Team:ETH Zurich/Infoproc
From 2013.igem.org
| Line 4: | Line 4: | ||
<h1>Information processing</h1> | <h1>Information processing</h1> | ||
<p align="justify">The agar minefield consists of colonies in a hexagonal grid with mine and non-mine colonies. We have two bacterial strains: Firstly the mine strain which provides the sender cells and secondly the non-mine strain which provides the receiver cells. The cells communicate through [https://2013.igem.org/Team:ETH_Zurich/Experiments_2#diffusion_experiment diffusion of OHHL] in the agar. The player pipets substrate on a colony, which leads to a change in the color of the colony. This gives the player information to logically carry out the next move in the game. The colonies remain white until any substrate is added. The left and the right click of the mouse is simulated with the addition of the multi-substrate mix and the single substrate respectively.</p>[[File:infoproc14.png|800px|center|thumb|<b>Figure 1: Signal transduction from secreted signaling molecule to colorimetric response.</b> The signal diffuses through the agar from sender cells (light blue) to receiver cells (dark grey). | <p align="justify">The agar minefield consists of colonies in a hexagonal grid with mine and non-mine colonies. We have two bacterial strains: Firstly the mine strain which provides the sender cells and secondly the non-mine strain which provides the receiver cells. The cells communicate through [https://2013.igem.org/Team:ETH_Zurich/Experiments_2#diffusion_experiment diffusion of OHHL] in the agar. The player pipets substrate on a colony, which leads to a change in the color of the colony. This gives the player information to logically carry out the next move in the game. The colonies remain white until any substrate is added. The left and the right click of the mouse is simulated with the addition of the multi-substrate mix and the single substrate respectively.</p>[[File:infoproc14.png|800px|center|thumb|<b>Figure 1: Signal transduction from secreted signaling molecule to colorimetric response.</b> The signal diffuses through the agar from sender cells (light blue) to receiver cells (dark grey). | ||
| - | The non-mine colonies are designed to distinguish between different concentrations of OHHL and translate this information into expression of different | + | The non-mine colonies are designed to distinguish between different concentrations of OHHL and translate this information into expression of different hydrolases. They are equipped with mutated LuxR promoters that show different OHHL sensitivities and serve as high pass filters. After an incubation time of 12 hours the player pipets a substrate on the colony. A hydrolase converts the substrate into a colored product which is visible by eye ]] |
<br clear="all"/> | <br clear="all"/> | ||
<p align="justify">The biology is explained here. The sender colony secretes the quorum sensing molecule 3-oxo-N-hexanoyl-L-homoserine lactone | <p align="justify">The biology is explained here. The sender colony secretes the quorum sensing molecule 3-oxo-N-hexanoyl-L-homoserine lactone | ||
Revision as of 23:31, 4 October 2013
Information processing
The agar minefield consists of colonies in a hexagonal grid with mine and non-mine colonies. We have two bacterial strains: Firstly the mine strain which provides the sender cells and secondly the non-mine strain which provides the receiver cells. The cells communicate through diffusion of OHHL in the agar. The player pipets substrate on a colony, which leads to a change in the color of the colony. This gives the player information to logically carry out the next move in the game. The colonies remain white until any substrate is added. The left and the right click of the mouse is simulated with the addition of the multi-substrate mix and the single substrate respectively.
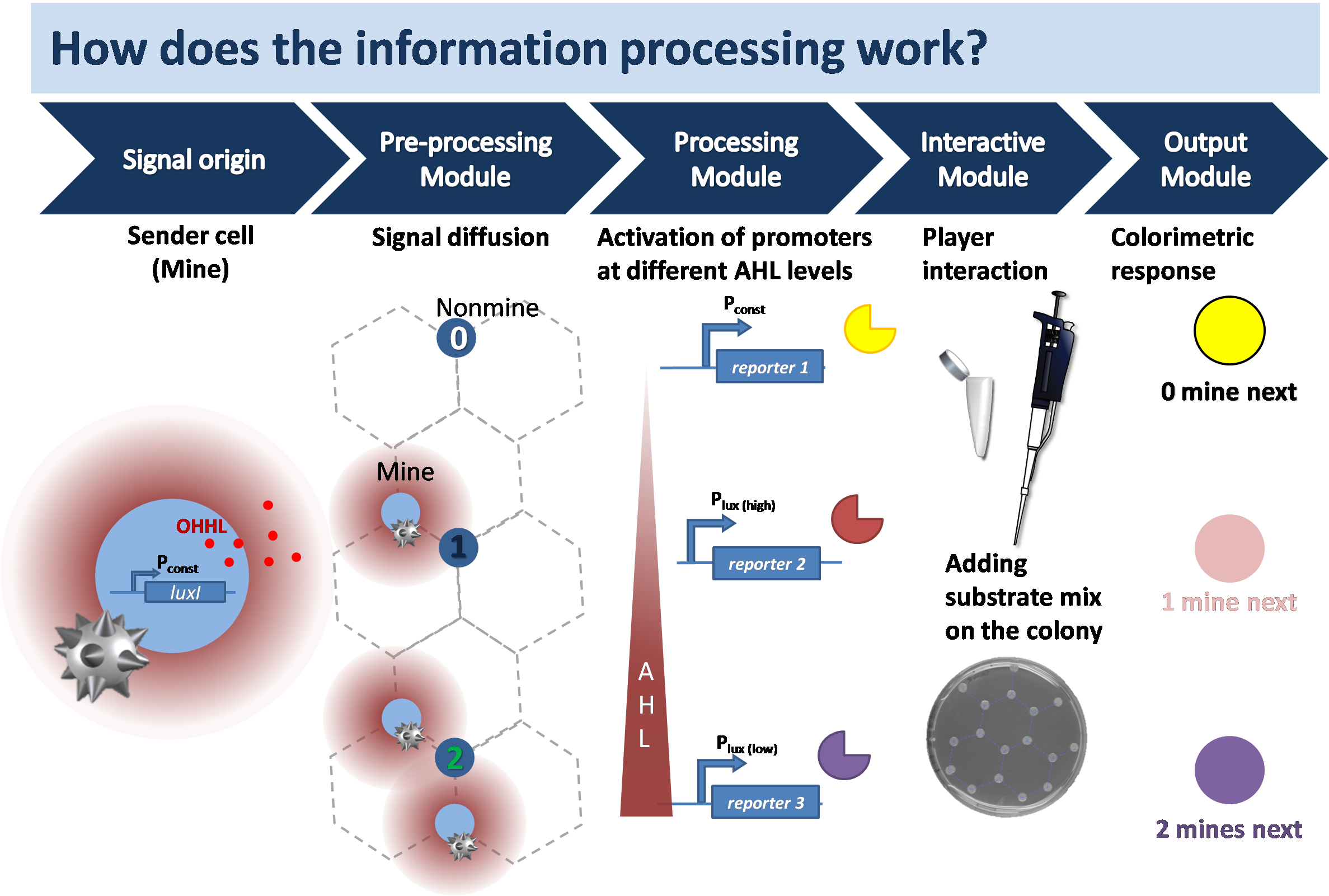
The biology is explained here. The sender colony secretes the quorum sensing molecule 3-oxo-N-hexanoyl-L-homoserine lactone (OHHL) that diffuses through the agar to the surrounding cells. In the receiver cells, the signaling molecule forms a complex with the inactive LuxR to activate it. The information is translated via mutated pLuxR promoters of different OHHL affinities which leads to the secretion of different hydrolases. Within minutes after the addition of substrate a change in color indicates the identity of the played colony and number of surrounding mine colonies.
 "
"





