Team:TU-Munich/Project/Safety
From 2013.igem.org
(→Transgene pollution) |
(→horizontal gene transfer) |
||
| Line 148: | Line 148: | ||
====horizontal gene transfer==== | ====horizontal gene transfer==== | ||
| + | Concerning horizontal gene transfer, the main safety threat would be the spread of transgenes from our moss to bacteria. The transfer of primary transgenes or marker genes could lead to new bacteria strains that express these genes and could pass them on to pathogenic strains. But while this process is very common in bacteria, apart from ''agrobacterium tumefaciens'', there are few examples of natural gene transfer between bacteria and higher eukaryotes [Quelle]. | ||
| - | + | For the transgene to remain in the bacterial population and thus in the environment, there has to be a strong selective pressure. This could be the case in highly contaminated environments, where the ability to break down a toxic pollutant is beneficial. We are aware that our moss carries transgenes that could be advantageous to bacteria living in these polluted environments. But since '''all''' of the effectors that we are testing actually come from bacteria, we do no think that this is the main problem. The genes for these effectors are already present in the bacteria population. Nevertheless, we need to have a closer look at antibiotic resistance genes such as ereB and the selection marker nptII. Antibiotic resistance genes are the focus of attention because of their potentially strong and general selective advantage in human pathogens. But in the case of ereB, which comes from ''E.coli'', the resistance gene is already present in the bacterial population on plasmids while in the moss it is controlled by a moss promotor and integrated in the moss genome. Therefore, it is much more likely that in case of ereB, the horizontal gene transfer occurs between bacteria and not between bacteria and moss. | |
| - | A possible way to eliminate superfluous DNA sequences after a successful transformation is the use of a two-component site-specific recombination system such as Cre-loxP (Dale and Ow, 1991). The recombinase Cre recognizes short sequences known (loxP) and is then able to excise DNA betweeen two loxP sites that are in the same orientation. Therefore, marker genes flanked by loxP sites can be efficiently excised from transgenic plants if Cre is present. | + | A possible way to eliminate superfluous DNA sequences such as the nptII marker after a successful transformation is the use of a two-component site-specific recombination system such as Cre-loxP (Dale and Ow, 1991). The recombinase Cre recognizes short sequences known (loxP) and is then able to excise DNA betweeen two loxP sites that are in the same orientation. Therefore, marker genes flanked by loxP sites can be efficiently excised from transgenic plants if Cre is present. |
====light-triggered Killswitch==== | ====light-triggered Killswitch==== | ||
Revision as of 18:05, 4 October 2013
Biosafety
Biosafety deals with the prevention of unintented exposure to pathogens and toxins, or their accidental release http://www.who.int/csr/resources/publications/biosafety/Biosafety7.pdf WHO.
Biosafety to us means minimizing the risks concering the team working in the lab, the general public and the environment. Physcomitrella itself does not pose any risks to the health of the researcher or the general public, since it is endemic to many parts of the world [Source]. In the lab, we are able to cultivate our moss in bioreactors in which the flow of substances is tightly controlled. They can even be used for the production of therapeutic proteins http://www.greenovation.com/ greenovation. For usage in a sewage plant, such reactors could be upscaled and special filter systems could be applied to ensure that no moss can escape into the environment. However, our long-term goal is to use moss filters in polluted environments but at the same time ensuring the highest level of biological safety.
We believe that the main biosafety-issues that have to be addressed regarding our project are
- toxicity of recombinant proteins
- toxicity of degradation products
- transgene pollution (vertical and horizontal gene transfer)
In order to minimize the aforementioned risks, we have taken several measures, including a light-triggered killswitch, non-sporulating Physcomitrella strains and the evaluation of the genetic parts that we are using in terms of safety.
Toxicity of recombinant proteins
The most obvious safety threat is that the recombinant proteins might be toxic and thus have a negative effect on the environment. This is the foundation of our safety consideration - that the products from all parts and genetic circuits must not cause harm by themselves. All our effector proteins (laccase, catechol-2,3-dioxygenase, erythromycin esterase B) are all from S1 organisms and are not toxic. For example, laccases are already used in the food industry http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2963825/ Osma et al., 2010. For the establishment of a eukaryotic expression system, we are also using parts of mammalian origin and from S2-organisms (see safety forms below). We have made the decisions to use parts from S2 organism carefully and only after consulting our safety officer. He could ensure us that the parts and circuits we are using do not pose any threat. When possible, we have tried to substitute parts from S2 organisms with parts from S1 organisms. All of the parts we are using (such as a polioviral internal ribosome entry site (IRES) and the Ig-Kappa secretion signal from mouse) are widely used in molecular biology laboratories all around the world and are considered safe.
Safety evaluation for used BioBricks and composite parts
Table 1: extended safety evaluation for biobricks of mammalian origin or from S2 organisms
| Part | BBa_K1159004 | BBa_K1159304 | BBa_K729004 |
|---|---|---|---|
| Organism name and strain name or number | Homo sapiens | Mus musculus (mammal) | Staphylococcus aureus |
| Risk Group | 1 | 1 | 2 |
| name of the part and brief description | Proteinphosphatase 1 is able to bind the cyanotoxin microcystin and is used in our bioaccumulation project. Our moss could thus filter the toxin from the water | Ig-Kappa is used as a signal peptide for our eucaryotic chassis Physcomitrella patens in order to secrete effector proteins | part BBa_K729004 (nuclease from Staphylococcus aureus, but we have performed a PCR to remove bacterial signal peptides) It causes genomic degradation and we are using it in our kill switch to kill the moss and destroy all genetic material |
| How did you physically acquire the organism or part? | Cooperation with iGEM Team Dundee 2013 | gene synthesis | parts registry |
| What potential safety/health risks to team members, other people at your institution, or the general public could arise from your use of this organism/part? | No safety risks | No potential safety/health risks of the part itself. Composite parts containing this signal sequence could be possibly dangerous when the signal sequence is fused to a dangerous protein. However, we are not using the part in combination with possibly dangerous other parts. | The protein produced by the part is not dangerous http://www.thermoscientificbio.com/uploadedFiles/Resources/en0181-usa-msds.pdf Thermo Scientific. However, S. aureus nuclease production could contribute to disease pathogenesis in vivo |
| What measures do you intend to take to ensure that your project is safe for team members, other people at your institution, and the general public? | We have all completed a biosafety course and we handle everything in the lab with the necessary precaution. We will not distribute any parts or organisms outside the lab, we are using moss that is not able to sporulate and have implemented a kill switch (see basic safety form) | ||
| Explain why you believe the part iself is not dangerous | The part is human-derived and should thus not be dangerous | It is a eukaryotic secretion signal which enables secretion of proteins that carry this signal sequence in mammals and other eukaryotes. Therefore the part itself is not dangerous | S. aureus nuclease production contributes to disease pathogenesis in vivo so it could possibly enhance pathogenesis in other microorganisms as well http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2982853/ Berends et al., 2010. It is therefore important to ensure that no genetic material of our moss gets into the environment |
| Why do you need to use this organism/part? Is there an organism/part from a less dangerous Risk Group that would accomplish the same purpose? | Part is needed to filter the dangerous cyanotoxin microcystin from the water. Since proteinphosphatase binds microcystin with a very high affinity it is the best choice. | Part is needed to be able to secrete our effector proteins. Since we are using Physcomitrella patens as a chassis, we have to use a eukaryotic secretion signal. The use of the Ig-kappa secretion signal from mouse (BSL 1) is widely established and does not pose any safety risks | Part is needed in our kill switch to ensure that the moss completely killed and that all genetic material is destroyed. |
| Is the organism/part listed under the Australia Group guidelines, or otherwise restricted for transport? | No | ||
| Please describe the BioSafety Level of the lab in which the team works | The lab we work in has BioSafety Level 1 and we are not using organisms with a BSL level greater than 1 | ||
Table 2: extended safety evaluation for biobricks of mammalian origin or from S2 organisms
| Part | BBa_K1159300 | BBa_K1159201 / BBa_K1159200 | BBa_K801030 |
|---|---|---|---|
| Organism name and strain name or number | Human poliovirus 1 strain Mahoney | Streptococcus pyogenes | Simean virus 40 |
| Risk Group | 2 | 2 | 2 |
| name of the part and brief description | Internal ribosome entry site (IRES) from poliovirus 1 allows for translation initiation in the middle of a messenger RNA (mRNA) sequence and thus enables to build constructs consisting of several parts controlled by one promoter | Spytag/Spycatcher part is used for posttranslational protein fusions (fusion of our effectors to membrane receptors).The two parts consists of a domain of streptococcus pyogenes fibronectin-binding protein FbaB. This domain was split and the fragments were rationally engineered http://www.ncbi.nlm.nih.gov/pubmed/22366317 Zakeri et al., 2012 | part BBa_K801030 is a nuclear localization sequence (NLS) from Simian virus 40, it enables targeting of proteins into the nucleus |
| How did you physically acquire the organism or part? | Cooperation with Prof. Dr. Fussenegger from ETH Zurich http://www.ncbi.nlm.nih.gov/pubmed/16026881 Hartenbach & Fussenegger, 2005 | gene synthesis | parts registry |
| What potential safety/health risks to team members, other people at your institution, or the general public could arise from your use of this organism/part? | Human poliovirus 1 is a human pathogen but the IRES part we are using is itself not harmful | We believe that the part itself is not dangerous. Since the binding of tag/catcher is very specific, the catcher cannot bind to other proteins and only binds proteins that are fused to the tag | the part itself consists of 7 amino acids and does not pose any safety or health risks |
| What measures do you intend to take to ensure that your project is safe for team members, other people at your institution, and the general public? | We have all completed a biosafety course and we handle everything in the lab with the necessary precaution. We will not distribute any parts or organisms outside the lab, we are using moss that is not able to sporulate and have implemented a kill switch (see basic safety form) | ||
| Explain why you believe the part iself is not dangerous | Internal ribosome entry sites are widely used as a means to accomplish polycistronic expression in eukaryotes | Since it was partly engineered, the parts itself is not dangerous in a way that it is connected to Streptococcus pyogenes. The fusion product tag-catcher itself is not dangerous but this depends on the proteins tag/catcher are fused to | the part is based on the plasmid pGADT7 AD from Clontech, so the NLS is a commercially available and widely used. We therefore consider it not dangerous |
| Why do you need to use this organism/part? Is there an organism/part from a less dangerous Risk Group that would accomplish the same purpose? | Without IRES we would have to put a promotor in front of every protein we want to express which would make cloning of large constructs much more difficult. There are IRES elements found in cellular mRNAs (e.g. in Fibroblast growth factor) however, they are not as established in expression systems | Part is needed for posttranslational fusion of our effectors to membrane receptors. There is no part from a less dangerous risk group that would accomplish this purpose this well | Part is needed to target the nuclease of our killswitch into the nucleus of our moss. Without a localization signal, the killswitch would not work. The NLS from SV40 is used in almost all commercially available vector systems and has been in use for a long time which is why we chose it |
| Is the organism/part listed under the Australia Group guidelines, or otherwise restricted for transport? | No | ||
| Please describe the BioSafety Level of the lab in which the team works | The lab we work in has BioSafety Level 1 and we are not using organisms with a BSL level greater than 1 | ||
Toxicity of degradation products
None of the transgenic plants that we have created is able to produce a toxic substance. However, degradation products of hormones, antibiotics and other pollutants could be potentially toxic. Therefore, the reaction products will be monitored using LC/MS analysis. For example for EreB, the transformation products of erythromycin possess no endocrine and mutagenic properties http://www.endetech.eu/sites/default/files/field/image/2013.06.07-Oral presentation ICWRER 2013.pdf endetech. In general, it is very difficult to find data on the toxicity of these degradation products.
Additionally, the substances used in bioaccumulation (Glutathion-S-transferase, Proteinphosphatase 1) are not harmful themselves, but moss that has bound toxic substances (e.g. microcystin) in high concentrations is potentially toxic and has to be disposed of separately and with caution.
Transgene pollution
Transgene pollution describes the spread of transgenes beyond the genetically modified species by natural gene flow mechanisms http://www.cabi.org/cabreviews/FullTextPDF/2003/20033177371.pdf Commandeur et al., 2003. Transgenes can spread from transgenic to non-transgenic populations of the same species or related wild species (vertical gene transfer) or to entirely different species such as bacteria (horizontal gene transfer). Because the impact of novel genes on other species is unpredictable, transgene pollution is undesirable and could pose a risk to the ecosystem. When talking about transgenes, one also has to differentiate between primary transgenes such as our effectors and superfluous DNA sequences such as antibiotic resistance markers.
preventing vertical gene transfer with non-sporulating Physcomitrella strains
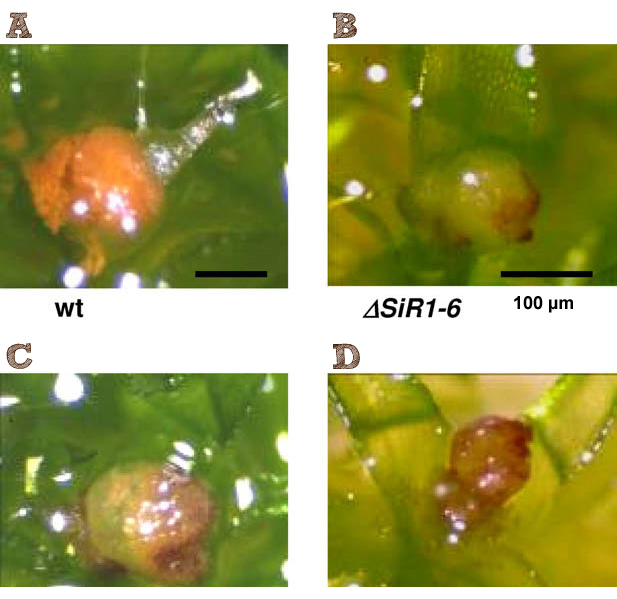
In order to prevent vertical gene transfer and to limit the spread of the moss in the environment, we looked for strains which are not able to form mature spores. P. patens is monoicous, meaning that male and female organs are produced in one plant. Normally, at the tips of adult gametophores the sexual organs, antheridia (male) and archegonia (female), are produced under inducing conditions. After fertilisation of the egg inside the archegonium a sporophyte develops which contains approx. 5000 spores. A possible way to inhibit successful germination of these spores is to knock out the enzyme sulfite reductase 1 (SiR1). This protein reduces sulfite to sulfide and is involved in sulfur metabolism. [http://www.sciencedirect.com/science/article/pii/S0014579310002462 Wiedemann et al, 2010] disrupted PpSiR1 by homologous recombination and found that ΔSiR1 plants showed strong developmental alterations and are unable to produce mature spores. In the ΔSiR1 lines, only one third of the number of sporophytes was formed, the spore capsules cracked open when the spores inside were still immature and these mutant spores did not germinate. Since it takes more time to establish a mature culture with the knock-out mutants and considering the limited time in iGEM competitions, we did not actually use this mutant in our experiments for iGEM. However, the mutant can easily be ordered from the [http://www.moss-stock-center.org/| International Moss Stock Center]. A consideration for the future would be to design the integration vector in a way that targeted gene knockouts in the disulfite reductase gene are possible, thus inihibitng sporulation and having a targeted integration of constructs.
horizontal gene transfer
Concerning horizontal gene transfer, the main safety threat would be the spread of transgenes from our moss to bacteria. The transfer of primary transgenes or marker genes could lead to new bacteria strains that express these genes and could pass them on to pathogenic strains. But while this process is very common in bacteria, apart from agrobacterium tumefaciens, there are few examples of natural gene transfer between bacteria and higher eukaryotes [Quelle].
For the transgene to remain in the bacterial population and thus in the environment, there has to be a strong selective pressure. This could be the case in highly contaminated environments, where the ability to break down a toxic pollutant is beneficial. We are aware that our moss carries transgenes that could be advantageous to bacteria living in these polluted environments. But since all of the effectors that we are testing actually come from bacteria, we do no think that this is the main problem. The genes for these effectors are already present in the bacteria population. Nevertheless, we need to have a closer look at antibiotic resistance genes such as ereB and the selection marker nptII. Antibiotic resistance genes are the focus of attention because of their potentially strong and general selective advantage in human pathogens. But in the case of ereB, which comes from E.coli, the resistance gene is already present in the bacterial population on plasmids while in the moss it is controlled by a moss promotor and integrated in the moss genome. Therefore, it is much more likely that in case of ereB, the horizontal gene transfer occurs between bacteria and not between bacteria and moss.
A possible way to eliminate superfluous DNA sequences such as the nptII marker after a successful transformation is the use of a two-component site-specific recombination system such as Cre-loxP (Dale and Ow, 1991). The recombinase Cre recognizes short sequences known (loxP) and is then able to excise DNA betweeen two loxP sites that are in the same orientation. Therefore, marker genes flanked by loxP sites can be efficiently excised from transgenic plants if Cre is present.
light-triggered Killswitch
In order to prevent our moss from escaping into the environment, we have included a . When exposed to red-light, a nuclease system is activated which destroys all genetic material, killing the moss and preventing the spread of genetic material of the moss. With this genetic circuit it would be possible to create an niche for the transgenic moss by just covering an area with a blue filter foil. For more information, look at the killswitch page
Biosecurity
Biosecurity is the prevention of loss, theft, misuse, diversion or intentional release of pathogens and toxins. Physcomitrella itself is in no way pathogenic and is endemic to many parts of the world. However, just like with every transgenic organism, there is the theoretical possibility to use Physcomitrella to cause harm, e.g. when it is used to secrete toxic substances. Nevertheless, other organisms which have for example shorter generation times or are pathogenic by nature seem to be more appropriate for such dual-use applications. Additionally, none of the transgenic plants (listed here) that we have created could be used to cause harm.
Labsafety
The lab we work in is classified as BSL 1 (biosafety level 1), according to the [http://eur-lex.europa.eu/LexUriServ/LexUriServ.do?uri=CELEX:32000L0054:en:NOT|European Union Directive 2000/54/EG] and the German "Gesetz zur Regelung der Gentechnik ([http://www.gesetze-im-internet.de/gentg/| GenTG])" (law for the regulation of genetic engineering). There is a total of four Biosafety levels, with BSL 1 being the lowest and BSL 4 being the highest. This classification of the respective Biosafety levels is very similar to the one given in the [http://www.who.int/csr/resources/publications/biosafety/en/Biosafety7.pdf| World Health Organization (WHO) Laboratory Biosafety Manual]. Work inside a BSL 1 lab, such as ours, involves no devices that are potentially harmful to the researchers if they act according to the general precautionary measures. Especially, no pathogenic organisms are used.
A regular safety briefing and a lecture about the legal basics concerning biotechnology and genetic engineering are basic elements of our education at TU Munich. In this context, the handling of biological material, dangerous aspects of chemicals and the circumstances and protocols at the lab we work in are explained. Additionally a special safety briefing was held for all iGEM students by Dr. Martin Schlapschy who is the responsible for lab safety in Prof. Skerras lab.
All of us have worked in laboratories before and have experience with biological parts and chemicals. When we are unsure about the safety measures that have to be taken when handling certain chemicals and devices, we always have the support of our instructors and the researchers working at Prof. Skerras lab.
Safety precautions during molecular biology experiments
Just like in every other biochemical laboratory, there are substances and devices in our lab which are potentially dangerous. Here are three examples of how these situations are handled in our lab.
1. In every laboratory of molecular biology, specific chemicals are required for the staining of DNA, in order to make it visible on agarose gels. Most of them directly intercalate into double-stranded DNA, making them carcinogenic. The substance we use is ethidium bromide. To prevent skin contact, we wear protective gloves made from nitrile rubber and change them frequently to prevent contamination. All gels and materials that come into contact with ethidium bromide are disposed of separately. This is done in order to prevent their unintended leakage into the environment with subsequent harm to humans, animals and plants.
2. Methods of molecular biology often require strong acids or bases, like hydrochloric acid, or toxic substances such as methanol. We handle them with extreme caution under a fume hood and dispose of them separately.
3. Many devices in the lab can be potentially dangerous towards researchers if they are used carelessly or in the wrong way. One example for this are lamps emitting ultraviolet radiation, which can cause damage to the eyes. When dealing with UV radiation, we always wear safety helmets made out of plexiglas. We are aware of the potential harm caused by devices that we are using and thus can protect ourselves appropriately.
Safety Forms
File:TUM13 Basic Safety Form.pdf Basic Safety Form
File:TUM13 Safety Form2.pdf Safety Form 2
References
http://www.ncbi.nlm.nih.gov/pubmed/6327079 Edens et al., 1984
- http://www.ncbi.nlm.nih.gov/pubmed/6327079 Edens et al., 1984 Edens, L., Bom, I., Ledeboer, A. M., Maat, J., Toonen, M. Y., Visser, C., and Verrips, C. T. (1984). Synthesis and processing of the plant protein thaumatin in yeast. Cell, 37(2):629–33.
 "
"



AutoAnnotator:
Follow us:
Address:
iGEM Team TU-Munich
Emil-Erlenmeyer-Forum 5
85354 Freising, Germany
Email: igem@wzw.tum.de
Phone: +49 8161 71-4351