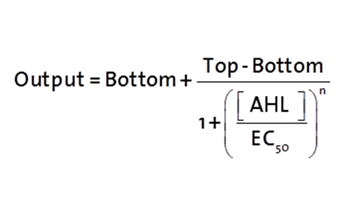Team:ETH Zurich/Experiments 5
From 2013.igem.org
| (85 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:ETH_Zurich/Templates}} | {{:Team:ETH_Zurich/Templates}} | ||
{{:Team:ETH_Zurich/Templates/stylesheet}} | {{:Team:ETH_Zurich/Templates/stylesheet}} | ||
| + | |||
<h1>High pass filters</h1> | <h1>High pass filters</h1> | ||
| - | + | [[File:Promotercomparisoncoli.png|500px|right|thumb|<b>Comparison of a chosen set of promoters.</b> The wild-type, P<sub>LuxR</sub> variant (G1, and the mutants created by rational design and partial sensitivity recovery SM1 and SM5.]] | |
| - | <p align="justify">The detection of different AHL levels depending on the number of mines requires filters in our detection system. We decided to create a high pass filter library by doing site directed mutagenesis of the wild type [http://parts.igem.org/Part:BBa_R0062 BBa_R0062 | + | <p align="justify">The detection of <b>different AHL levels</b> depending on the number of mines requires filters in our detection system. We decided to create a <b>high pass filter library</b> by doing site directed mutagenesis of the wild type [http://parts.igem.org/Part:BBa_R0062 BBa_R0062] P<sub>LuxR</sub>. The sites for mutagenesis were chosen from literature ([https://2013.igem.org/Team:ETH_Zurich/Experiments_5#referencespromoter 2]) (see <i>Figure 1</i>). If you want to know more about the site directed saturation mutagenesis please read the theoretical part [https://2013.igem.org/Team:ETH_Zurich/Processing_2 here].<br> |
| - | We were able to isolate a promoter having a lower expression level and being less sensitive than the wild type promoter (<i>Figure 2</i>). The promoter is called [http://parts.igem.org/Part:BBa_K1216007 | + | We were able to isolate a promoter having a lower expression level and being less sensitive than the wild type promoter (<i>Figure 2</i>). The promoter is called P<sub>LuxR</sub> variant [http://parts.igem.org/Part:BBa_K1216007 BBa_K1216007] (G1, according to its initial position in the deep well plate).<br> Those two different promoters were used to create two high pass filter to detect different AHL concentrations. The two promoters were analyzed not only in <b>liquid culture</b> by 96-well plate assays and single cell analysis (FACS) but also for'' E.coli'' on <b>agar plate</b>. Interestingly the EC<sub>50</sub> sensitivity (half maximal effective concentration, EC<sub>50</sub>) is different between cells in liquid culture and cells on agar plates. We then used the P<sub>LuxR</sub> variant (G1) to do <b>partial sensitivity recover</b>y by backmutating the two mutated nucleotides of the variant. <b>Rational design and oligomers</b> allow us to create a library of <b>7 different sensitive promoters</b>. See the figure on the left, and the full results in the <i>second library</i> part below.</p> |
<br clear="all"/> | <br clear="all"/> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| + | <html><div id="firstmutant" class="frog"></div></html> | ||
| + | <h1><b>First mutant:</b> Preliminary tests and dose response curve single cell analysis of the wild-type P<sub>LuxR</sub> and P<sub>LuxR</sub> varaint (G1)</h1> | ||
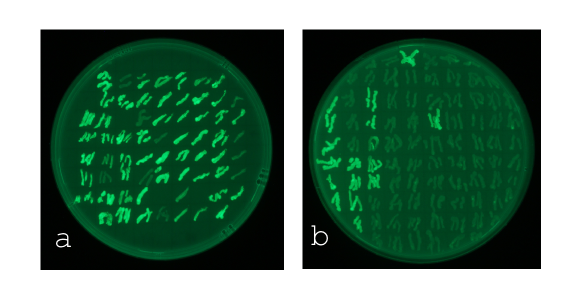
| + | [[File:posneg_selection.png|left|450px|thumb| <b>Figure 1: Two sample plates used in the selection process.</b> <b>a</b> Plate containing AHL (100 nM). All bright colonies have either a promoter that is sensitive to AHL or a constitutive one. <b>b</b> Plate without AHL. All bright colonies have a constitutive promoter and can be ruled out.]] | ||
| + | <br><b>Positive and negative selection of the cells transformed with mutated P<sub>LuxR</sub></b><br><br> | ||
| + | In this experiment the 'luxbox' of the [http://parts.igem.org/Part:BBa_R0062 BBa_R0062] was mutated by site directed saturation muatgenisis in the P<sub>Lac</sub>-LuxR-P<sub>LuxR</sub> [http://parts.igem.org/Part:BBa_J09855 BBa_J09855] construct using GFP as a reporter. <br>At first we incubated the transformed cells on an AHL containing plate to rule out all AHL-insensitive mutations. All colonies expressing GFP were then restreaked on a plate which contained no AHL, thus allowing us to rule out constitutive promoters. The remaining clones were then grown in luquid cultures on 96-well plates and tested for sensitivity over a broad range of AHL concentrations. Promising candidates were then measured in a flow cytometer to check for bistable promoters. | ||
<br clear="all"/> | <br clear="all"/> | ||
| - | + | [[File:c6wtreader.jpg|380px|right|thumb|<b>Figure 2: Sensivity curve of the wild type promoter and G1 mutant.</b> The AHL concentration range is plotted against the standarized fluorescence (au).<br> For the wild type: EC<sub>50</sub>=5.86 nM, R<sub>2</sub>= 0.87,n=1.7<br> Fore the G1 mutant: EC<sub>50</sub>=1'341 nM, R<sub>2</sub>= 0.98,n=0.8. All assays were carried out in triplicates, results are presented as mean ± standard deviation.]] | |
| - | + | <b>Preliminary tests on microtiter plate reader screening </b> | |
| - | + | <p align="justify"><br>First of all we did a fast and preliminary screening of the mutated P<sub>LuxR</sub> promoters with the microtiter plate reader. Thus, we carried out dose response curves using the Tecan Infinite M200 plate reader. The test range was inspired from literature ([https://2013.igem.org/Team:ETH_Zurich/Experiments_5#referencespromoter 1]). Afterwards we did <b>flow cytometry</b> of the most promissing promoters to have high quality data and exclude bistable promoters. <br><br> The results are shown in Figure 2.</p> | |
| - | [[File: | + | |
| - | < | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | n | + | |
| - | EC<sub>50</sub> | + | |
| - | + | ||
<br clear="all"/> | <br clear="all"/> | ||
| - | + | <b>Dose response curve single cell analysis of the wild-type P<sub>LuxR</sub> and the first P<sub>LuxR</sub> variant (G1)</b> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
<b>Liquid culture and agar plate AHL detection comparison</b><br><br> | <b>Liquid culture and agar plate AHL detection comparison</b><br><br> | ||
| - | <p align="justify">Here we employed single cell flow cytometry to obtain high quality fluorescence data of each promoter under different AHL concentrations.we started by analyzing cells grown in liquid culture, then we shifted to agar plate format in order to fit to our project. We obtained different EC<sub>50</sub> values between cells in liquid culture and in agar plates for each luxR promoter (see Figure | + | <p align="justify">Here we employed single cell flow cytometry to obtain high quality fluorescence data of each promoter under different AHL concentrations.we started by analyzing cells grown in liquid culture, then we shifted to agar plate format in order to fit to our project. We obtained different EC<sub>50</sub> values between cells in liquid culture and in agar plates for each luxR promoter (see Figure 4 and 5). See [https://2013.igem.org/wiki/index.php?title=Team:ETH_Zurich/Materials#facs_data methods] for the protocol</p> |
<br clear="all"/> | <br clear="all"/> | ||
| - | [[File:WTliquidagar.jpg|400px|left|thumb|<b>Figure 3: Sensitivity curve of the wild type BBa_R0062 on agar plates and liquid culture</b> For the liquid culture we got :EC<sub>50</sub>=0.02 nM | + | [[File:WTliquidagar.jpg|400px|left|thumb|<b>Figure 3: Sensitivity curve of the wild type BBa_R0062 on agar plates and liquid culture</b> For the liquid culture we got: EC<sub>50</sub>=0.02 nM |
| - | nM, R<sup>2</sup>=0.84, n=1.7; For the agar plates we got :EC<sub>50</sub>=4.45 nM, R<sup>2</sup>=0.80, n=1.7 | + | nM, R<sup>2</sup>=0.84, n=1.7; For the agar plates we got: EC<sub>50</sub>=4.45 nM, R<sup>2</sup>=0.80, n=1.7. All assays were carried out in duplicates, results are presented as mean ± standard deviation.]] |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | [[File:G1_plate.png|400px|right|thumb|<b>Figure 4: Sensitivity curve of the G1 (P<sub>LuxR</sub> variant BBa_K1216007) on agar plates and liquid culture</b><br>For the liquid culture we got: EC<sub>50</sub>=6482 nM, R<sup>2</sup>=0.97, n=0.8;<br> For the agar plates we got: EC<sub>50</sub>=13'704nM, R<sup>2</sup>=0.93, n=0.8. All assays were carried out in duplicates, results are presented as mean ± standard deviation.]] | ||
<br clear="all"/> | <br clear="all"/> | ||
<b>In liquid culture</b><br><br> | <b>In liquid culture</b><br><br> | ||
| - | <p align="justify">The direct plot of both sensitivity curves (Figure | + | <p align="justify">The direct plot of both sensitivity curves (Figure 5) shows the shifted sensitivity of the luxR promoter in comparison to the wild type BBa_R0062 promoter. The EC<sub>50</sub> shifted from 0.02nM for the wild type BBA_R0062 to 6'250 nM for the luxR variant promoter which is equal to a 300'000 fold increase.</p> |
<br clear="all"/> | <br clear="all"/> | ||
| - | [[File:Comparison.png|left|400px|thumb|<b>Figure | + | [[File:Comparison.png|left|400px|thumb|<b>Figure 5: Comparison of the Wild Type P<sub>LuxR</sub> and the P<sub>LuxR</sub> mutant sensitivity curves in liquid culture</b> using the BD LSRFortessa™ Flow Cytometer System<br>For the wild type: EC<sub>50</sub>=0.02 nM, R<sup>2</sup>=0.84, n=1.7<br>For G1 we got: EC<sub>50</sub>=6482 nM, R<sup>2</sup>=0.97, n=0.8. All assays were carried out in duplicates, results are presented as mean ± standard deviation.]] |
<br clear="all"/> | <br clear="all"/> | ||
<p align="justify"><b>On agar plates</b> | <p align="justify"><b>On agar plates</b> | ||
| - | <br><br>The direct plot of both sensitivity curves (Figure 6) shows the shifted sensitivity of the luxR promoter in comparison to the wild type BBa_R0062 promoter. The EC<sub>50</sub> shifted from 4.45 nM for the wild type BBA_R0062 to | + | <br><br>The direct plot of both sensitivity curves (Figure 6) shows the shifted sensitivity of the P<sub>luxR</sub> promoter in comparison to the wild type BBa_R0062 promoter. The EC<sub>50</sub> shifted from 4.45 nM for the wild type BBA_R0062 to 13'704 nM for the luxR variant promoter which is equal to a 2800 fold increase. Therefore, comparing with the results from cells growing in liquid culture an increase of 220 and 2 times for wild type BBa_R0062 and P<sub>LuxR</sub> variant respectively. If you want to know more about the methods please click [https://2013.igem.org/wiki/index.php?title=Team:ETH_Zurich/Materials#micro_plate here]<br><br></p> |
| - | [[File:WT_and_G1_on_agar_plates.jpg|400px|left|thumb|<b>Figure | + | [[File:WT_and_G1_on_agar_plates.jpg|400px|left|thumb|<b>Figure 6: Comparison of the sensivity of the wild type promoter and the mutated luxR promoter on agar plates</b><br>For the wild type we got :EC<sub>50</sub>=4.45 nM, R2=0.80, n=1.7<br>For the P<sub>LuxR</sub> variant we got :EC<sub>50</sub>=13'704 nM, R2=0.93, n=0.8. All assays were carried out in duplicates, results are presented as mean ± standard deviation.]] |
| + | [[File:Dot plot.png|right|620px|thumb|<b>Figure 7: Flow cytometry dot plot of the wild type BBa_R0062 promoter vs. the P<sub>luxR</sub> mutant</b> The histogram plots of eGFP levels for each tested AHL concentration, culturing condition and corresponding promoter. On the top the data is the wild type luxR promoter. On the bottom the data of the luxR promoter variant.]] | ||
<br clear="all"/> | <br clear="all"/> | ||
| - | <h1>Rational design of additional | + | |
| + | <html><div id="secondlibrary" class="frog"></div></html> | ||
| + | <h1><b> Second library : </b> Back mutation and single cell analysis of the first P<sub>LuxR</sub> varaint (G1) by rational design and partial sensitivity recovery.</h1> | ||
| + | |||
| + | Rational design and single cell analysis of additional P<sub>LuxR</sub> variants with partial sensitivity recovery based on the first P<sub>LuxR</sub> variant (G1). | ||
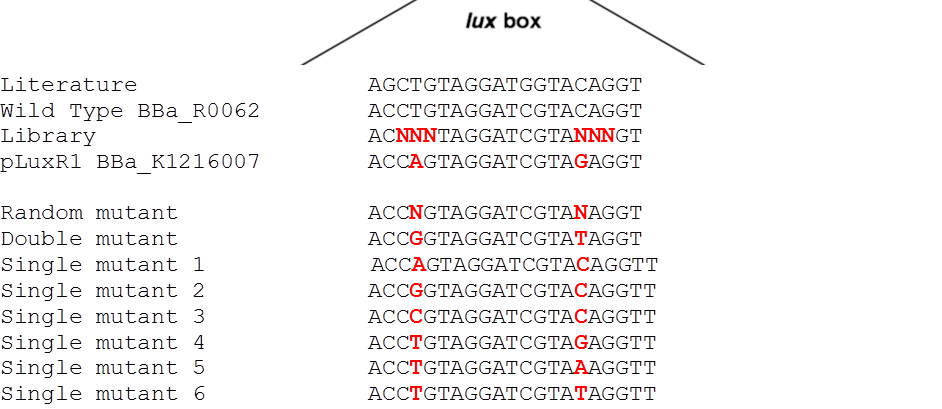
| + | [[File:G1VariantLibrary.png|600px|left|thumb|<b>Figure 8: Sequence alignment of the additional luxR variants.</b>]] | ||
<p>According to initial model predictions confirmed by successive [https://2013.igem.org/Team:ETH_Zurich/Experiments_6 experimental validation] the G1 promoter sensitivity is too low to drive a significant response in the concentration gradient established by our sender cells.<br> | <p>According to initial model predictions confirmed by successive [https://2013.igem.org/Team:ETH_Zurich/Experiments_6 experimental validation] the G1 promoter sensitivity is too low to drive a significant response in the concentration gradient established by our sender cells.<br> | ||
| - | We need a collection of promoters with a set of | + | We need a collection of promoters with a set of EC<sub>50</sub> values between the wild type and G1 itself. |
Since G1 has been obtained through two random mutations: 4T>A and 16C>G reverting one of the two, we reasoned, should result in a LuxR binding strength closer to the wild type. | Since G1 has been obtained through two random mutations: 4T>A and 16C>G reverting one of the two, we reasoned, should result in a LuxR binding strength closer to the wild type. | ||
| - | To test this hypothesis we ordered oligos encompassing all the possible combination of single position mutants (See list) and plan to experimentally characterize, and eventually deposit in the registry, all the resulting | + | To test this hypothesis we ordered oligos encompassing all the possible combination of single position mutants (See list) and plan to experimentally characterize, and eventually deposit in the registry, all the resulting P<sub>LuxR</sub> variants. In parallel we used degenerate oligos to generate a small library (16 members) containing all the combination of double mutants for the two G1 key position to verify if all the double mutants show similar EC<sub>50</sub>.</p> |
| - | [[File: | + | <br clear="all"> |
| - | </p> | + | |
| + | <b>Finally characterized promoter library</b> | ||
| + | <html><a id="promLibrary" class="frog"></a></html> | ||
| + | [[File:Promtoer library.jpg|1100px|center|thumb|<b>Figure 9.Dose response curve experiment plates of the promoter library with an RFP reporter.</b> ON the picture you can see 11 plates having different AHL concentrations in the agar( 10^-5 nM, 10^-4 nM, 10^-3 nM,10^-2 nM,10^-1 nM,10^0 nM,10^1 nM,10^2 nM,10^3 nM,10^4 nM,10^5 nM.) of the backmutations SM1, SM2, SM3, SM3, S, SM5, SM6 and DM. (SM = Single mutation, DM = Double mutation.]] | ||
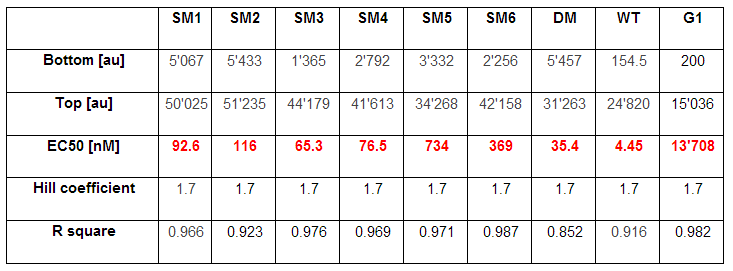
| + | <p>As explained above we created a library of promoters originating from P<sub>LuxR</sub> variant (G1). We characterized the dose response curve to AHL on agar plates of all 7 promoters by following the same protocol as for the P<sub>LuxR</sub> variant characterization. See the different shifted dose response curves in Figure 10. and the characteristics of the library in Table 1.<br><br> | ||
| + | Thanks to the <b>analytical solution of the model</b> (see [https://2013.igem.org/Team:ETH_Zurich/Modeling/Analytical_Approximations here]) predicting the needed EC<sub>50</sub> of 100 nM of the promoter to fit to our system we choose the SM1 mutant for our final set-up. (We did not choose SM2 because when we choose a promoter, according to the prediction of the model, the experiments were not done in duplicates at this time, and the characteristics changed of course as soon as we had duplicates). This promoter gets activated when two mines are nearby the receiver cell. | ||
| + | [[File:Promoterlibratablecoli.png|700px|center|thumb|<b>Table 1. Characteristics of the promoter library </b>]]</p> | ||
| + | <br clear="all"> | ||
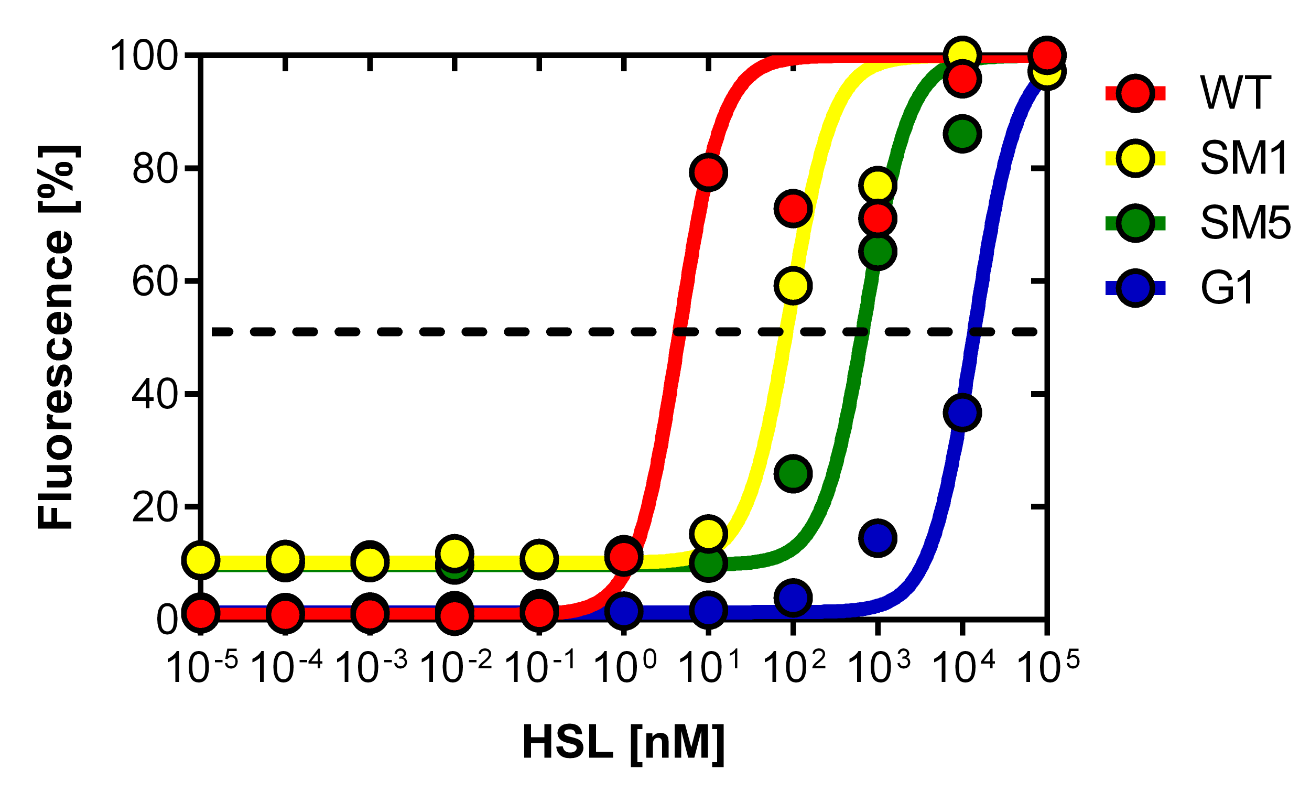
| + | [[File:promotercomparisonfinal.png|left|500px|thumb|<b>Figure 10. Dose response curve of the promoter library.</b> 7 different promoter were created by partial sensitivity recovery of the P<sub>LuxR</sub> variant. The colonies have the P<sub>Lac</sub>-LuxR-P<sub>LuxR</sub> BBa_J09855 construct with the different mutated promoter as well as a RFP reporter to analyze the dose response in the flow cytometer. The colonies were grown on 11 different plates with 11 different concentrtions as explained in Figure 9. Please see table 1 for the characteristics of the promoter library.]] | ||
| + | [[File:Promotercomparisoncoli.png|500px|right|thumb|<b>Figure 11. Comparison of a choosen set of promoters.</b> The wild-type, P<sub>LuxR</sub> variant (G1, and the mutants created by rational design and partial sensitivity recovery SM1 and SM5.]] | ||
| + | <br clear="all"/> | ||
| + | Figure 10. shows the relative fluorescence over the [AHL] of the dose-response curves of the whole library exepted the G1 and the wild type P<sub>LuxR</sub> promoter. Figure 11. is a comparison of a choosen set of promissing promoter we tried in different constructs for the final gameplay according to the prediction of the model. | ||
| + | |||
| + | <br clear="all"/> | ||
| + | |||
| + | <h1>Fluorescence data analysis</h1> | ||
| + | [[File:Formulapluxr1.png|left|300px]] | ||
| + | <p>The fitting of the following graphs was performed using this equation :<br><br> | ||
| + | Output = eGFP levels [au]<br> | ||
| + | Top = maximal eGFP level [au] ("full induction")<br> | ||
| + | Bottom = minimal eGFP level [au] (“leakiness”)<br> | ||
| + | n = Hill coefficient (“cooperativity”)<br> | ||
| + | EC<sub>50</sub> = Half-maximal effective concentration (“sensitivity”)<br> | ||
| + | [AHL]=AHL concentration [nM]</p> | ||
<br clear="all"/> | <br clear="all"/> | ||
| + | <html><div id="referencespromoter" class="frog"></div></html> | ||
<h1>References</h1> | <h1>References</h1> | ||
Latest revision as of 20:05, 15 November 2013
High pass filters
The detection of different AHL levels depending on the number of mines requires filters in our detection system. We decided to create a high pass filter library by doing site directed mutagenesis of the wild type [http://parts.igem.org/Part:BBa_R0062 BBa_R0062] PLuxR. The sites for mutagenesis were chosen from literature (2) (see Figure 1). If you want to know more about the site directed saturation mutagenesis please read the theoretical part here.
We were able to isolate a promoter having a lower expression level and being less sensitive than the wild type promoter (Figure 2). The promoter is called PLuxR variant [http://parts.igem.org/Part:BBa_K1216007 BBa_K1216007] (G1, according to its initial position in the deep well plate).
Those two different promoters were used to create two high pass filter to detect different AHL concentrations. The two promoters were analyzed not only in liquid culture by 96-well plate assays and single cell analysis (FACS) but also for E.coli on agar plate. Interestingly the EC50 sensitivity (half maximal effective concentration, EC50) is different between cells in liquid culture and cells on agar plates. We then used the PLuxR variant (G1) to do partial sensitivity recovery by backmutating the two mutated nucleotides of the variant. Rational design and oligomers allow us to create a library of 7 different sensitive promoters. See the figure on the left, and the full results in the second library part below.
First mutant: Preliminary tests and dose response curve single cell analysis of the wild-type PLuxR and PLuxR varaint (G1)
Positive and negative selection of the cells transformed with mutated PLuxR
In this experiment the 'luxbox' of the [http://parts.igem.org/Part:BBa_R0062 BBa_R0062] was mutated by site directed saturation muatgenisis in the PLac-LuxR-PLuxR [http://parts.igem.org/Part:BBa_J09855 BBa_J09855] construct using GFP as a reporter.
At first we incubated the transformed cells on an AHL containing plate to rule out all AHL-insensitive mutations. All colonies expressing GFP were then restreaked on a plate which contained no AHL, thus allowing us to rule out constitutive promoters. The remaining clones were then grown in luquid cultures on 96-well plates and tested for sensitivity over a broad range of AHL concentrations. Promising candidates were then measured in a flow cytometer to check for bistable promoters.
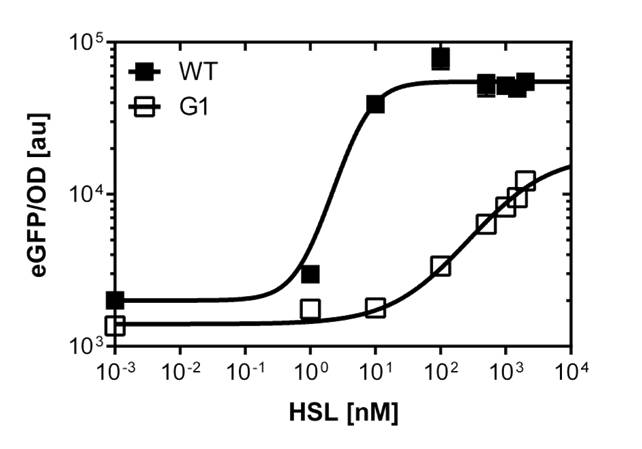
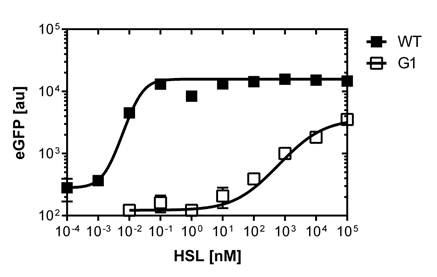
For the wild type: EC50=5.86 nM, R2= 0.87,n=1.7
Fore the G1 mutant: EC50=1'341 nM, R2= 0.98,n=0.8. All assays were carried out in triplicates, results are presented as mean ± standard deviation.
Preliminary tests on microtiter plate reader screening
First of all we did a fast and preliminary screening of the mutated PLuxR promoters with the microtiter plate reader. Thus, we carried out dose response curves using the Tecan Infinite M200 plate reader. The test range was inspired from literature (1). Afterwards we did flow cytometry of the most promissing promoters to have high quality data and exclude bistable promoters.
The results are shown in Figure 2.
Dose response curve single cell analysis of the wild-type PLuxR and the first PLuxR variant (G1)
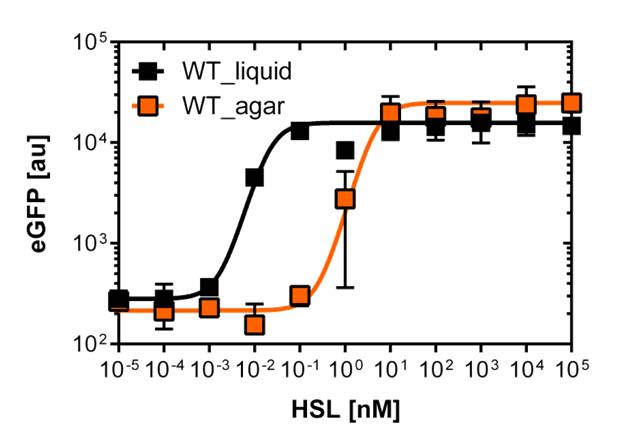
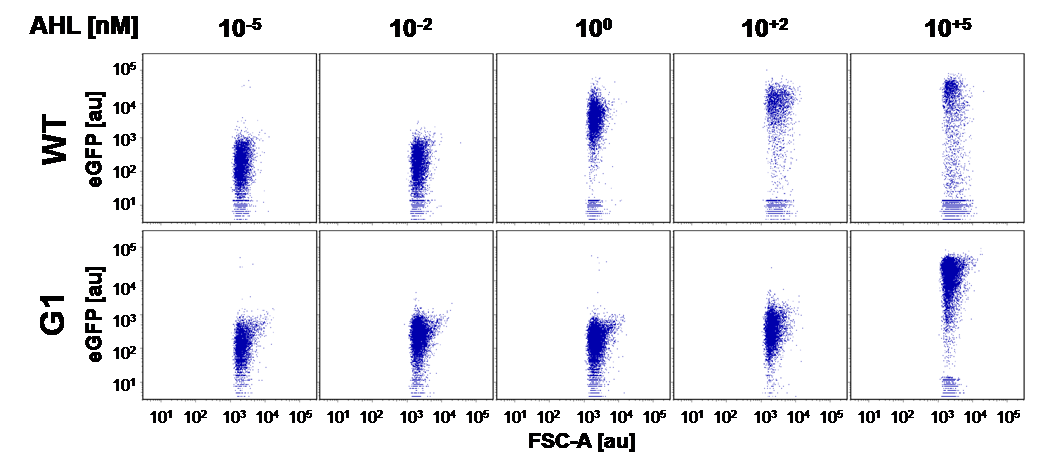
Liquid culture and agar plate AHL detection comparison
Here we employed single cell flow cytometry to obtain high quality fluorescence data of each promoter under different AHL concentrations.we started by analyzing cells grown in liquid culture, then we shifted to agar plate format in order to fit to our project. We obtained different EC50 values between cells in liquid culture and in agar plates for each luxR promoter (see Figure 4 and 5). See methods for the protocol
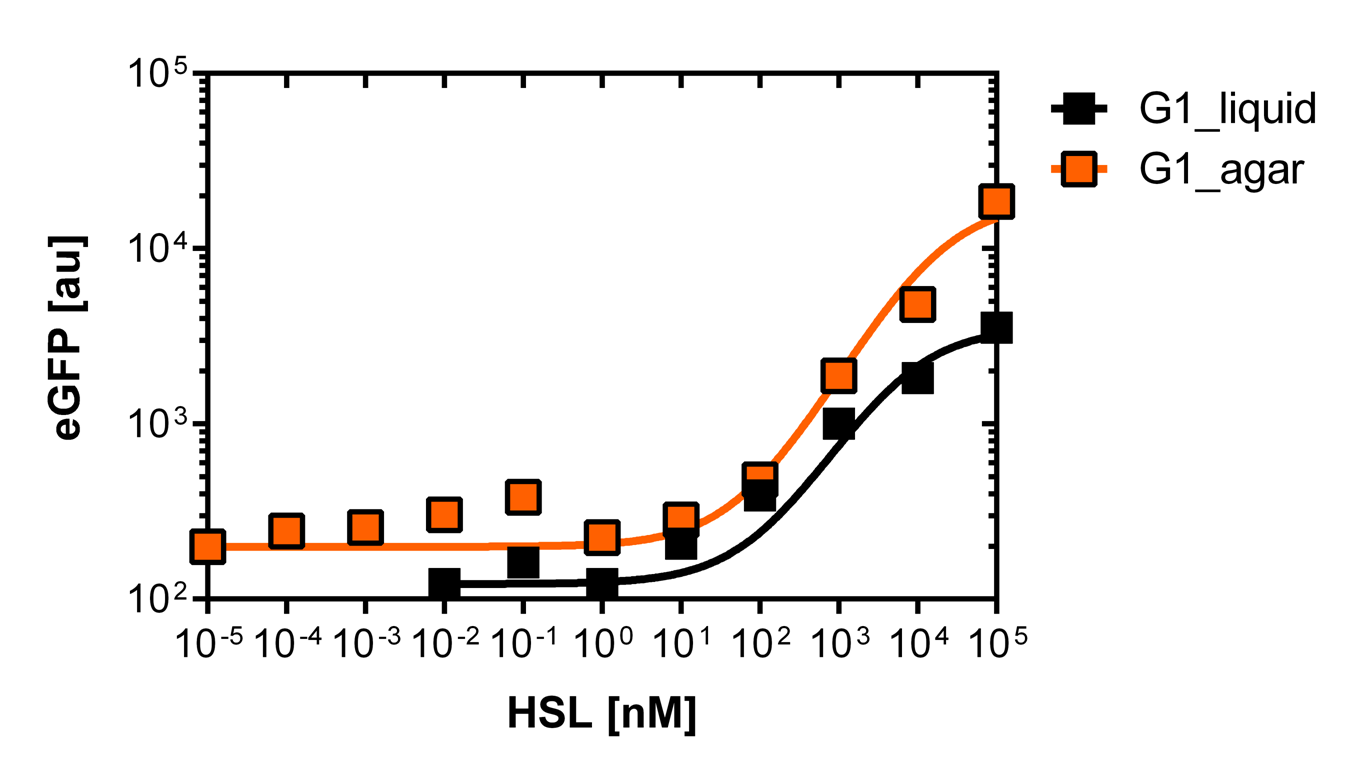
For the liquid culture we got: EC50=6482 nM, R2=0.97, n=0.8;
For the agar plates we got: EC50=13'704nM, R2=0.93, n=0.8. All assays were carried out in duplicates, results are presented as mean ± standard deviation.
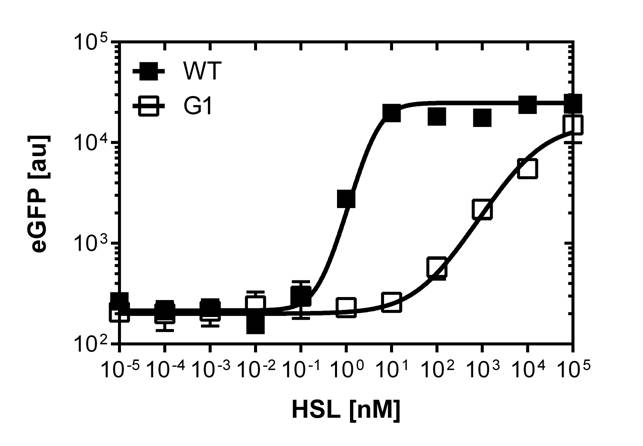
In liquid culture
The direct plot of both sensitivity curves (Figure 5) shows the shifted sensitivity of the luxR promoter in comparison to the wild type BBa_R0062 promoter. The EC50 shifted from 0.02nM for the wild type BBA_R0062 to 6'250 nM for the luxR variant promoter which is equal to a 300'000 fold increase.
For the wild type: EC50=0.02 nM, R2=0.84, n=1.7
For G1 we got: EC50=6482 nM, R2=0.97, n=0.8. All assays were carried out in duplicates, results are presented as mean ± standard deviation.
On agar plates
The direct plot of both sensitivity curves (Figure 6) shows the shifted sensitivity of the PluxR promoter in comparison to the wild type BBa_R0062 promoter. The EC50 shifted from 4.45 nM for the wild type BBA_R0062 to 13'704 nM for the luxR variant promoter which is equal to a 2800 fold increase. Therefore, comparing with the results from cells growing in liquid culture an increase of 220 and 2 times for wild type BBa_R0062 and PLuxR variant respectively. If you want to know more about the methods please click here
For the wild type we got :EC50=4.45 nM, R2=0.80, n=1.7
For the PLuxR variant we got :EC50=13'704 nM, R2=0.93, n=0.8. All assays were carried out in duplicates, results are presented as mean ± standard deviation.
Second library : Back mutation and single cell analysis of the first PLuxR varaint (G1) by rational design and partial sensitivity recovery.
Rational design and single cell analysis of additional PLuxR variants with partial sensitivity recovery based on the first PLuxR variant (G1).
According to initial model predictions confirmed by successive experimental validation the G1 promoter sensitivity is too low to drive a significant response in the concentration gradient established by our sender cells.
We need a collection of promoters with a set of EC50 values between the wild type and G1 itself.
Since G1 has been obtained through two random mutations: 4T>A and 16C>G reverting one of the two, we reasoned, should result in a LuxR binding strength closer to the wild type.
To test this hypothesis we ordered oligos encompassing all the possible combination of single position mutants (See list) and plan to experimentally characterize, and eventually deposit in the registry, all the resulting PLuxR variants. In parallel we used degenerate oligos to generate a small library (16 members) containing all the combination of double mutants for the two G1 key position to verify if all the double mutants show similar EC50.
Finally characterized promoter library
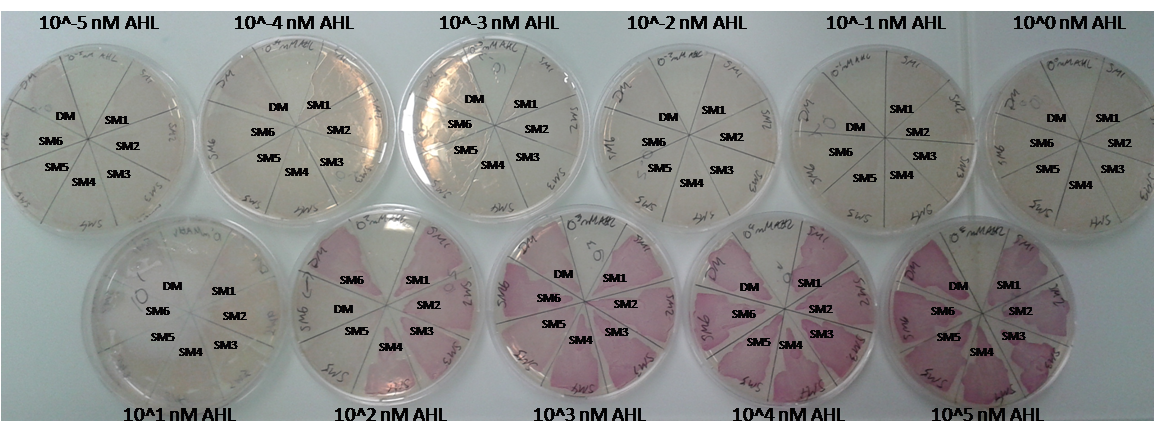
As explained above we created a library of promoters originating from PLuxR variant (G1). We characterized the dose response curve to AHL on agar plates of all 7 promoters by following the same protocol as for the PLuxR variant characterization. See the different shifted dose response curves in Figure 10. and the characteristics of the library in Table 1.
Thanks to the analytical solution of the model (see here) predicting the needed EC50 of 100 nM of the promoter to fit to our system we choose the SM1 mutant for our final set-up. (We did not choose SM2 because when we choose a promoter, according to the prediction of the model, the experiments were not done in duplicates at this time, and the characteristics changed of course as soon as we had duplicates). This promoter gets activated when two mines are nearby the receiver cell.
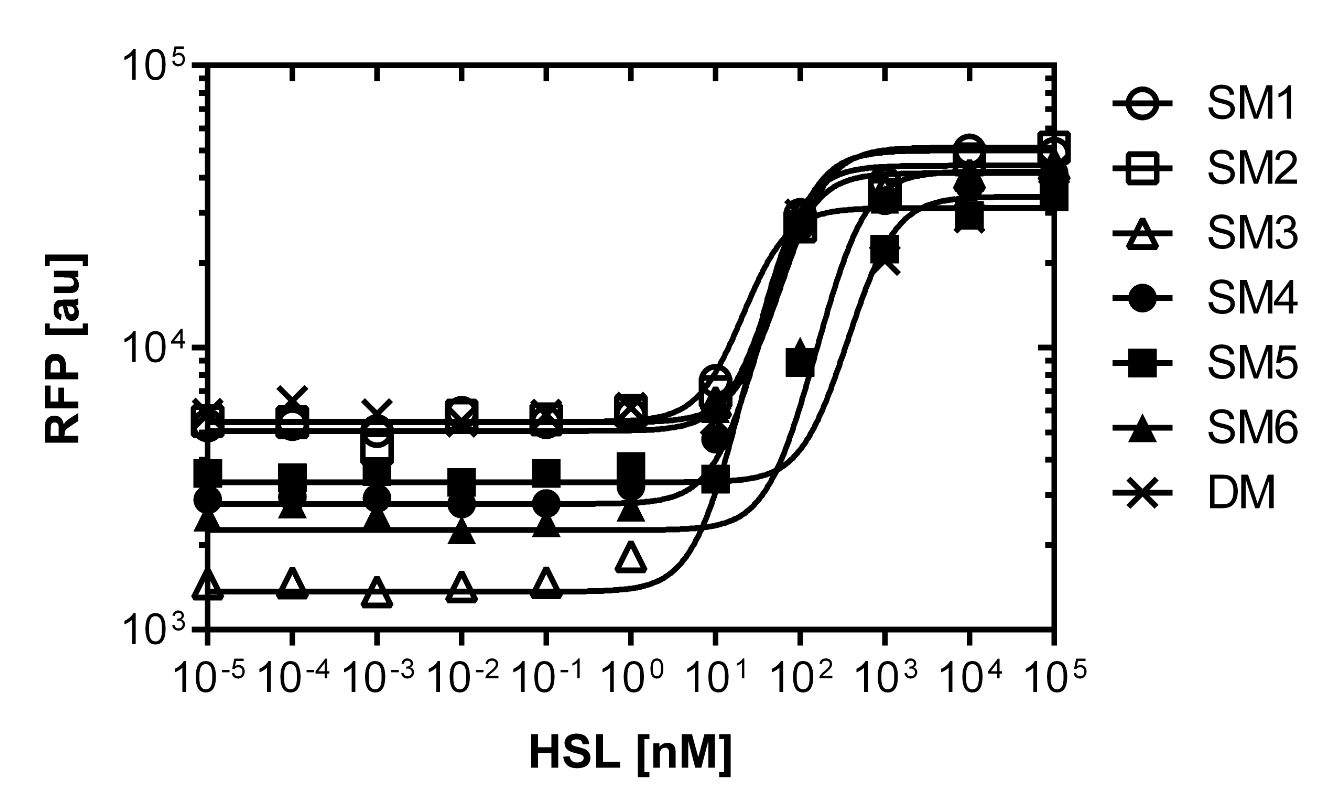
Figure 10. shows the relative fluorescence over the [AHL] of the dose-response curves of the whole library exepted the G1 and the wild type PLuxR promoter. Figure 11. is a comparison of a choosen set of promissing promoter we tried in different constructs for the final gameplay according to the prediction of the model.
Fluorescence data analysis
The fitting of the following graphs was performed using this equation :
Output = eGFP levels [au]
Top = maximal eGFP level [au] ("full induction")
Bottom = minimal eGFP level [au] (“leakiness”)
n = Hill coefficient (“cooperativity”)
EC50 = Half-maximal effective concentration (“sensitivity”)
[AHL]=AHL concentration [nM]
References
(1) M Geske G.D.,Evaluation of a focused library of N-aryl L-homoserine lactones reveals a new set of potent quorum sensing modulators.
(2) Luis Caetano A Mutational Analysis Defines Vibrio fischeri LuxR Binding Sites
 "
"