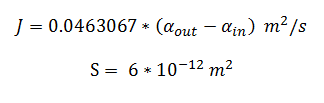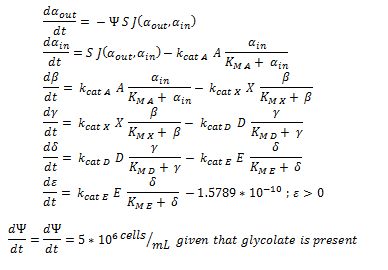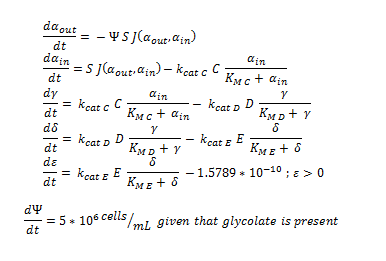Team:SydneyUni Australia/Modelling Results
From 2013.igem.org
| Line 61: | Line 61: | ||
=='''The Non-monooxygenase Pathway'''== | =='''The Non-monooxygenase Pathway'''== | ||
| - | ODE overview: | + | '''ODE overview:''' |
[[File: Igem_ode_11.png|450px]] | [[File: Igem_ode_11.png|450px]] | ||
| Line 88: | Line 88: | ||
if y(6) > 0.0005 | if y(6) > 0.0005 | ||
| - | if y(7) > 1. | + | if y(7) > 1.6*10^11 |
dy(7)=0 | dy(7)=0 | ||
else | else | ||
| Line 129: | Line 129: | ||
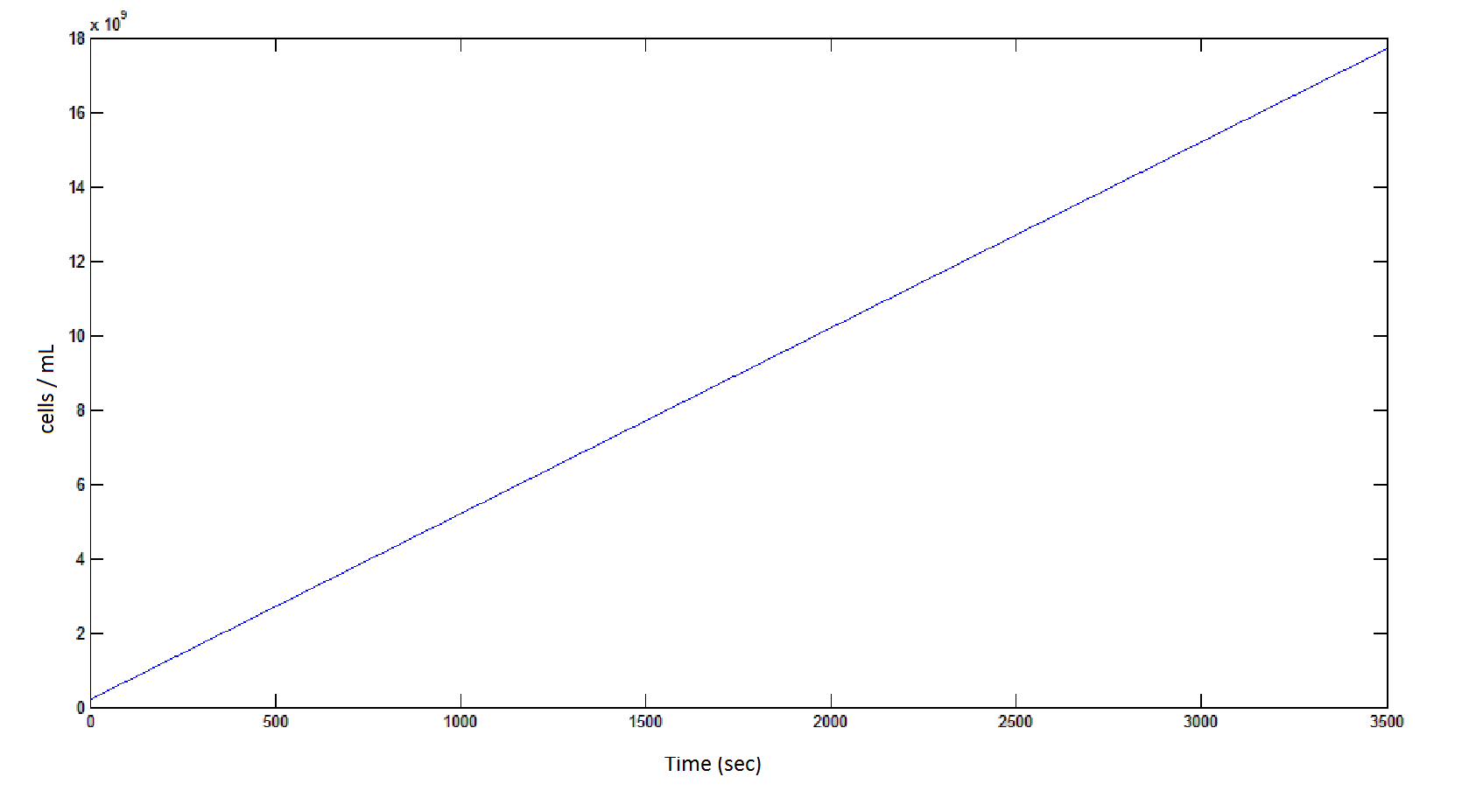
Graph 4: the blue line represents the linear increase of cells due to the presence of intracellular glycolate. | Graph 4: the blue line represents the linear increase of cells due to the presence of intracellular glycolate. | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | =='''The Non-monooxygenase Pathway'''== | ||
| + | |||
| + | '''ODE overview''': | ||
| + | |||
| + | [[File: Igem_ode_22.png|450px]] | ||
| + | |||
| + | |||
| + | |||
| + | '''Raw MATLAB code:''' | ||
| + | |||
| + | <nowiki> | ||
| + | function dy = p450(t,y) | ||
| + | |||
| + | dy=zeros(6,1); | ||
| + | |||
| + | dy(1)= -y(6)*(6*10^(-12))*0.0463067*(y(1)-y(2)); | ||
| + | dy(2)= ((6*10^(-12))*0.0463067*(y(1)-y(2))) - 0.0113*25.55*(y(2)/(.12+y(2))); | ||
| + | dy(3) = 0.0113*25.55*(y(2)/(.12+y(2))) - 0.6*25.55*(y(3)/(0.16+y(3))); | ||
| + | dy(4)= 0.6*25.55*(y(3)/(0.16+y(3))) - 25.4*25.55*(y(4)/(20+y(4))); | ||
| + | |||
| + | |||
| + | if y(5) > 2*10^(-10) | ||
| + | dy(5)= 25.4*25.55*(y(4)/(20+y(4))) -1.5789*10^(-10) | ||
| + | else | ||
| + | dy(5) = 25.4*25.55*(y(4)/(20+y(4))) | ||
| + | end | ||
| + | |||
| + | if y(5) > 0.0005 | ||
| + | if y(6) > 1.6*10^11 | ||
| + | dy(6)=0 | ||
| + | else | ||
| + | dy(6) = 5*10^6 | ||
| + | end | ||
| + | else | ||
| + | dy(6) = 0 | ||
| + | end | ||
| + | |||
| + | end | ||
| + | |||
| + | </nowiki> | ||
| + | |||
| + | '''MATLAB output:''' | ||
| + | |||
| + | '''The rate at which DCA is removed in solution:''' | ||
| + | |||
| + | [[File: DCAcominin1.png|950px]] | ||
| + | |||
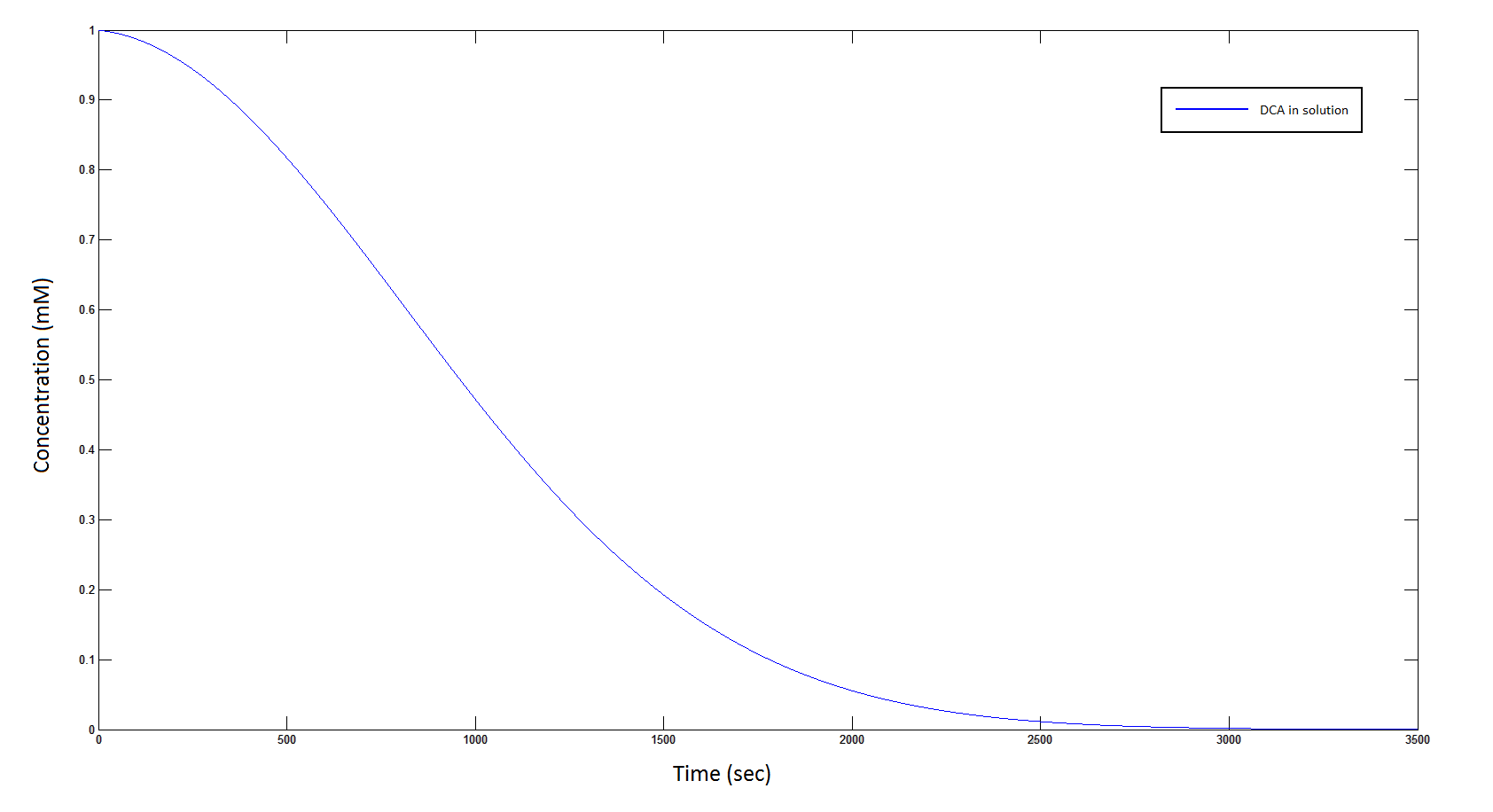
| + | Graph 1: The blue line represents how the concentration of DCA in solution decreases due to the action of our DCA degraders. The red line represents the intracellular concentions of the metabolites (disregard in this graph). One can see that an initial concentration of 2E8 cells/mL completely removes the DCA with a concentration 1mM in (roughly) 150mins. | ||
| + | |||
| + | '''The Rate at which the intracellular concentration of the metabolites change over time:''' | ||
| + | |||
| + | [[File: intermediates111.png|950px]] | ||
| + | |||
| + | Graph 2: Each line represents the concentration of each of the metabolites . This graph is simply a rescaling of graph 1. Note: the glycolate won’t accumulate in the cell – it is metabolised – the model had glycolate as the final product. It is used to show that the presence of glycolate can attribute to cell growth. | ||
| + | |||
| + | '''Rescaling the graph once again: the rate at which the metabolic intermediate change over time.''' | ||
| + | |||
| + | [[File: intermediates22222.png|950px]] | ||
| + | |||
| + | Graph 3: Each line represents the concentration of each of the metabolites . This graph is simply a rescaling of graph 1 and 2. | ||
| + | |||
| + | |||
| + | '''The rate at which the cells grow over time:''' | ||
| + | |||
| + | [[File: cellscellscelllls.png|950px]] | ||
| + | |||
| + | Graph 4: the blue line represents the linear increase of cells due to the presence of intracellular glycolate. | ||
| + | |||
| + | |||
| + | |||
Revision as of 04:51, 24 October 2013


Running the Model
The modelling for both the pathays had initial conditions of:
center>
| Constant | Value | Comment | ||||
|---|---|---|---|---|---|---|
| KM A | 0.530mM | From literature [] | ||||
| KM A | 0.530mM | From literature [] | ||||
| KM A | 0.530mM | From literature [] | ||||
| KM A | 0.530mM | From literature [] | ||||
| KM A | 0.530mM | From literature [] | ||||
| KM A | 0.530mM | From literature [] | ||||
| KM A | 0.530mM | From literature [] | ||||
| KM A | 0.530mM | From literature [] | ||||
| KM A | 0.530mM | From literature [] | ||||
| KM A | 0.530mM | From literature [] | ||||
| Alcohol Dehydrogenase | Adhlb 1/2* | B | ||||
| p450 | p450 | C | KM C = .120 mM, kcat C = 0.0113 s-1 | DCA | chloroacetaldehyde | [3] |
| Chloroacetaldehyde Dehydrogenase | aldA | D | KM D = 0.06mM, kcat D = 0.60 s-1 | chloroacetaldehyde | chloroacetate | [4] |
| Haloacetate Dehydrogenase | dhlB | E | KM E = 20 mM, kcat E = 25.4 s-1 | chloroacetate | glycolate | [5] |
</center>
and using the constants above the flux, J, took the value:
The Non-monooxygenase Pathway
ODE overview:
Raw MATLAB code:
function dy = nop450(t,y)
dy=zeros(7,1);
dy(1)= -y(7)*(6*10^(-12))*0.0463067*(y(1)-y(2));
dy(2)= ((6*10^(-12))*0.0463067*(y(1)-y(2)))-3.3*25.55*(y(2)/(0.53+y(2)));
dy(3)= 3.3*25.55*(y(2)/(0.53+y(2)))-0.0871*25.55*(y(3)/(0.94+y(3)));
dy(4)= .0871*25.55*(y(3)/(0.94+y(3)))- 0.6*25.55*(y(4)/(0.16+y(4)));
dy(5)= 0.6*25.55*(y(4)/(0.16+y(4))) - 25.4*25.55*(y(5)/(20+y(5)));
if y(6) > 2*10^(-10)
dy(6)= 25.4*25.55*(y(5)/(20+y(5))) -1.5789*10^(-10)
else
dy(6) = 25.4*25.55*(y(5)/(20+y(5)))
end
if y(6) > 0.0005
if y(7) > 1.6*10^11
dy(7)=0
else
dy(7) = 5*10^6
end
else
dy(7) = 0
end
end
MATLAB output:
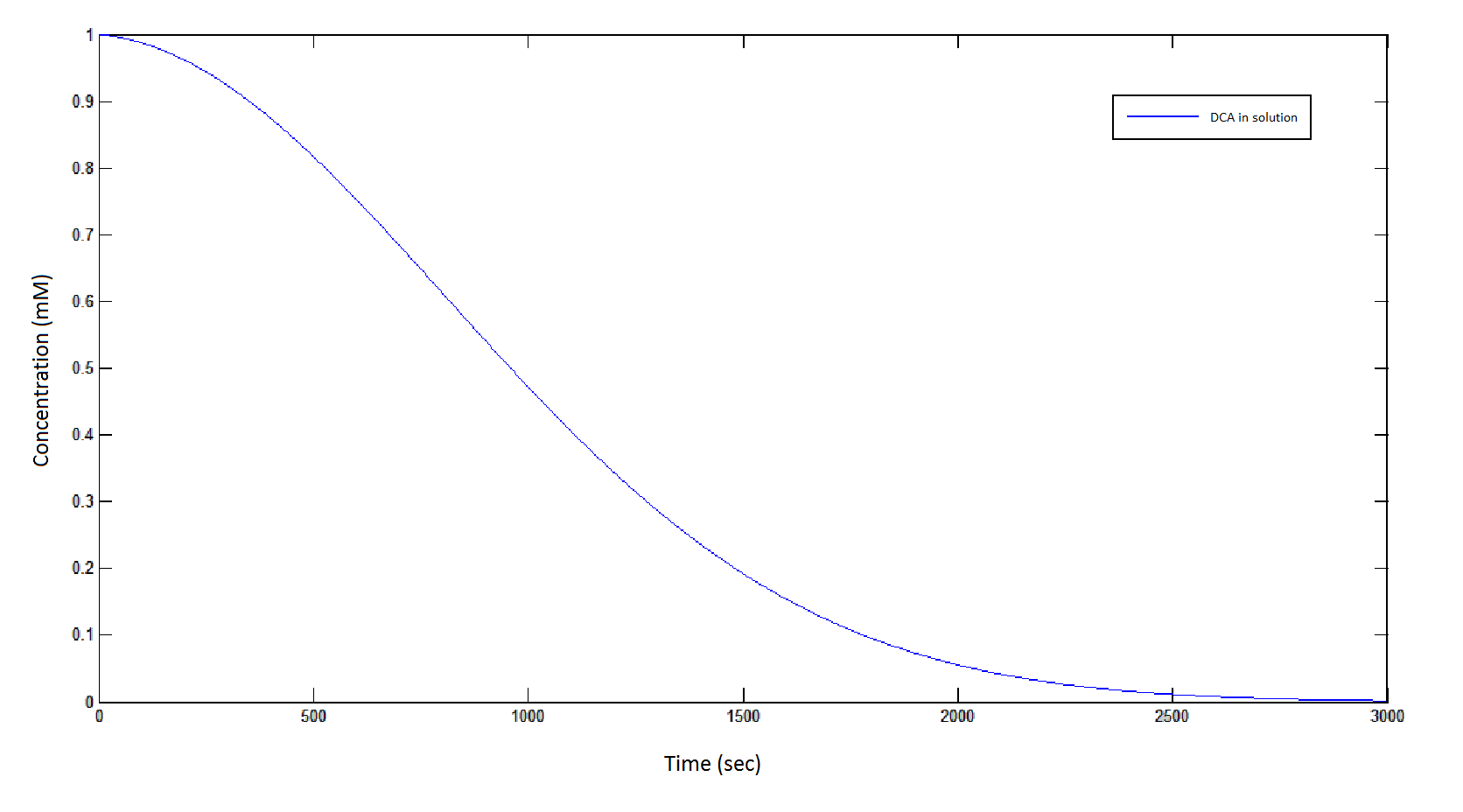
The rate at which DCA is removed in solution:
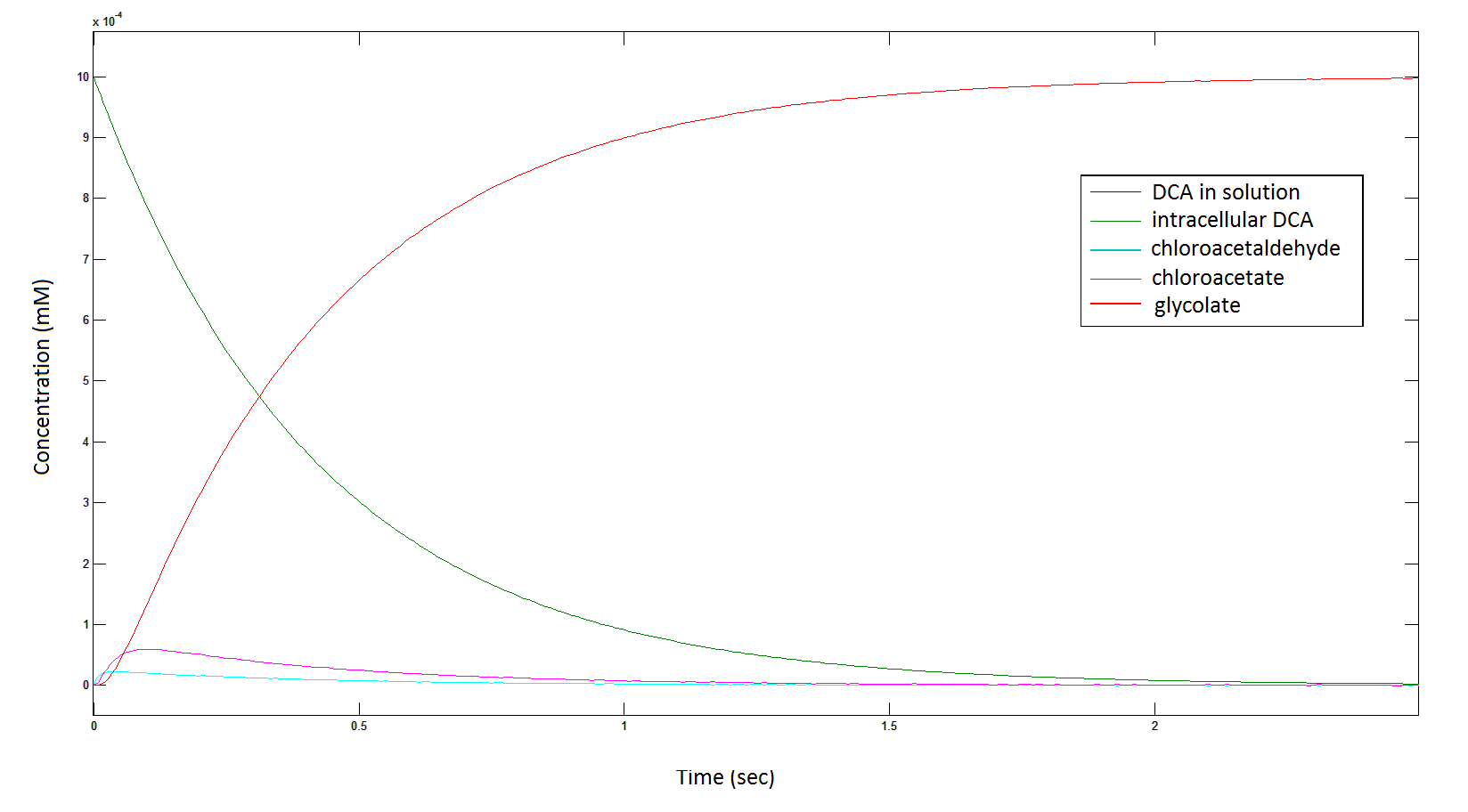
Graph 1: The blue line represents how the concentration of DCA in solution decreases due to the action of our DCA degraders. The red line represents the intracellular concentions of the metabolites (disregard in this graph). One can see that an initial concentration of 2E8 cells/mL completely removes the DCA with a concentration 1mM in (roughly) 150mins.
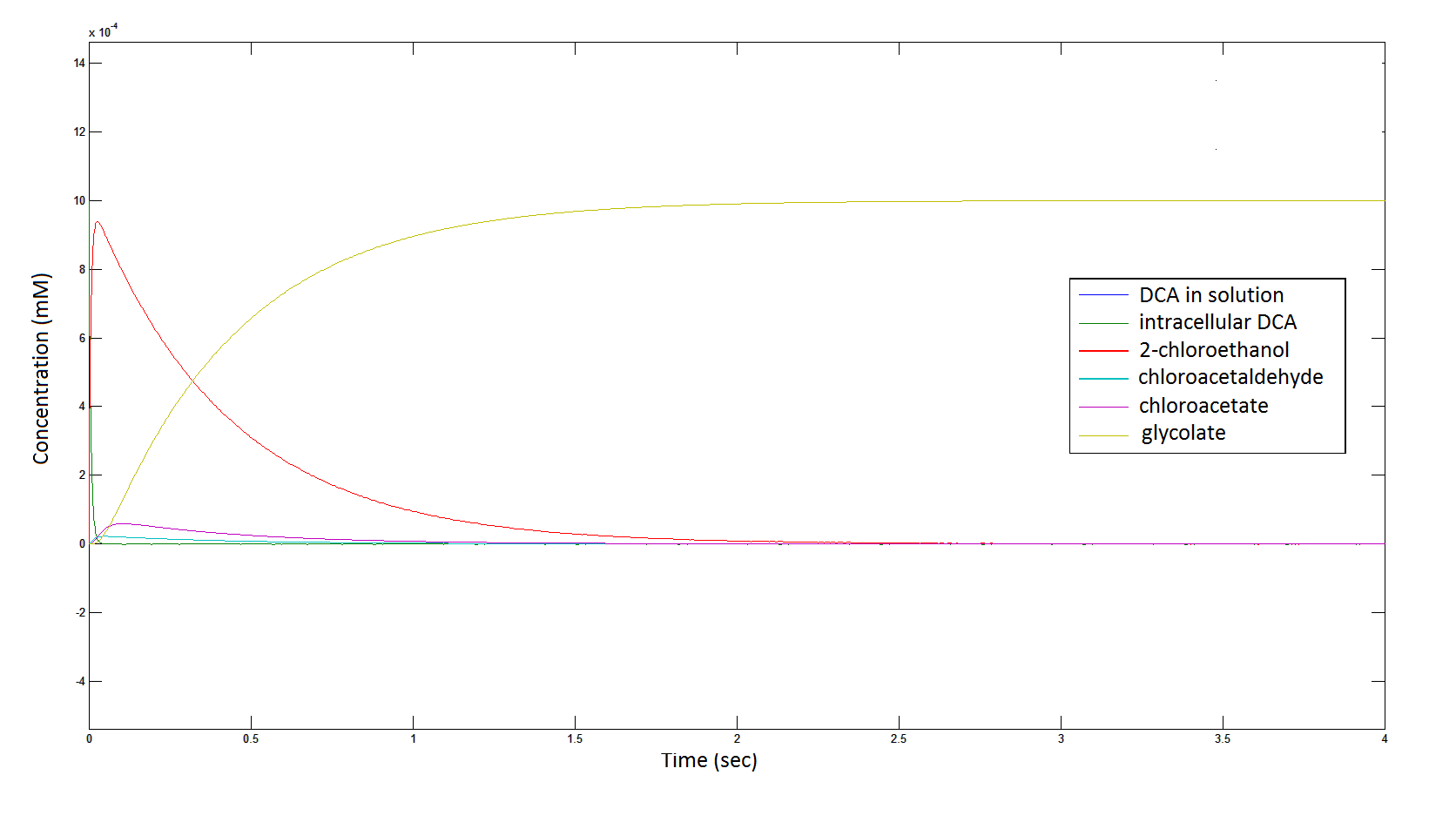
The Rate at which the intracellular concentration of the metabolites change over time:
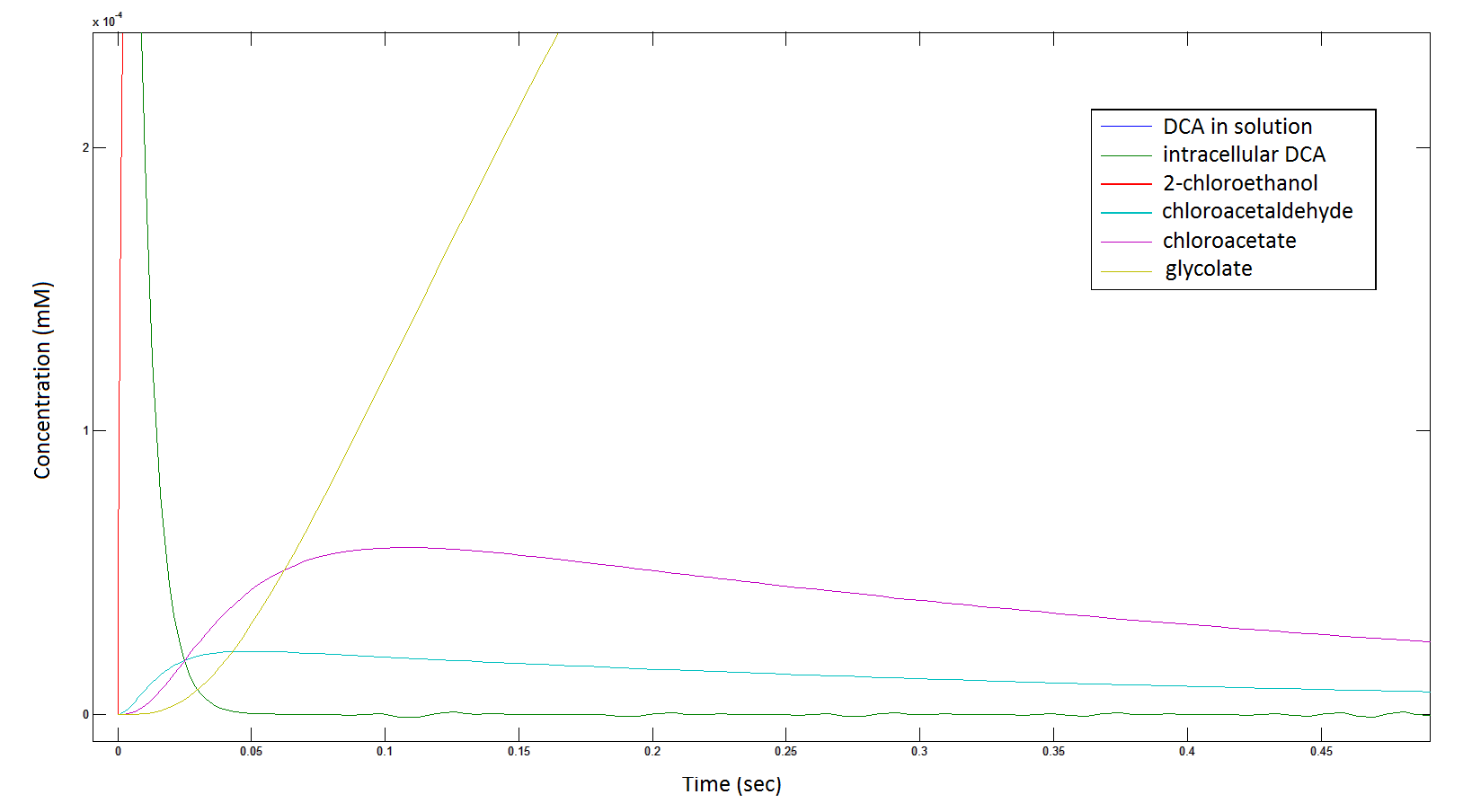
Graph 2: Each line represents the concentration of each of the metabolites . This graph is simply a rescaling of graph 1. Note: the glycolate won’t accumulate in the cell – it is metabolised – the model had glycolate as the final product. It is used to show that the presence of glycolate can attribute to cell growth.
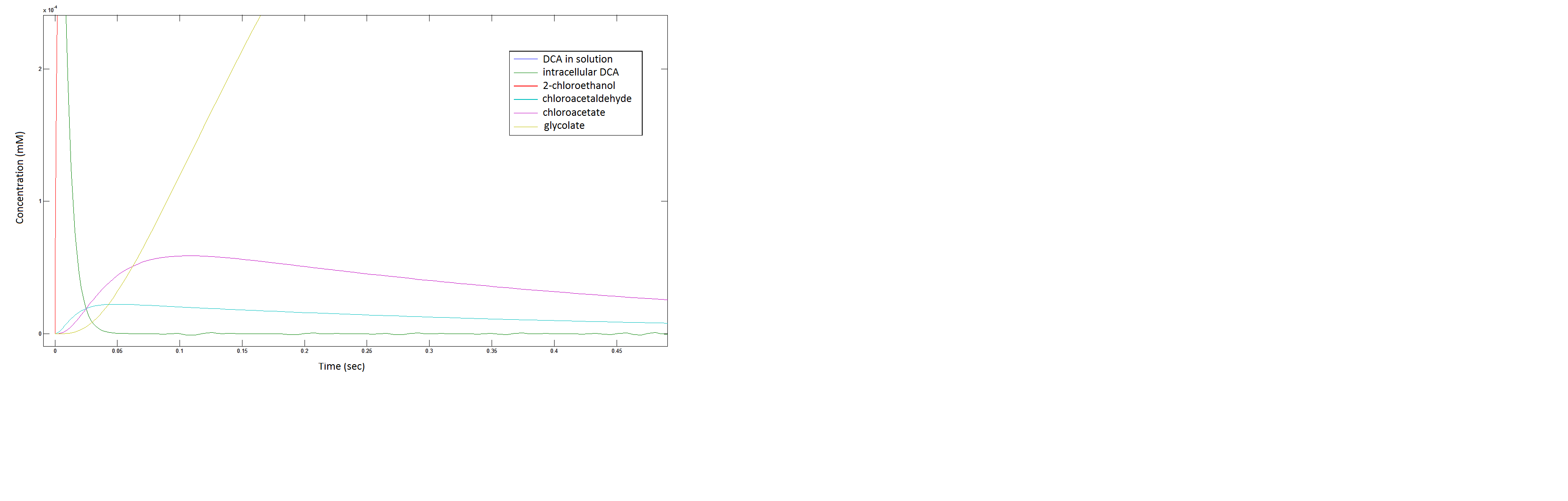
Rescaling the graph once again: the rate at which the metabolic intermediate change over time.
Graph 3: Each line represents the concentration of each of the metabolites . This graph is simply a rescaling of graph 1 and 2.
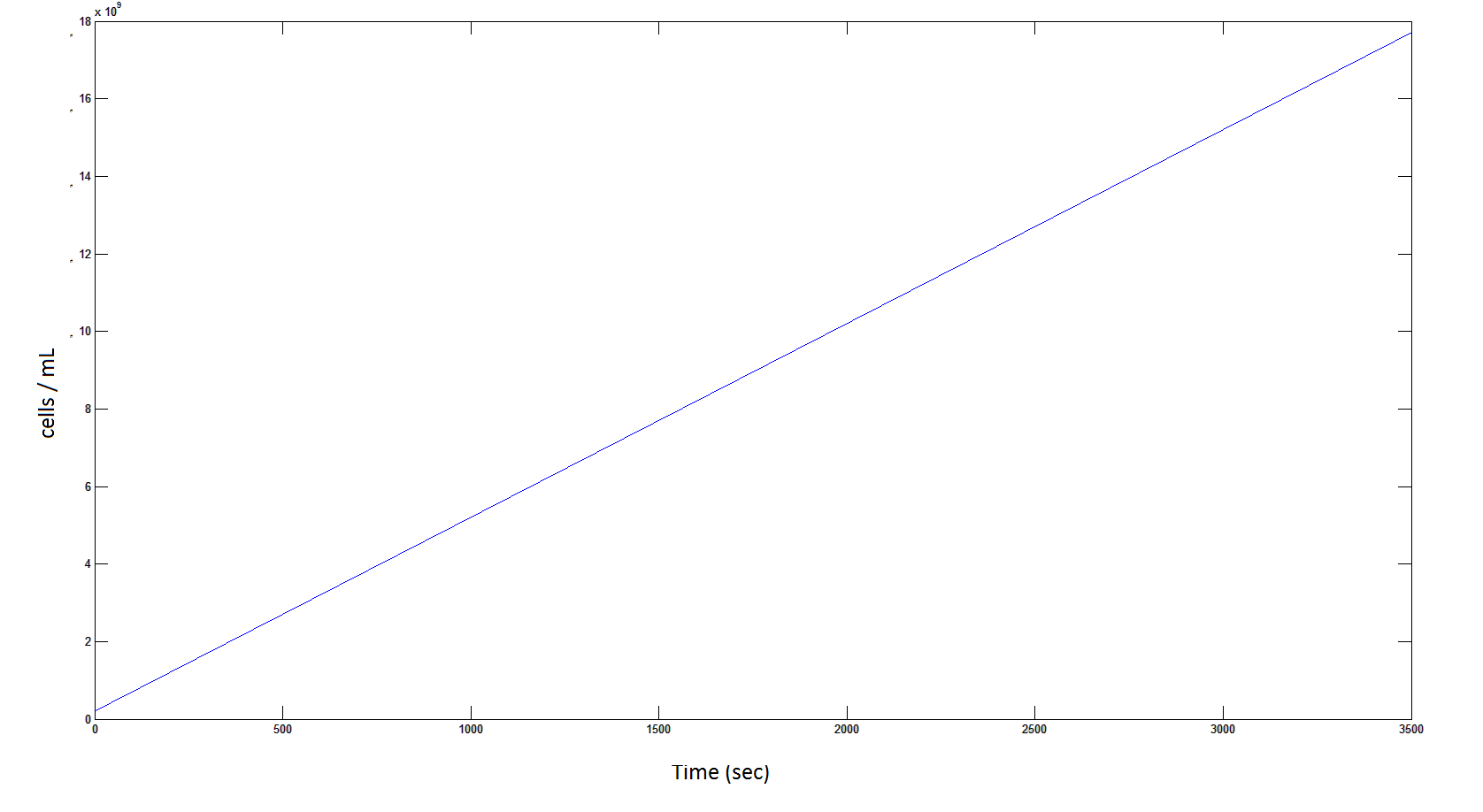
The rate at which the cells grow over time:
Graph 4: the blue line represents the linear increase of cells due to the presence of intracellular glycolate.
The Non-monooxygenase Pathway
ODE overview:
Raw MATLAB code:
function dy = p450(t,y)
dy=zeros(6,1);
dy(1)= -y(6)*(6*10^(-12))*0.0463067*(y(1)-y(2));
dy(2)= ((6*10^(-12))*0.0463067*(y(1)-y(2))) - 0.0113*25.55*(y(2)/(.12+y(2)));
dy(3) = 0.0113*25.55*(y(2)/(.12+y(2))) - 0.6*25.55*(y(3)/(0.16+y(3)));
dy(4)= 0.6*25.55*(y(3)/(0.16+y(3))) - 25.4*25.55*(y(4)/(20+y(4)));
if y(5) > 2*10^(-10)
dy(5)= 25.4*25.55*(y(4)/(20+y(4))) -1.5789*10^(-10)
else
dy(5) = 25.4*25.55*(y(4)/(20+y(4)))
end
if y(5) > 0.0005
if y(6) > 1.6*10^11
dy(6)=0
else
dy(6) = 5*10^6
end
else
dy(6) = 0
end
end
MATLAB output:
The rate at which DCA is removed in solution:
Graph 1: The blue line represents how the concentration of DCA in solution decreases due to the action of our DCA degraders. The red line represents the intracellular concentions of the metabolites (disregard in this graph). One can see that an initial concentration of 2E8 cells/mL completely removes the DCA with a concentration 1mM in (roughly) 150mins.
The Rate at which the intracellular concentration of the metabolites change over time:
Graph 2: Each line represents the concentration of each of the metabolites . This graph is simply a rescaling of graph 1. Note: the glycolate won’t accumulate in the cell – it is metabolised – the model had glycolate as the final product. It is used to show that the presence of glycolate can attribute to cell growth.
Rescaling the graph once again: the rate at which the metabolic intermediate change over time.
Graph 3: Each line represents the concentration of each of the metabolites . This graph is simply a rescaling of graph 1 and 2.
The rate at which the cells grow over time:
Graph 4: the blue line represents the linear increase of cells due to the presence of intracellular glycolate.
References:
[1] Krooshof, G. H., I. S. Ridder, et al. (1998). "Kinetic Analysis and X-ray Structure of Haloalkane Dehalogenase with a Modified Halide-Binding Site†." Biochemistry 37(43): 15013-15023.
[2] Janecki, D. J., K. G. Bemis, et al. (2007). "A multiple reaction monitoring method for absolute quantification of the human liver alcohol dehydrogenase ADH1C1 isoenzyme." Analytical Biochemistry 369(1): 18-26.
[3] Pandey, A. V. and C. E. Flück (2013). "NADPH P450 oxidoreductase: Structure, function, and pathology of diseases." Pharmacology & Therapeutics 138(2): 229-254.
[4] van der Ploeg, J., Shmidt, M. P., Landa, A. S., and Janssen, D. B. (1994). "Identification of Chloroacetaldehyde Dehydrogenase Involved in 1,2-Dichloroethane Degradation." Applied Environmental Microbiology (60(5): 1699-1605.
[5] van der Ploeg, J., van Hall, G., Janssen, D. B. (1991) "Characterization of the haloacid dehalogenase from Xanthobacter autotrophicus GJ10 and sequencing of the dhlB gene." Journal of Bacteriology 173(24):7925-33.
[6] Sinensky, Mi “Homeoviscous Adaption – A Homeostatic Process that Regulates the Viscosity of Membrane Lipids in Escheria coli” Proceedings from the National Academy of Science 71(2): 522-525.
[7] CyberCell Database
 "
"