Team:Paris Bettencourt/Project/Infiltrate
From 2013.igem.org

Background
: Latent tuberculosis persists inside macrophages of the lungs, where it is partially protected from both the host immune system and conventional antibiotics.
Aim:
To create an E. coli strain capable of entering the macrophage cytosol and delivering a lytic enzyme to kill mycobacteria.
Results:
- We expressed the enzyme Trehalose Dimycolate Hydrolase (TDMH) in E.coli and showed that it is highly toxic to mycobacteria in culture.
- We expressed the lysteriolyin O (LLO) gene in E. coli and showed that it is capable of entering the macrophage cytosol.
- We co-infected macrophages with both mycobacteria and our engineered E. coli to characterize the resulting phagocytosis and killing.
BioBricks:
-List of biobricks with links.
Introduction
Mycobacterium tuberculosis (Mtb), the bacterium responsible for tuberculosis (TB), spreads by aerosol and infects its host through the airways. The bacterium is phagocytosed by macrophages in the lung, yet often evades death in the lysosome. Mtb can persist for years or even decades inside macrophages by inhibiting phagosome/lysosome fusion and supressing the normal acidification of the lysosome.
An efficient treatment for persistent TB must enter infected macrophages and kill the pathogen there. In our system, E. coli is both the vector and the therapeutic agent agent by expressing the gene LLO to enter macrophages and TDMH to kill mycobacteria.
Experimental design
The killing assay in liquid media
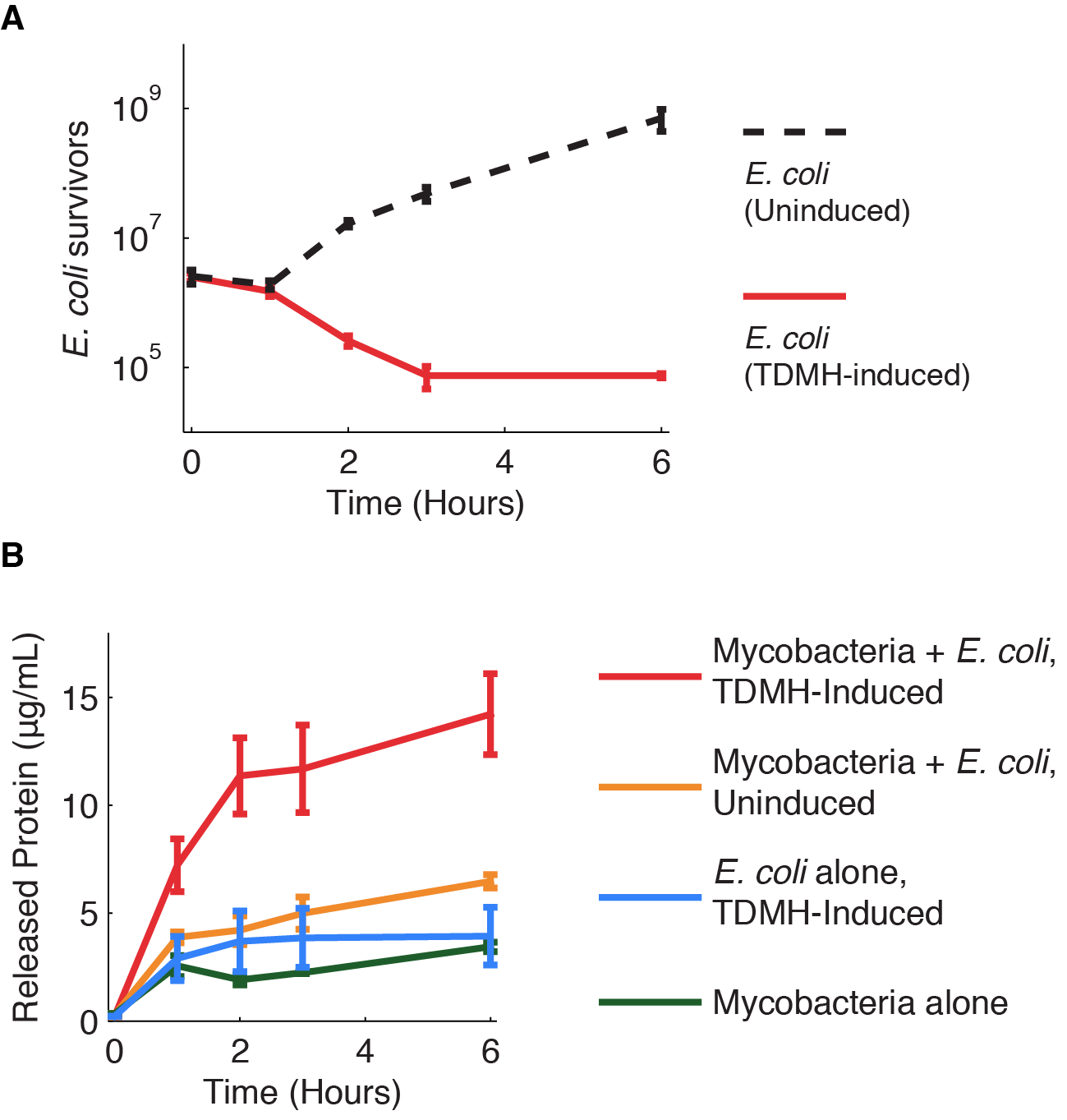
At first we wanted to investigate how TDMH producing E. coli is efficient in killing mycobacteria. E. coli BL21 was transformed with the TDMH carrying plasmid (pET21b) and with the pColA Duet plasmid carrying an RFP cassette with a constitutive Anderson promoter (BBa_J23102). The killing efficiency was tested on Mycobacterium smegmatis MC2, a non-phatogenic rapidly growing strain of mycobacteria. We cultivated E. coli BL21 and M. smegmatis MC2 in LB media, with different ratios of the two strains:
• 1 E. coli for 1 M. smegmatis
• 1 E. coli for 10 M. smegmatis
• 1 E. coli for 100 M. smegmatis
In these cultures E. coli was induced with 10 mM IPTG, which led to overexpression and release of TDMH in the media. The growth of both strains was measured for different time points (0, 1, 2, 3 and 6 h) by standard dilution series procedure and was expressed in CFUs/mL. Nalidixic acid plates (3 µg/mL) were used for following the growth of M. smegmatis m while the growth of E. coli was followed on kanamycin plates. For each time point the total amount of proteins was followed by the Bradford assay. Different control groups were also designed – a group where M. smgematis is grown without E. coli , a group where E. coli is induced and grown without M. smegmatis , and a group where M. smegmatis is was cultivated with the un-induced strain of E. coli .
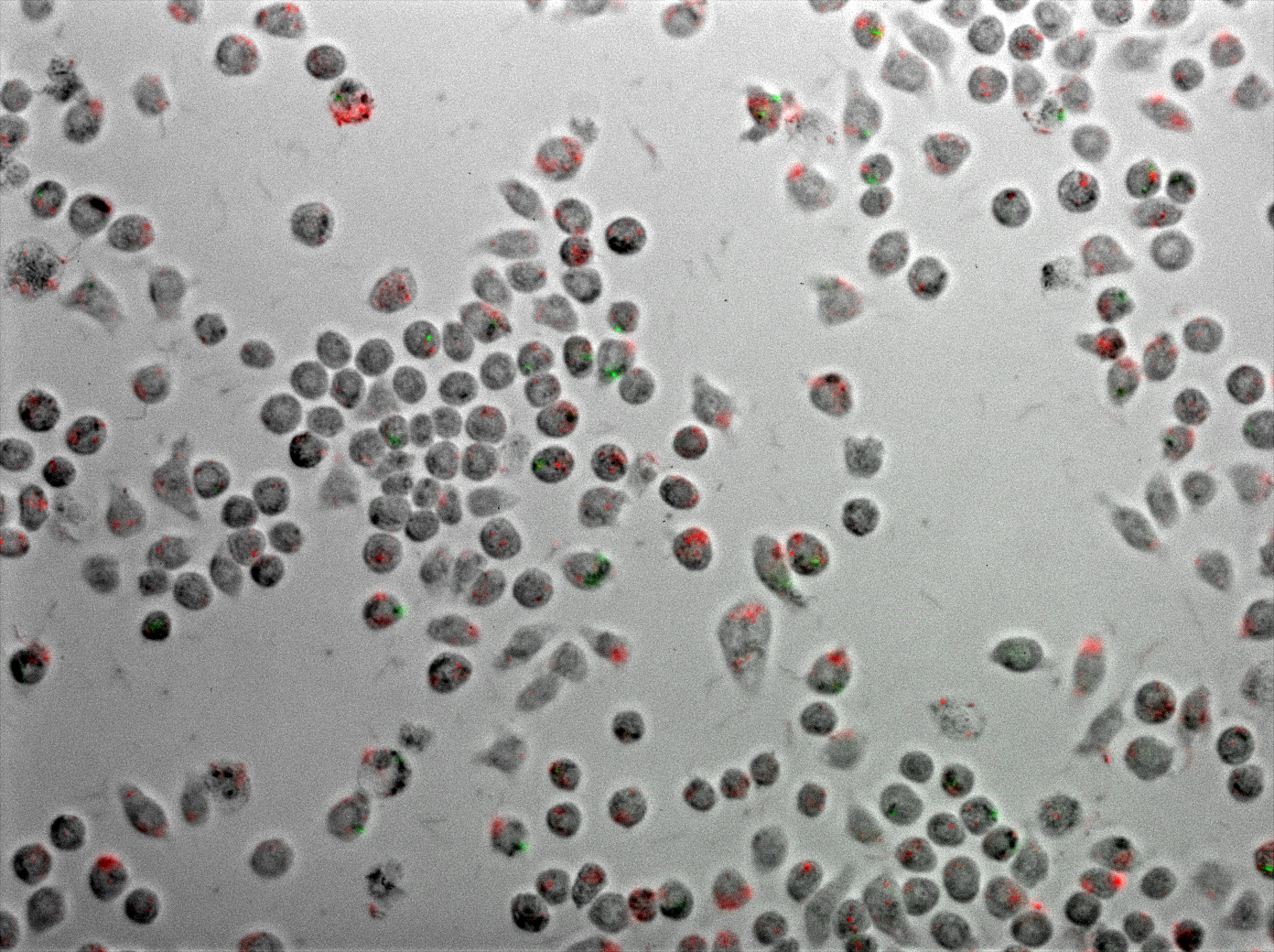
The killing assay inside the macrophages
To test if E. coli carrying TDMH could kill mycobacteria which are inside of the macrophages a J774 macrophage cell line was used. M. smegmatis MC2 was transformed with pMyco-gfp plasmid (Alibaud et al. 2011). E. coli BL21 was transformed with 3 plasmids: pET21b + TDMH, pACYC184 + hly and pColA + BBa_J23102. The macrophages were grown in 24 well plates in RPMI 1640 media supplemented with 1 % glutamine, 10% of fetal bovine serum, 1% HEPES, 1% sodium pyruvate. When around 90 % of confluence was reached, the cells were infected with M. smegmatis . Mycobacteria from an overnight culture were resuspended in RPMI and set to the final O.D. of 0.1. After infection, cells were incubated for 1 h (37 oC, 5% CO2) and then washed 10 times with PBS. After washing the cells new RPMI media was added and the cells were infected with an overnight culture of E. coli which was induced with 10 µM IPTG. E. coli were resuspended in RPMI where the final O.D. was set to 0.1. Then the infected cells were incubated for 1 h (37 oC, 5% CO2), washed 10 times with PBS and fresh RPMI media was added. Macrophages were then observed under the fluorescent microscope and after 3 h of incubation. We counted and compared the percentage of the macrophages which were infected with mycobacteria for these two time points. We also formed control groups: macrophages which were infected only with mycobacteria or only with E. coli so we could see the infection rate for these two strains; macrophages which were infected with M. smegmatis and with E. coli which wasn’t induced with IPTG. We also performed the same experiment where in the first infection step the cells were infected with E. coli and in the second one they were infected with mycobacteria.
Main results
Killing assay outside the macrophages
2 ratios were successful regarding the killing of M.smegmatis:
1 to 10 and 1 to 1.Whereas in the 1 to 100 ratio we observed a recovery of M.smegmatis.
In the figure 1 you can see in blue the behavior of M.smegmatis alone, in red the behavior of M.smegmatis when we add E.coli uninduced and in green the behavior of M.smegmatis when we add E.coli and we induced to produce TDMH.
We decided that we will use the 1/1 ratio for the rest of our experiments.
Killing assay inside the macrophages: microscopy and video movie
Discussion
Our results show an indeniable role of the protein TDMH in the lysis of M.smegmatis. This target is independant of the Mycobacteria’s growth making it a molecule of choice in the war against tuberculosis.
Using E.coli as a vector allows us to target the macrophages using a natural process: the phagocytosis. We showed that E.coli can enter a macrophage where there is already M.smegmatis. They didn’t seem to be in the same phagosome but the TDMH produced by E.coli can reach M.smegmatis. This is possible thanks to the lysteriolysin that lysed E.coli’s phagosome.
Perspectives
It will be interesting to study the stability of TDMH at the phagosome’s pH and to imagine a protein delivery system that can target the macrophages without using a living organism (for example like they do with imiglucerase). The problem is our protein is not glycosylated so it will not be that easy.
Bibliography
1. Yang Y, Bhatti A, Ke D, Gonzalez-Juarrero M, Lenaerts A, Kremer L, Guerardel Y, Zhang P, Ojha AK (2012) : Exposure to a cutinase-like serine esterase triggers rapid lysis of multiple mycobacterial species. J Biol Chem. 2013 Jan 4;288(1):382-92.
2. Rajesh Jayachandran, Varadharajan Sundaramurthy, Benoit Combaluzier , Philipp Mueller, Hannelie Korf, Kris Huygen, Toru Miyazaki, Imke Albrecht, Jan Massner, Jean Pieters (2007) : Survival of Mycobacteria in Macrophages Is Mediated by Coronin 1-Dependent Activation of Calcineurin. Cell, Volume 130, Issue 1, 13 July 2007, Pages 12-14



 "
"







 +33 1 44 41 25 22/25
+33 1 44 41 25 22/25


