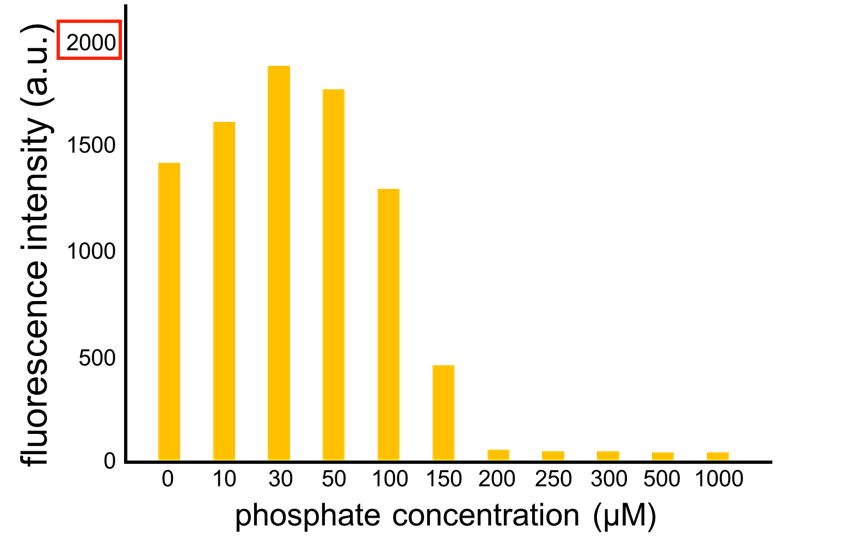Team:Tokyo Tech/Submitted Parts
From 2013.igem.org
| Line 75: | Line 75: | ||
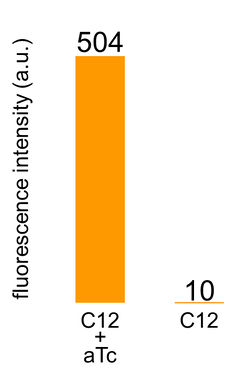
[[Image:Titech2013_Submitted_Parts_Fig_5-1-6_K1139110.jpg|200px|thumb|right|Fig. 5-1-6. The result of crosstalk circumvention]] | [[Image:Titech2013_Submitted_Parts_Fig_5-1-6_K1139110.jpg|200px|thumb|right|Fig. 5-1-6. The result of crosstalk circumvention]] | ||
</h2> | </h2> | ||
| + | <br><br> | ||
<h3>Best new BioBrick part (engineered): [http://parts.igem.org/Part:BBa_K1139110 BBa_K1139110]</h3> | <h3>Best new BioBrick part (engineered): [http://parts.igem.org/Part:BBa_K1139110 BBa_K1139110]</h3> | ||
<h2> | <h2> | ||
| Line 81: | Line 82: | ||
</p> | </p> | ||
</h2> | </h2> | ||
| - | <br><br><br><br> | + | <br><br><br><br><br><br> |
</div><br> | </div><br> | ||
</div> | </div> | ||
Revision as of 09:22, 27 September 2013
Parts submitted to the Registry
Contents |
For a brief overview of our main results, please have a look at our Main Results page. Go to “Home”
Favorite Tokyo_Tech 2013 iGEM Team Parts
| Name | Type | Description | Designer | Length | |
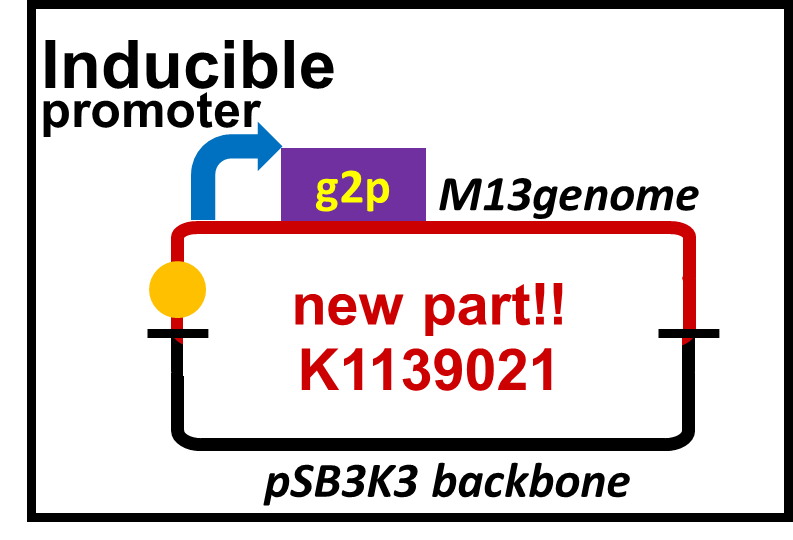
| W | BBa_K1139021 | Composite | Plux-M13-Plac-GFP | Naoki Watarai | 7705 |
| W | BBa_K1139110 | Composite | Pcon-LasR-Plux/tet-GFP | Naoki Watarai | 1888 |
| W | BBa_K1139201 | Measurement | PphoA-GFP-TT | Sara Ogino | 1017 |
Tokyo_Tech 2013 iGEM Team Parts
| Name | Type | Description | Designer | Length | |
| BBa_K1139019 | Composite | Promoterless-M13 | Naoki Watarai | 6400 | |
| W | BBa_K1139020 | Composite | PlacIQ-M13-Plac-GFP | Naoki Watarai | 7406 |
| BBa_K1139150 | Measurement | PRM/lac-GFP-TT | Naoki Watarai | 1032 |
Best new BioBrick part (natural and engineered): BBa_K1139021
M13 is a filamentous phage that infects only F+ strains of E. coli, which does not kill the host cell. This biobrick is extracted from M13mp18 phage vector by PCR. It includes 11 ORFs, M13 origin, a packaging sequence and lac promoter. The promoter on the upstream of g2 (gene 2) is altered to lux promoter. A phage particle is formed only when the host cell receives AHL signal (3OC6HSL, C6) because g2p (gene 2 protein) is an endonuclease needed for a plasmid to be replicated by M13 origin, and to be packaged into the phage particle. As a reporter, GFP is inserted on the downstream of the lac promoter.
Best new BioBrick part (natural): BBa_K1139020
We confirmed that M13 genome with two modifications related to our design kept plaque forming activity. One is replacement of the promoter for G2p protein with a constitutive promoter, PlacIq (BBa_I14032). The other is accommodation of pSB3K3 backbone. Even though the plasmid has two different types of replication origins, M13 origin and pSB3 origin, this plasmid (BBa_K1139020, Fig. 5-1-2) formed plaque. In contrast, construction intermediates without a promoter for g2p coding sequence (Promoterless-M13 + Plac, Promoterless-M13 + Plac-GFP BBa_K1139022, Fig. 5-1-3) could not form plaque.
Fig. 5-1-2. PlacIQ-M13-Plac-GFP on pSB3 (BBa_K1139020) |
Fig. 5-1-3. Promoterless-M13-Plac-GFP on pSB3 (BBa_K1139022) |
Best improved part and Best new BioBrick part (natural):
BBa_K1139201
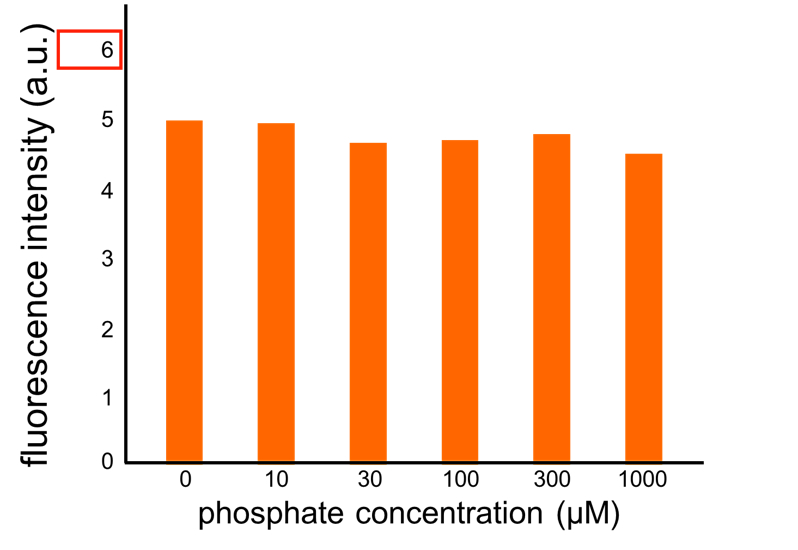
We improved a phosphate sensor part since the existing phosphate sensor part (OUC-China 2012, BBa_K737024) did not have sufficient data. We constructed this part by amplifying the phoA promoter region of E. coli (MG1655) and ligating it upstream of GFP part. Compared to OUC-China’s phosphate sensor part including phoB promoter (Fig. 5-1-5), our phosphate sensor part shows clearer result (Fig. 5-1-4).
Best new BioBrick part (engineered): BBa_K1139110
We constructed BBa_K1139110 by combining Pcon-lasR (BBa_K553003) and Plux/tet-GFP (BBa_K934025). This is the first Biobrick part that succeeded in confirming circumvention of crosstalk of LasR to lux promoter. Using this part with plasmid that is constitutively expressing luxR and tetR, we succeeded in confirming circumvention of crosstalk LasR to lux promoter. To know more about crosstalk circumvention assay, please see here.
 "
"