Team:Paris Saclay/Modeling
From 2013.igem.org
(→What does our model look like?) |
(→What does our model look like?) |
||
| Line 39: | Line 39: | ||
==''What does our model look like?''== | ==''What does our model look like?''== | ||
| - | <p>In our model, we establish a virtual | + | <p>In our model, we establish a virtual cell. Inside of this cell, we simulate some different reactions about different molecules. We observe those molecules reacting in some scenarios which are prepared for testing our system. Of cause, we also reveal our experiments in the model. </p> |
{| align="center" | {| align="center" | ||
|- | |- | ||
Revision as of 19:32, 4 October 2013
Contents |
Introduction
Those words could give you the first impression of our modeling.
What do we model?
The degradation of PCBs demands multi bio-chemical reactions performing successively in aerobic or anaerobic condition. The regulator FNR is the key to control the processus. In different condition, FNR activated or desactivated, and foster or prevent some gene expression. So our super bacterium PCBs buster can be regulated by our will, and does the right thing at the right time.
This modeling is about the simulation of FNR aero/anaerobic regulation system that we have successfully achieved in our experiments. Briefly, we try to build bio-informatically a model which can reveal the alternate expression of FNR and its reporter, a virtuel system include bio-chemical phenomenons from the translation of fnr to the production of reporter protein.
What’s our purpose for this modeling?
The main aim of our modeling is to recreate the aero/anaerobic reporter protein expression system. By alternating the oxygen concentration, active or inactive FNR combines (or not) with potential reporter sensor gene sequence and make green or red color. Our job is to see if we can observe the alternating red and green curve when we change the oxygen condition.
How do we model?
As the research of FNR regulation system is a well-studied territory, we found some excellent scientific paper in library especially [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2867928/ Regulation of Aerobic to Anaerobic Transitions by the FNR cycle in Escherichia coli by Dean A.Tolla and Michael A.Savageau]. They have built successfully a differential equation system. And in our modeling we decided to redo what they have done but using a different method: a stochastic system created by a new modeling tool: Hsim. We also programe, in additional a online simulator, this simulator can be performed on the web site without installing the program in computer.
What does our model look like?
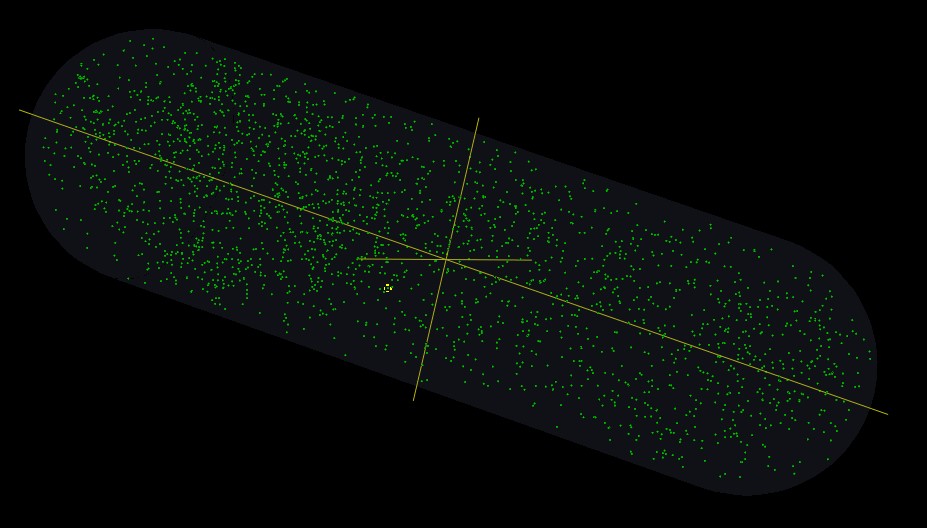
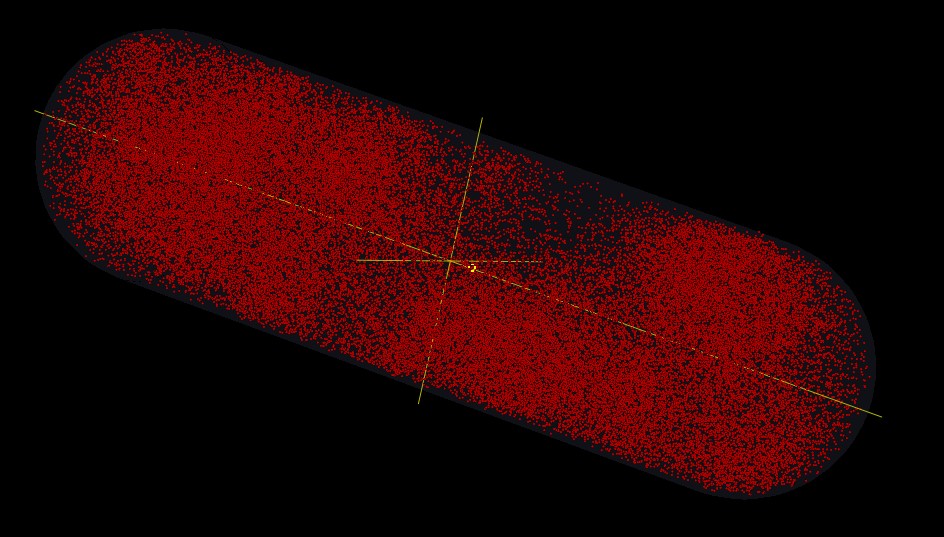
In our model, we establish a virtual cell. Inside of this cell, we simulate some different reactions about different molecules. We observe those molecules reacting in some scenarios which are prepared for testing our system. Of cause, we also reveal our experiments in the model.
| 
|
In the model here we only reveal 2 kinds of reporter protein (green and red, respectivelly regulated by activator and repressor promotor). In anaerobic condition, active FNR leads to the production of the green reporter protein and suppress the expression of the red one. In aerobic condition, FNR is inactivated by oxygen, there is no production of green reporter protein while the red reporter protein is synthetised.
 "
"