Team:Tokyo Tech
From 2013.igem.org
| Line 89: | Line 89: | ||
<br><br> | <br><br> | ||
<div id="marginbox"> | <div id="marginbox"> | ||
| - | + | <h3 style="color:#d3381c;">Click a step shown below to know what happens in our circuit!</h3> | |
<table align="center" border=0> | <table align="center" border=0> | ||
<tr><td> | <tr><td> | ||
Revision as of 06:35, 28 October 2013
Project BackgroundIn this iGEM Competition, we intended to tell the public the development of synthetic biology, especially about the network programming, as well as we enjoyed our activity for iGEM. Tokyo Tech 2013 assisted with an experiment workshop for high school students, participated in a poster session and collected feedback from public people as human practice (Fig. 1-1-1). Now we know that an interesting story makes public people easily understand the importance of programming genetic circuits in synthetic biology. To respond to the public's expectations further, we also address a farming issue. Thus we aimed to catch this story into E. coli, the life of ninja: battle and farming. |
 [Fig. 1-1-1. Poster session at the university of Tokyo]
|
|||||||
[Fig. 1-1-2. Ninja vs. Samurai in Tokyo Tech]
|
StoryNinja is a Japan’s ancient spy-warrior. Ninja usually mimics himself as an ordinary civilian. Once he detects samurai who is the target of assassination, he immediately gets ready for battle. He attacks samurai with shuriken, throwing knives (Fig. 1-1-2). |
|||||||
Click a step shown below to know what happens in our circuit!
|
||||||||
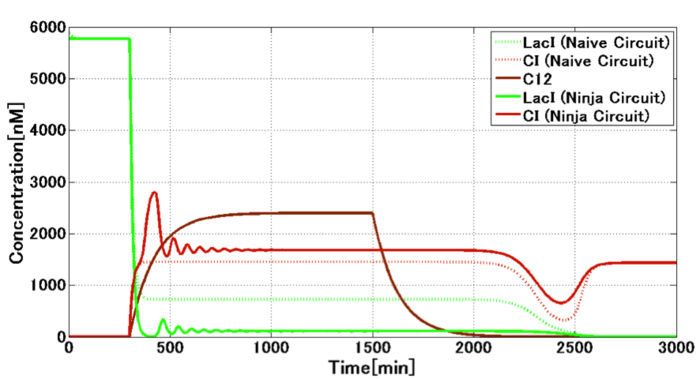
Project OverviewIn our programming of artificial genetic circuit, E. ninja heads casts. In response to E. civilian signal or E. samurai signal, E. ninja changes its state: “Mimic state” or “Attack state.” The circuit of E. ninja contains a bi-stable switch part and a signal dependent switching part. We decided to use 3OC6HSL and 3OC12HSL as the signals. The crosstalk between these two signals is well known as a significant problem in synthetic biology. To realize an accurate switching, we designed circuit which achieved the circumvention of the crosstalk that occurs in bacterial cell-cell communication system (Fig. 1-1-4). |
[Fig. 1-1-3. The result of our wet experiment for the circumvention of the crosstalk] |
|||||||
 [Fig. 1-1-4 Our designed circuit for circumvention of the crosstalk]
|
||||||||
|
Our wet experiment results showed that the combination of lux/tet hybrid promoter and TetR protein circumvented the crosstalk by preventing the LasR protein from acting on LuxR-binding sequences (Fig. 1-1-3). Our mathematical model based on these results showed the circumvention of the crosstalk in the circuit including toggle switch and crosstalk circumvention system(Fig. 1-1-5). |
||||||||
[Fig. 1-1-5. Our mathematical model for the circuit of E. ninja] |
||||||||
 [Fig. 1-1-6. Our new part for inducible phage release]
|
 [Fig. 1-1-7. Distribution of plaques and analysis] |
In addition, E. ninja releases M13 phage, which corresponds to shuriken, when E. ninja receives E. samurai signal. The inducible phage release will open new way in synthetic biology by achieving programmed DNA messaging (Fig. 1-1-6). |
||||||
|
In the second-life story, E. ninja starts farming in a peaceful village. He can increase plant growth by synthesizing several plant hormones depending on the soil environment. We constructed an improved phosphate sensor (phoA promoter, BBa_K1139201). Also, we learned methods for quantitative analysis of cytokinin, one of the plant hormone, through a bioassay of cucumber seed sprouts (Fig. 1-1-8). Towards further consideration of farming with microbes, we have also continued the human practice investigation through some interviews with science foundations and organizations (Fig. 1-1-8). Future Works |
 [Fig. 1-1-8. Our bioassay of cucumber seed sprouts]
|
|||||||
|
We believe that our results can also contribute to various fields. First, our crosstalk circumvention system gives more flexibility to design genetic circuits. Because, by adding only a few genes, you can circumvent crosstalk of AHL. Second, our inducible phage release system can make DNA messaging more complex and more diverse. Moreover, for bioremediation, we can search for new M13 phage hosts by using the M13 phage we have designed. We hope to contribute to spreading the importance and the great possibilities of synthetic biology through the public. Our farming project is expected to act as a pioneering trail toward new approaches in farming. Especially, our strategy to produce plant hormones in temporal patterns in E. coli will be applied to studying the plants’ response to external plant hormones. |
||||||||
 "
"