Team:Yale/Project Validate
From 2013.igem.org
(Difference between revisions)
(→Synthesizing and Assembling the Heterologous Enzymes) |
(→Synthesizing and Assembling the Heterologous Enzymes) |
||
| Line 35: | Line 35: | ||
|- | |- | ||
|style="padding-right: 20px;"|[[File:Step 1 outline.jpg|400px]] | |style="padding-right: 20px;"|[[File:Step 1 outline.jpg|400px]] | ||
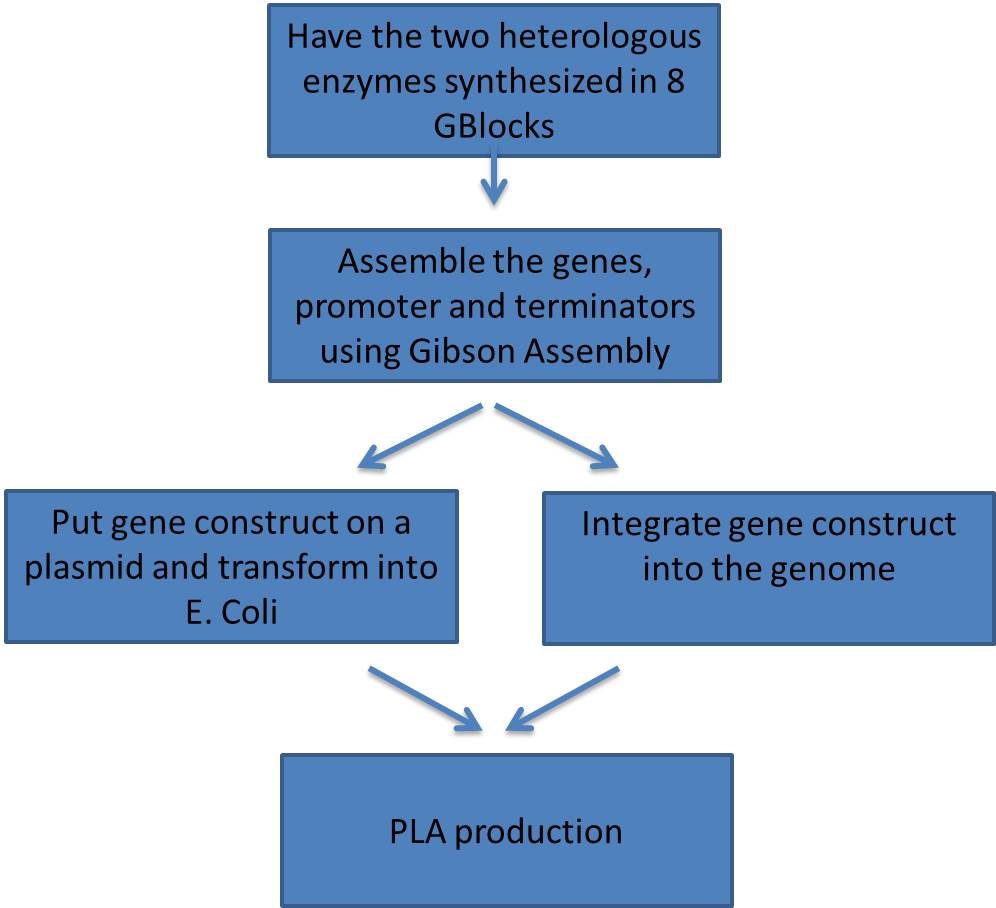
| - | |This was our plan in order to validate PLA synthesis. | + | |This was our plan in order to validate PLA synthesis. To save money we ordered each of the two heterologous genes in four pieces from IDT giving us 8 GBlocks. An inducible promoter preceded each gene so we could tightly regulate the expression of the enzyme. The terminators were amplified from other sources and given homology to the appropriate Gblocks. |
|} | |} | ||
<br> | <br> | ||
| Line 43: | Line 43: | ||
<center>[[File:GblockPLA.png|800px]]</center><br> | <center>[[File:GblockPLA.png|800px]]</center><br> | ||
<br> | <br> | ||
| - | Each fragment was amplified using PCR | + | Each fragment was amplified using PCR to give homology to adjacent fragments. Using Gibson assembly, our plan was to assemble all 10 fragments into one construct. <br><br> |
<center>[[File:PLAgenes.png|800px]]</center><br> | <center>[[File:PLAgenes.png|800px]]</center><br> | ||
<br> | <br> | ||
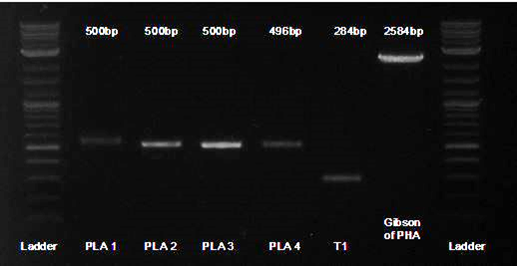
| - | + | To facilitate the process, we used Gibson assembly on each gene separately. Here is a gel of the first 4 Gblocks (labeled PLA1-4), along with the first terminator (labeled T1), and then the 5 pieces assembled together.<br> | |
<br> <center>[[File:PHAgel.JPG|600px]] [[File:PHA.png|200px]]</center><br> | <br> <center>[[File:PHAgel.JPG|600px]] [[File:PHA.png|200px]]</center><br> | ||
Revision as of 01:49, 6 September 2013
| Project Overview | Validate PLA synthesis | Develop bioassay | Apply MAGE | Introduce export system | Make a bioplastic |
|---|
Contents |
Aims for the Project
- Engineer strains of E. coli to validate PLA synthesis
- Develop bioassay to screen PLA production
- Apply MAGE to optimize PLA production, guided by FBA
- Introduce type 1 secretion system to export and extract PLA
- Make a bioplastic
Engineer strains of E. coli to validate PLA synthesis
Synthesizing and Assembling the Heterologous Enzymes
- In order to reproduce the results of the Lee group we needed to insert the two heterologous genes.
- Clostridum propionicum - propionate CoA transferase (denoted PCT)
- Pseudomonas resinovorans - polyhydroxyalkanoate synthase (denoted PHA)
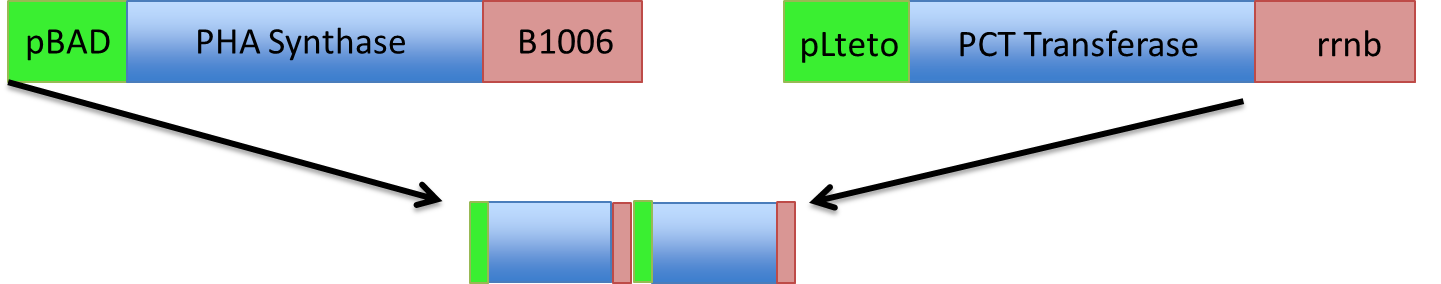
Here is a schematic of what the entire construct looks like with both promoter, genes and terminators.
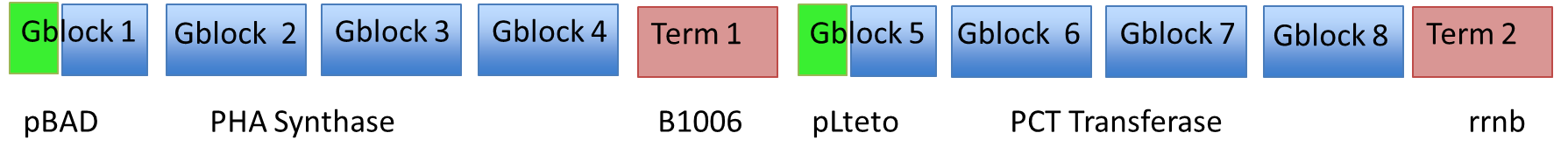
Each fragment was amplified using PCR to give homology to adjacent fragments. Using Gibson assembly, our plan was to assemble all 10 fragments into one construct.

To facilitate the process, we used Gibson assembly on each gene separately. Here is a gel of the first 4 Gblocks (labeled PLA1-4), along with the first terminator (labeled T1), and then the 5 pieces assembled together.

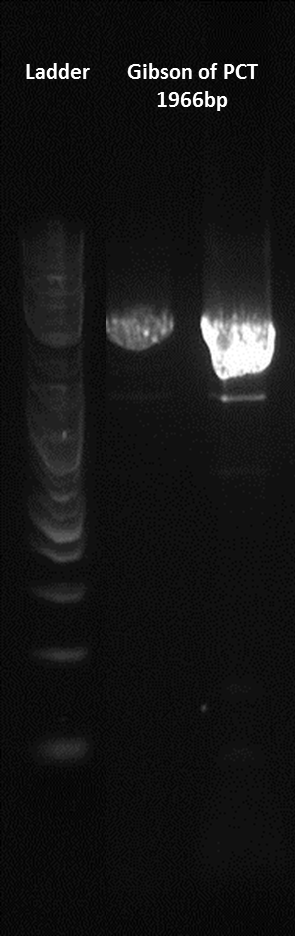
Here is a gel of the assembly of the last 4 Gblocks, along with the second terminator.

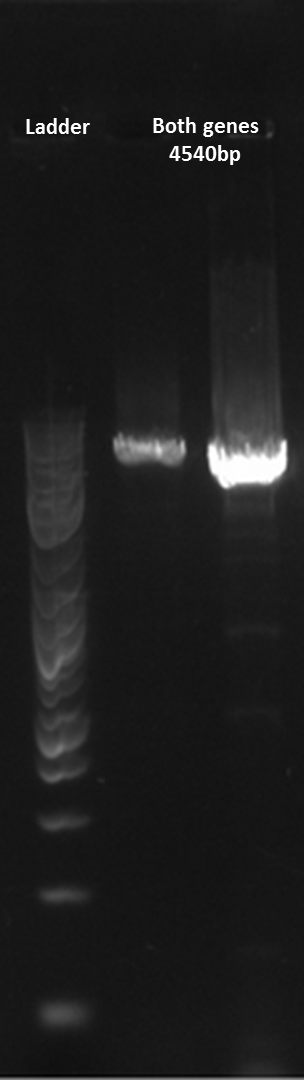
Here is a gel of the assembly of the entire construct.
Sequencing the construct
- Keck Biotechnology Resource Laboratory kindly sequenced our construct for us
- Using our Geneious license we aligned the sequencing results with the desired sequence of our construct
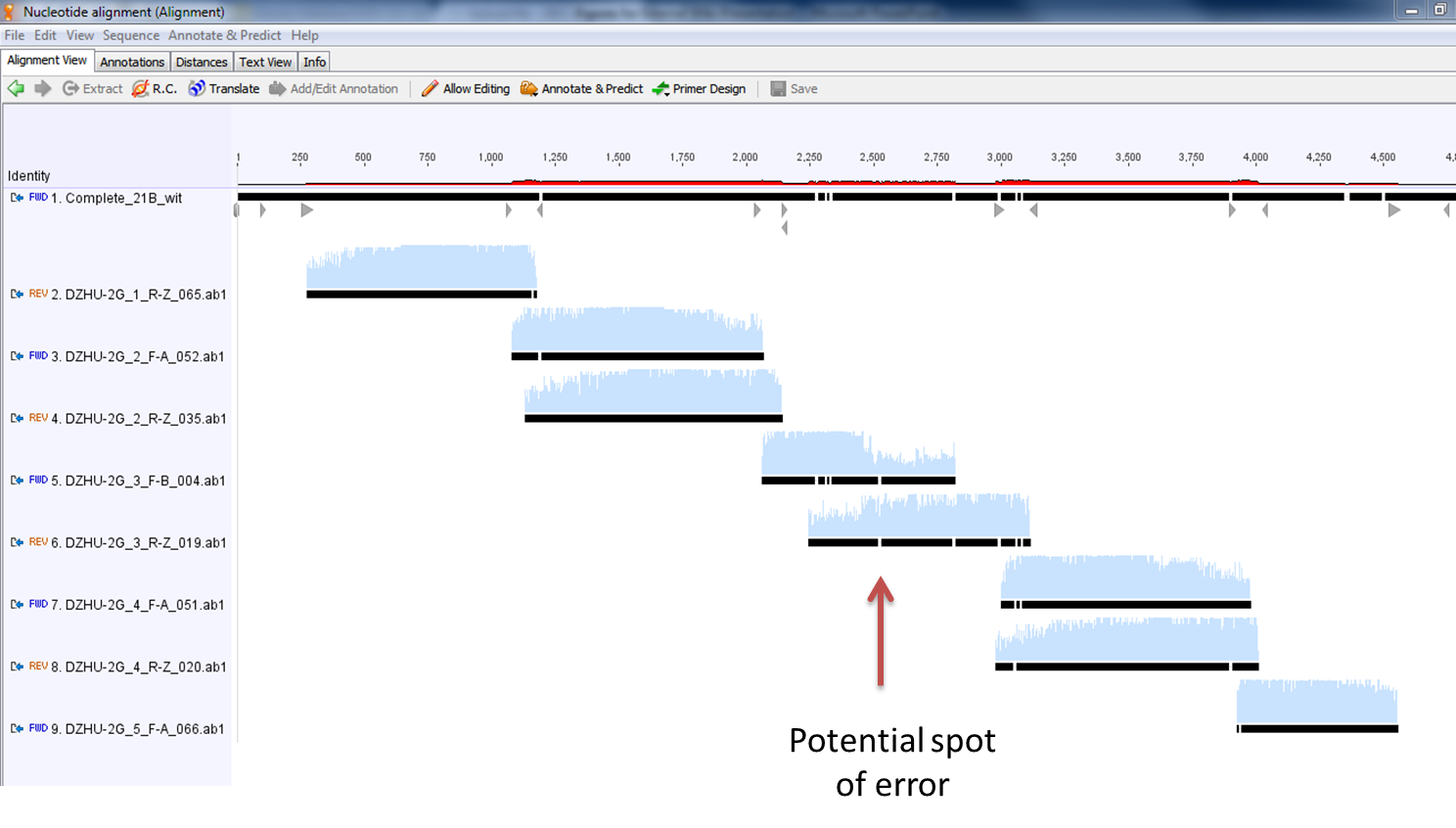
- Here is a zoomed-in picture of the results

- These are the results of the alignment of all 8 fragments with the desired sequence. Ignoring the mismatches at the beginning and end of the sequencing fragments, along with those mismatches covered by the complementary fragment, there appears to only be one legitimate error in our construct. It is denoted below with the red arrow. Fortunately, this happens to fall in a noncoding region of the construct (after the first terminator and before the second promoter).
Inserting via Plasmid
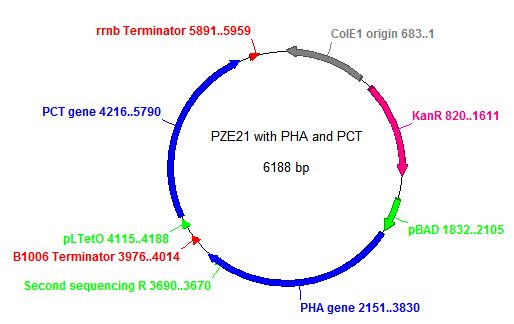
- In order to insert the two heterologous genes into E. coli, we used Gibson assembly to add our construct onto a plasmid with KanR as a selectable marker.
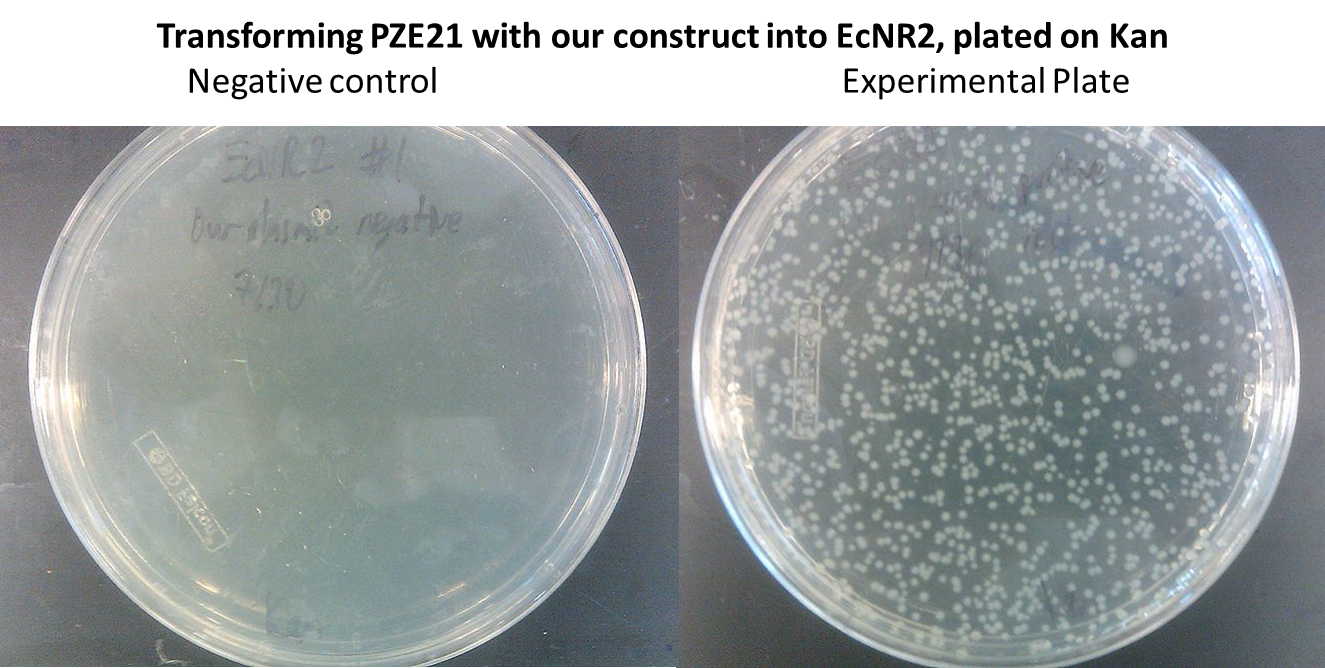
Inserting into the Genome
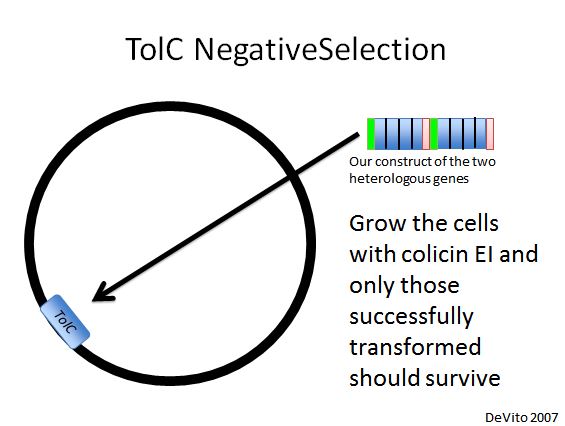
TolC Negative Selection
- We used a strain with TolC located at 21B, a highly recombinogenic site in the genome (Isaacs et al. 2011). Our plan was to replace TolC with our construct using double stranded DNA recombination, then use colicin E1 negative selection to pick desired cells (DeVito 2007).
Positive Selection with KanR
- Since we had trouble with the TolC negative selection, we decided to pursue an alternative option, which was positive selection.
- We planned to add kanamycin resistance to the end of our construct and use double-stranded recombination to integrate it into the genome again at site 21B.

 "
"