Team:ETH Zurich/Experiments 3
From 2013.igem.org
| Line 4: | Line 4: | ||
<h1><b>What about those <i>hydrolases</i> ?</b></h1> | <h1><b>What about those <i>hydrolases</i> ?</b></h1> | ||
| - | We use hydrolases | + | We use hydrolases to realize a colorimetric response by conversion of a specific substrate. We will use five hydrolases : Beta-Galactosidase (lacZ),Beta-Glucuronidase (GusA); alkaline phosphatase (phoA);Glycoside hydrolase (NagZ) and the acetyl esterase (aes).<br><br> On this page you will find all relevant information on the hydrolases and enzyme substrate reactions based on papers and our results.<br> |
<h1><b>Enzyme-substrate reactions</b></h1> | <h1><b>Enzyme-substrate reactions</b></h1> | ||
| - | <p>We have cloned fluorescent receiver systems as backup for our circuit in case the hydrolase reaction do not work properly.<br>The enzyme substrate reactions take less than 5 minutes and are visible by eye.<br><br> Our minesweeper | + | <p>We have cloned fluorescent receiver systems as backup for our circuit in case the hydrolase reaction do not work properly.<br>The enzyme substrate reactions take less than 5 minutes and are visible by eye.<br><br> Our minesweeper is becoming better and better so keep on track for updates !</p> |
<table style="float:left;margin-top:10px"> | <table style="float:left;margin-top:10px"> | ||
| Line 47: | Line 47: | ||
<tr> | <tr> | ||
<td>Aes</td> | <td>Aes</td> | ||
| - | <td> | + | <td>Acetyl esterase</td> |
<td>Magenta butyrate</td> | <td>Magenta butyrate</td> | ||
<td>5-bromo-6-chloro-3-indoxyl butyrate</td> | <td>5-bromo-6-chloro-3-indoxyl butyrate</td> | ||
| Line 55: | Line 55: | ||
<td> NagZ</td> | <td> NagZ</td> | ||
<td>Glycoside hydrolase</td> | <td>Glycoside hydrolase</td> | ||
| - | <td>X-glucosaminide X- | + | <td>X-glucosaminide X-Glucnac</td> |
<td>5-bromo-4-chloro-3-indolyl-N-acetyl-beta-D-glucosaminide</td> | <td>5-bromo-4-chloro-3-indolyl-N-acetyl-beta-D-glucosaminide</td> | ||
<td style="color:Lightblue">Blue</td> | <td style="color:Lightblue">Blue</td> | ||
| Line 65: | Line 65: | ||
| - | What about the hydolases ? How do they work and where do they come from ? Why do we use hydrolases ? | + | What about the hydolases? How do they work and where do they come from? Why do we use hydrolases? |
<br><br> | <br><br> | ||
<h1><b>Beta-galactosidase</b> (LacZ)</h1> | <h1><b>Beta-galactosidase</b> (LacZ)</h1> | ||
| Line 76: | Line 76: | ||
<br clear="all"/> | <br clear="all"/> | ||
| - | <h1><b> | + | <h1><b>Acetyl esterase</b> (Aes)</h1> |
[[File:Magenta_butyrate_on_aes_t7_strain.JPG|left|248px|thumb|<b>Fig.1: Magenta butyrate on Aes expressing colony</b>]] | [[File:Magenta_butyrate_on_aes_t7_strain.JPG|left|248px|thumb|<b>Fig.1: Magenta butyrate on Aes expressing colony</b>]] | ||
<br clear="all"/> | <br clear="all"/> | ||
Revision as of 07:34, 7 September 2013
Contents |
What about those hydrolases ?
We use hydrolases to realize a colorimetric response by conversion of a specific substrate. We will use five hydrolases : Beta-Galactosidase (lacZ),Beta-Glucuronidase (GusA); alkaline phosphatase (phoA);Glycoside hydrolase (NagZ) and the acetyl esterase (aes).
On this page you will find all relevant information on the hydrolases and enzyme substrate reactions based on papers and our results.
Enzyme-substrate reactions
We have cloned fluorescent receiver systems as backup for our circuit in case the hydrolase reaction do not work properly.
The enzyme substrate reactions take less than 5 minutes and are visible by eye.
Our minesweeper is becoming better and better so keep on track for updates !
| Hydrolase | Complementary substrate / IUPAC name | Visible color | Concentration[M] | Time(s)for colorimetric response | ||
|---|---|---|---|---|---|---|
| LacZ | Beta-Galactosidase | X-Gal | 5-Bromo-4chloro-3-indolyl-beta-galactopyranoside | Blue | ||
| LacZ | Beta-Galactosidase | Green-beta-D-Gal | N-Methyl-3-indolyl-beta-D_galactopyranoside | Green | ||
| GusA | Beta-glucuronidase | Magenta glucuronide | 6-chloro-3-indolyl-beta-D-glucuronide-cycloheylammonium salt | Red | ||
| PhoA | Alkaline phosphatase | pNPP | 4-Nitrophenylphosphatedi(tris) salt | Yellow | ||
| Aes | Acetyl esterase | Magenta butyrate | 5-bromo-6-chloro-3-indoxyl butyrate | Magenta | ||
| NagZ | Glycoside hydrolase | X-glucosaminide X-Glucnac | 5-bromo-4-chloro-3-indolyl-N-acetyl-beta-D-glucosaminide | Blue | ||
What about the hydolases? How do they work and where do they come from? Why do we use hydrolases?
Beta-galactosidase (LacZ)
Alkaline phosphatase(PhoA)
Acetyl esterase (Aes)
Glycoside hydrolase (NagZ)
Beta-glucuronidase (GusA)
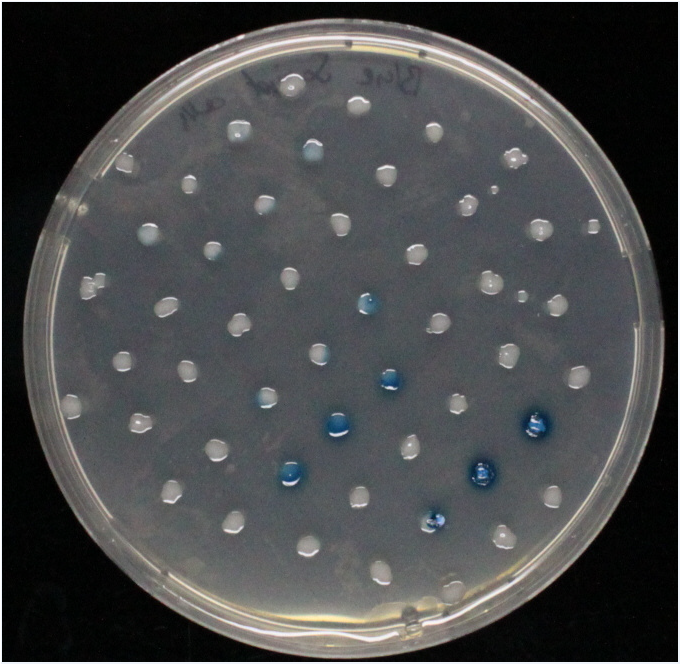
Do the substrates and enzymes cross-react ?
An enzyme-substrate test matrix (Figure 6) was established to test each substrate against each enzyme. The results were as expected (Figure 6.2) and no cross reaction is visible. The NagZ-X - glucosaminide X-Glunac reveal some difficulties in the liquid culture as well as on the agar plate.
 "
"