Team:UGent/Experiments
From 2013.igem.org
| Line 6: | Line 6: | ||
<h2> Experiment 1 </h2> | <h2> Experiment 1 </h2> | ||
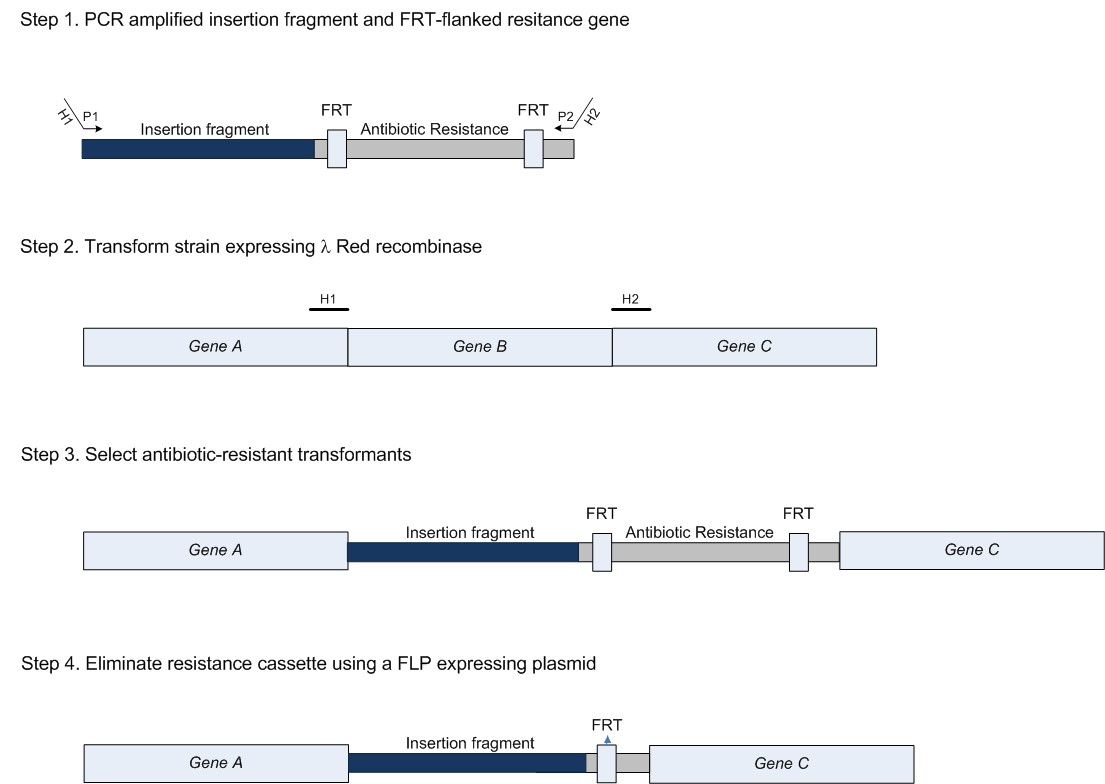
<p> Experiment 1 is the <b>knock-in</b> (KI) of the construct containing ccdA, GFP and the homologous regions (this was constructed beforehand). This will be done by using the method of Datsenko & Wanner [PNAS 2000], based on homologous recombination. The principle of the KO/KI method is depicted below: </p> | <p> Experiment 1 is the <b>knock-in</b> (KI) of the construct containing ccdA, GFP and the homologous regions (this was constructed beforehand). This will be done by using the method of Datsenko & Wanner [PNAS 2000], based on homologous recombination. The principle of the KO/KI method is depicted below: </p> | ||
| - | </html> | + | |
| + | <table> | ||
| + | <tr> | ||
| + | <td></html> | ||
[[File:UGent_2013_Datsenko-Wanner.jpg|thumb|400px|center|Datsenko-Wanner, 2000]] | [[File:UGent_2013_Datsenko-Wanner.jpg|thumb|400px|center|Datsenko-Wanner, 2000]] | ||
| - | <html> | + | <html></td> |
| - | < | + | <td><img src="https://static.igem.org/mediawiki/2013/4/44/UGent_2013_Exp1.jpg" width="500"> |
| - | < | + | </td> |
| - | </ | + | /tr> |
| + | </table> | ||
| + | |||
| + | <A HREF="https://static.igem.org/mediawiki/2013/f/ff/UGent_2013_Experiment_1_KI_ccdA-Pmb1-GFP.pdf" target="_blank">Protocol experiment 1</A> | ||
<br> | <br> | ||
Revision as of 14:08, 1 October 2013
|
ExperimentsTo test our idea we are conducting 6 experiments. These are described below. Experiment 1Experiment 1 is the knock-in (KI) of the construct containing ccdA, GFP and the homologous regions (this was constructed beforehand). This will be done by using the method of Datsenko & Wanner [PNAS 2000], based on homologous recombination. The principle of the KO/KI method is depicted below:
Experiment 2Next is finding a way to administer the toxin to the cells. This will be done by putting the gene coding for the toxin on a plasmid (a linear piece of DNA) and transferring this plasmid into the bacterial cell. In experiment 2 a plasmid containing ccdB (toxin) under control of a T7 promotor will be constructed. The T7 promoter allows us to control the expression of ccdB by regulating the amount of IPTG added to the cells. Protocol experiment 2Experiment 3Once we have constructed the plasmid with ccdB and a T7 promoter, it has to be transferred into the cells in which CIChE will be performed. This “transformation” (the process of putting DNA into a cell) will be carried out in experiment 3. Protocol experiment 3Experiment 4Strains constructed in experiment 3 will be used to perform CIChE. Tandem gene replication of reporter protein GFP will be induced by replicating the antitoxin ccdA as a response on titration of the toxin ccdB. Titration of ccdB under inducible T7-promoter will be accomplished by different levels of IPTG [0.01 mM – 0.5mM] and different plasmid copy numbers [p5, p10 and p20]. Protocol experiment 4Experiment 5Experiment 6In this experiment, a new part, called BBa_K1105000, will be constructed by cloning Laciq-T7ccdB with standard Biobrick prefix and suffix in pSB1C3. Laciq-T7ccdB is derived from the plasmid p10-LacIq-T7ccdB, which originally comes from a mini F-plasmid positive strains (such as E. coli F+). The function of this part is to produce CcdB (in control of a T7 promotor), which interferes with the topoisomerase unit gyrA. Protocol experiment 6
|

Tweets van @iGEM_UGent |
||
|
|
 "
"