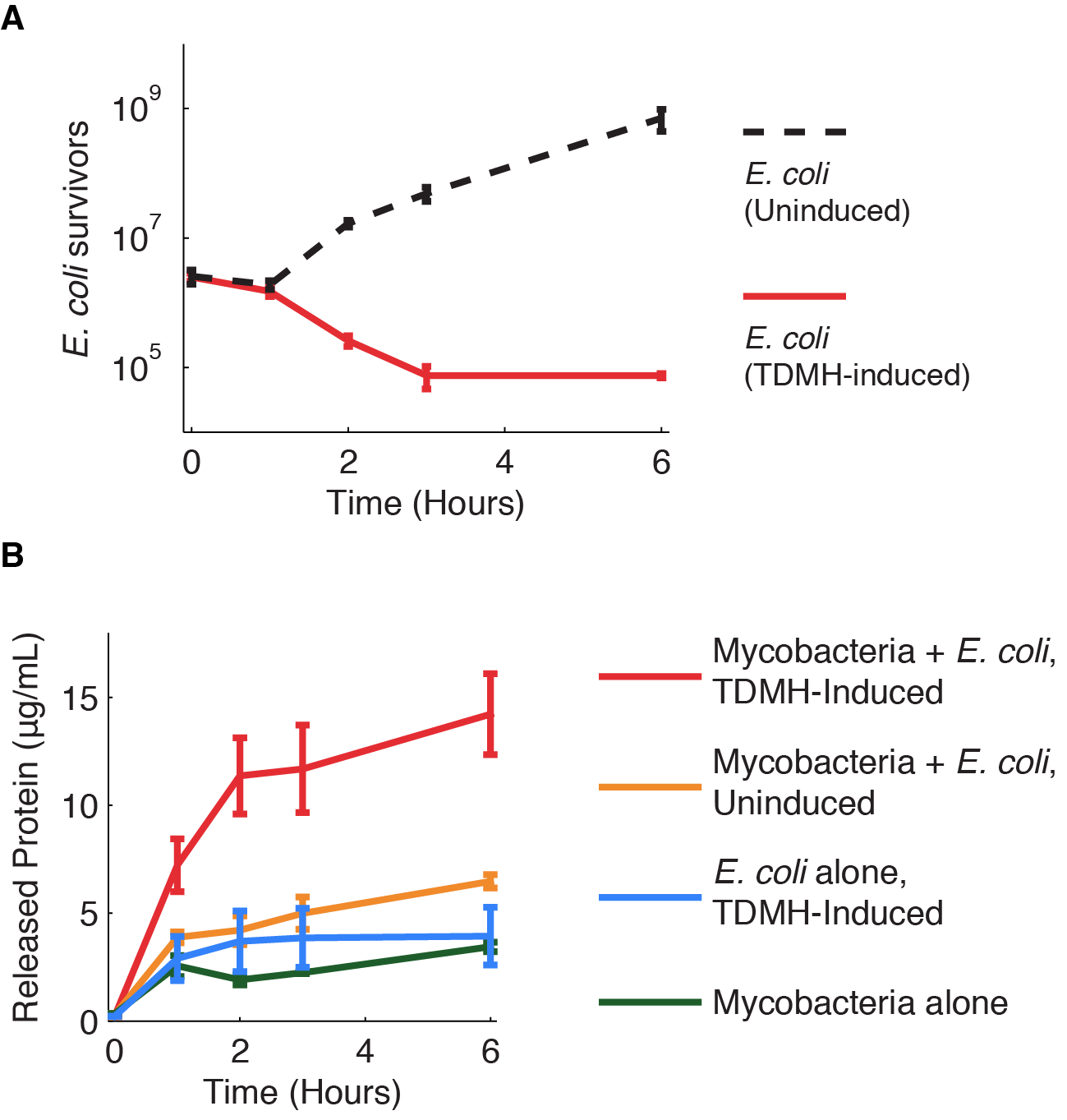
| - | <p><i>Figure 1</i> shows the killing of M. smegmatis with TDMH expressed by <i>E. coli</i>. We mixed liquid cultures of E. coli and M. smegmatis at equal cell densities as determined by plating assays. When expression of TMDH was induced with IPTG, nearly 99% of the M. smegmatis were killed within six hours. We saw no change in viability in cultures of M. smegmatis alone or when mixed with uninduced E. coli.</ | + | <p> <i>Figure 1</i> shows the killing of M. smegmatis with TDMH expressed by <i>E. coli</i>. We mixed liquid cultures of E. coli and M. smegmatis at equal cell densities as determined by plating assays. When expression of TMDH was induced with IPTG, nearly 99% of the M. smegmatis were killed within six hours. We saw no change in viability in cultures of M. smegmatis alone or when mixed with uninduced E. coli.<br> |
| + | <br> | ||
| + | We next sought to quantify the effectiveness of TDMH killing. We mixed induced<i> E. coli</i> and mycobacteria in different ratios and used plating assays to measure viability. As shown in <i>figure 2</i> mycobacterial killing displayed dose dependence on the <i>E. coli</i> cell density. Small numbers of <i>E. coli</i> could kill many mycobacteria. For example, in mixed populations with 100 mycobactera for each <i>E. coli</i>, we still observed >50% mycobacterial killing after 2 hours. This indicates that, on average, each <i>E. coli</i> produced enough TDMH to kill 50 mycobacterial. We reason that this killing may be even more effective inside macrophages, where constrained volumes will increase the effective TDMH concentration. | ||
Revision as of 18:58, 4 October 2013

Background:
Latent tuberculosis persists inside macrophages of the lungs, where it is partially protected from both the host immune system and conventional antibiotics.
Aim:
To create an E. coli strain capable of entering the macrophage cytosol and delivering a lytic enzyme to kill mycobacteria.
Results:
- We expressed the enzyme Trehalose Dimycolate Hydrolase (TDMH) in E.coli and showed that it is highly toxic to mycobacteria in culture.
- We expressed the lysteriolyin O (LLO) gene in E. coli and showed that it is capable of entering the macrophage cytosol.
- We co-infected macrophages with both mycobacteria and our engineered E. coli to characterize the resulting phagocytosis and killing.
BioBricks:
-BBa_K1137008 (TDMH)
Introduction
Mycobacterium tuberculosis (Mtb), the bacterium responsible for tuberculosis (TB), spreads by aerosol and infects its host through the airways. The bacterium is phagocytosed by macrophages in the lung, yet often evades death in the lysosome. Mtb can persist for years or even decades inside macrophages by inhibiting phagosome/lysosome fusion and supressing the normal acidification of the lysosome.
An efficient treatment for persistent TB must enter infected macrophages and kill the pathogen there. In our system, E. coli is both the vector and the therapeutic agent agent by expressing the gene LLO to enter macrophages and TDMH to kill mycobacteria.
TDMH and the mycobacterial cell wall
Mycobacterium species share a characteristic cell wall: thick, waxy, hydrophobic, and rich in mycolic acids. The low permeability of the envelope to hydrophilic solutes contributes to the intrinsic drug tolerance in mycobacteria.
Trehalose Dimycolate Hydrolase (TDMH) is a cutinase-like serine esterase that triggers rapid lysis of the mycobacterial cell wall by degrading the mycolate layer. The enzyme was first isolated from Mycobacterium smegmatis and subsequently shown to hydrolyze purified TDM from various mycobacterial species. Exposure to TDMH triggers an immediate release of free mycolic acids, ultimately leading to lysis of many mycobacteria including Mtb (Yang et al. 2012).
We have used M. smegmatis as a model system because Mtb are highly pathogenic and difficult to culture in the lab. M. smegmatis is a close relative of Mtb, shares many of its membrane properties, and is commonly used as a stand-in for Mtb physiology in the lab.
In our system, described in the methods below, we used E. coli BL21 (DE3) as a chassis to express TDMH from an IPTG-inducible strong T7 promoter.
Killing mycobacteria with TDMH
Figure 1 shows the killing of M. smegmatis with TDMH expressed by E. coli. We mixed liquid cultures of E. coli and M. smegmatis at equal cell densities as determined by plating assays. When expression of TMDH was induced with IPTG, nearly 99% of the M. smegmatis were killed within six hours. We saw no change in viability in cultures of M. smegmatis alone or when mixed with uninduced E. coli.
We next sought to quantify the effectiveness of TDMH killing. We mixed induced E. coli and mycobacteria in different ratios and used plating assays to measure viability. As shown in figure 2 mycobacterial killing displayed dose dependence on the E. coli cell density. Small numbers of E. coli could kill many mycobacteria. For example, in mixed populations with 100 mycobactera for each E. coli, we still observed >50% mycobacterial killing after 2 hours. This indicates that, on average, each E. coli produced enough TDMH to kill 50 mycobacterial. We reason that this killing may be even more effective inside macrophages, where constrained volumes will increase the effective TDMH concentration.
Bibliography
1. Yang Y, Bhatti A, Ke D, Gonzalez-Juarrero M, Lenaerts A, Kremer L, Guerardel Y, Zhang P, Ojha AK (2012) : Exposure to a cutinase-like serine esterase triggers rapid lysis of multiple mycobacterial species. J Biol Chem. 2013 Jan 4;288(1):382-92.
2. Rajesh Jayachandran, Varadharajan Sundaramurthy, Benoit Combaluzier , Philipp Mueller, Hannelie Korf, Kris Huygen, Toru Miyazaki, Imke Albrecht, Jan Massner, Jean Pieters (2007) : Survival of Mycobacteria in Macrophages Is Mediated by Coronin 1-Dependent Activation of Calcineurin. Cell, Volume 130, Issue 1, 13 July 2007, Pages 12-14



 "
"







 +33 1 44 41 25 22/25
+33 1 44 41 25 22/25


