Team:ETH Zurich/Experiments 3
From 2013.igem.org
| (52 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:ETH_Zurich/Templates}} | {{:Team:ETH_Zurich/Templates}} | ||
{{:Team:ETH_Zurich/Templates/stylesheet}} | {{:Team:ETH_Zurich/Templates/stylesheet}} | ||
| - | |||
<h1>Enzyme-substrate reactions</h1> | <h1>Enzyme-substrate reactions</h1> | ||
| Line 7: | Line 6: | ||
[[File:2013-09-16 09.30 (1).png|400px|right|thumb|<b>Figure 1: Colorimetric output.</b> This image shows liquid cultures of our mutant ''Escherichia coli'' strain expressing the enzymes PhoA, NagZ, LacZ, GusA or Aes to convert specific chromogenic substrates into colored outputs.]] | [[File:2013-09-16 09.30 (1).png|400px|right|thumb|<b>Figure 1: Colorimetric output.</b> This image shows liquid cultures of our mutant ''Escherichia coli'' strain expressing the enzymes PhoA, NagZ, LacZ, GusA or Aes to convert specific chromogenic substrates into colored outputs.]] | ||
| - | <p align="justify">To generate visible output by adding substrates to colonies in Colisweeper, we made use of orthogonal enzyme-substrate reactions. A set of chromogenic substrates was chosen to produce different colors depending on the abundant hydrolases and thereby to uncover the identity of each colony to the player.<br> | + | <p align="justify">To generate <b>visible output</b> by adding substrates to colonies in Colisweeper, we made use of orthogonal enzyme-substrate reactions. A set of <b>chromogenic substrates</b> was chosen to produce different colors depending on the abundant hydrolases and thereby to uncover the identity of each colony to the player.<br> |
The set of enzyme-substrate pairs chosen for the Colisweeper project, and characterizations of their reactions, are described below. Information on other possible substrates that can be used for the enzymes of the Colisweeper reporter system can be found in the [https://2013.igem.org/Team:ETH_Zurich/Experiments_7 reporter system] section.<br> | The set of enzyme-substrate pairs chosen for the Colisweeper project, and characterizations of their reactions, are described below. Information on other possible substrates that can be used for the enzymes of the Colisweeper reporter system can be found in the [https://2013.igem.org/Team:ETH_Zurich/Experiments_7 reporter system] section.<br> | ||
| - | To characterize the hydrolases used in Colisweeper, we conducted <html><a href="https://2013.igem.org/Team:ETH_Zurich/Materials#substrate test" >substrate tests</a></html> in liquid cultures as well as on colonies, <html><a href="https://2013.igem.org/Team:ETH_Zurich/Materials#enzyme_kinetics" >enzyme kinetics</a></html> and detection of HIS-tagged protein by Western Blot. Additionally, crosstalk assays and color overlay tests (multiple enzyme-substrate reactions together) have been performed for this project. A listing of chromogenic substrates used can be found in the | + | To characterize the hydrolases used in Colisweeper, we conducted <html><a href="https://2013.igem.org/Team:ETH_Zurich/Materials#substrate test" >substrate tests</a></html> in liquid cultures as well as on colonies, <html><a href="https://2013.igem.org/Team:ETH_Zurich/Materials#enzyme_kinetics" >enzyme kinetics</a></html> and detection of HIS-tagged protein by Western Blot. Additionally, <b>crosstalk</b> assays and <b>color overlay</b> tests (multiple enzyme-substrate reactions together) have been performed with the triple knockout Δ''aes''Δ''gusA''Δ''nagZ'' for this project. A listing of chromogenic and fluorogenic substrates used can be found in the overview table below. |
</p> | </p> | ||
<br clear="all"/> | <br clear="all"/> | ||
| - | <h1> | + | <h1>Overview of enzyme-substrate reactions</h1> |
<table style="float:left;margin-top:10px;font-size:12px;font-family:verdana"> | <table style="float:left;margin-top:10px;font-size:12px;font-family:verdana"> | ||
<tr> | <tr> | ||
| + | <th width="100px" >Hydrolase</th> | ||
<th width="225px" >Substrate</th> | <th width="225px" >Substrate</th> | ||
| - | + | <th width="125px" >Absorption λ<sub>max</sub> or Excitation/Emission</th> | |
| - | <th width="125px" | + | <th width="150px" >Stock solution</th> |
| - | <th width="150px" >Stock</th> | + | <th width="150px" >Liquid culture: final concentration</th> |
| - | <th width="150px" >Liquid culture</th> | + | <th width="150px" >Colonies:<br>1.5 μl of substrate solution</th> |
| - | <th width="150px" >Colonies</th> | + | |
<th width="150px" >Response time on ''E. coli'' colonies, at RT</th> | <th width="150px" >Response time on ''E. coli'' colonies, at RT</th> | ||
</tr> | </tr> | ||
<tr> | <tr> | ||
| + | <td rowspan="2" align="center">[https://2013.igem.org/Team:ETH_Zurich/Experiments_3#lacZ LacZ]</td> | ||
<td bgcolor="#F0FAFF">5-Bromo-4-Chloro-3-indoxyl-β-D-galactopyranoside (X-Gal)</td> | <td bgcolor="#F0FAFF">5-Bromo-4-Chloro-3-indoxyl-β-D-galactopyranoside (X-Gal)</td> | ||
| - | |||
| - | |||
<td>Blue,<br>615 nm</td> | <td>Blue,<br>615 nm</td> | ||
<td>0.5 M in DMSO</td> | <td>0.5 M in DMSO</td> | ||
<td>1 mM</td> | <td>1 mM</td> | ||
<td>50 mM</td> | <td>50 mM</td> | ||
| - | <td>~ 10 minutes</td> | + | <td><b>~ 10 minutes</b></td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
<td bgcolor="#E6FFCC">N-Methyl-3-indolyl-β-D-galactopyranoside (Green-Gal)</td> | <td bgcolor="#E6FFCC">N-Methyl-3-indolyl-β-D-galactopyranoside (Green-Gal)</td> | ||
| - | |||
| - | |||
<td>Green,<br>665 nm</td> | <td>Green,<br>665 nm</td> | ||
<td>0.1 M in DMSO</td> | <td>0.1 M in DMSO</td> | ||
<td>1 mM</td> | <td>1 mM</td> | ||
<td>50 mM</td> | <td>50 mM</td> | ||
| - | <td>~ 15 minutes</td> | + | <td><b>~ 15 minutes</b></td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| + | <td rowspan="2" align="center">[https://2013.igem.org/Team:ETH_Zurich/Experiments_3#gusA GusA]</td> | ||
<td bgcolor="#FFEEEE">6-Chloro-3-indolyl-β-D-glucuronide (Salmon-Glc)</td> | <td bgcolor="#FFEEEE">6-Chloro-3-indolyl-β-D-glucuronide (Salmon-Glc)</td> | ||
| - | |||
| - | |||
<td>Salmon,<br>540 nm</td> | <td>Salmon,<br>540 nm</td> | ||
<td>0.3 M in DMSO</td> | <td>0.3 M in DMSO</td> | ||
<td>1.5 mM</td> | <td>1.5 mM</td> | ||
<td>0.1 M</td> | <td>0.1 M</td> | ||
| - | <td>~ 5 minutes</td> | + | <td><b>~ 5 minutes</b></td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| + | <td> 4-MU-β-D-Glucuronide</td> | ||
| + | <td>Blue (fluorescent),<br>372 nm (λ<sub>Ex</sub>), <br>445 nm (λ<sub>Em</sub>)</td> | ||
| + | <td>50 mM in DMSO</td> | ||
| + | <td>100 μM</td> | ||
| + | <td>-</td> | ||
| + | <td><b>~ 5 minutes</b></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td rowspan="3" align="center">[https://2013.igem.org/Team:ETH_Zurich/Experiments_3#phoA PhoA]</td> | ||
<td bgcolor="#FFFFCC">4-Nitrophenoyl-phosphate (pNPP)</td> | <td bgcolor="#FFFFCC">4-Nitrophenoyl-phosphate (pNPP)</td> | ||
| - | |||
| - | |||
<td>Yellow,<br>405 nm</td> | <td>Yellow,<br>405 nm</td> | ||
<td>0.5 M in DEA</td> | <td>0.5 M in DEA</td> | ||
<td>50 mM</td> | <td>50 mM</td> | ||
<td>0.5 M</td> | <td>0.5 M</td> | ||
| - | <td>~ 1 minute</td> | + | <td><b>~ 1 minute</b></td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
<td bgcolor="#F0FAFF">5-Bromo-4-Chloro-3-indolyl phosphate (BCIP)</td> | <td bgcolor="#F0FAFF">5-Bromo-4-Chloro-3-indolyl phosphate (BCIP)</td> | ||
| - | |||
| - | |||
<td>Blue,<br>615 nm</td> | <td>Blue,<br>615 nm</td> | ||
<td>0.1 M in H<sub>2</sub>O</td> | <td>0.1 M in H<sub>2</sub>O</td> | ||
<td>1 mM</td> | <td>1 mM</td> | ||
<td>50 mM</td> | <td>50 mM</td> | ||
| - | <td>~ 30 minutes</td> | + | <td><b>~ 30 minutes</b></td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <td bgcolor="#F5CCE0">5-Bromo-6-Chloro-3-indoxyl butyrate (Magenta butyrate) | + | <td> 4-MU-phosphate</td> |
| - | + | <td>Blue (fluorescent),<br>372 nm (λ<sub>Ex</sub>), <br>445 nm (λ<sub>Em</sub>)</td> | |
| - | + | <td>50 mM in DMSO</td> | |
| + | <td>50 μM</td> | ||
| + | <td>-</td> | ||
| + | <td><b>~ 5 minutes</b></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td rowspan="3" align="center">[https://2013.igem.org/Team:ETH_Zurich/Experiments_3#aes Aes]</td> | ||
| + | <td bgcolor="#F5CCE0">5-Bromo-6-Chloro-3-indoxyl butyrate (Magenta butyrate)</td> | ||
<td>Magenta,<br>565 nm</td> | <td>Magenta,<br>565 nm</td> | ||
<td>0.5 M in Acetone</td> | <td>0.5 M in Acetone</td> | ||
<td>0.1 mM</td> | <td>0.1 mM</td> | ||
<td>20 mM</td> | <td>20 mM</td> | ||
| - | <td>~ 2 minutes</td> | + | <td><b>~ 2 minutes</b></td> |
</tr> | </tr> | ||
| - | |||
<tr> | <tr> | ||
<td bgcolor="#F5CCE0">5-Bromo-6-Chloro-3-indoxyl caprylate (Magenta caprylate) | <td bgcolor="#F5CCE0">5-Bromo-6-Chloro-3-indoxyl caprylate (Magenta caprylate) | ||
</td> | </td> | ||
| - | |||
<td>Magenta,<br>565 nm</td> | <td>Magenta,<br>565 nm</td> | ||
<td>0.2 M in Acetone</td> | <td>0.2 M in Acetone</td> | ||
<td>1 mM</td> | <td>1 mM</td> | ||
<td>0.2 M</td> | <td>0.2 M</td> | ||
| - | <td>overnight</td> | + | <td><b>overnight</b></td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
| - | <td bgcolor="#FFFFCC">4-Nitrophenyl- N-acetyl-β-D-glucosaminide (pNP-GluNAc) | + | <td> 4-MU-butyrate</td> |
| - | + | <td>Blue (fluorescent),<br>372 nm (λ<sub>Ex</sub>), <br>445 nm (λ<sub>Em</sub>)</td> | |
| - | + | <td>50 mM in DMSO</td> | |
| + | <td>150 μM</td> | ||
| + | <td>-</td> | ||
| + | <td><b>~ 5 minutes</b></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td rowspan="3" align="center">[https://2013.igem.org/Team:ETH_Zurich/Experiments_3#nagZ NagZ]</td> | ||
| + | <td bgcolor="#FFFFCC">4-Nitrophenyl- N-acetyl-β-D-glucosaminide (pNP-GluNAc)</td> | ||
<td>Yellow,<br>405 nm</td> | <td>Yellow,<br>405 nm</td> | ||
<td>15 mM in H<sub>2</sub>O</td> | <td>15 mM in H<sub>2</sub>O</td> | ||
<td>0.01 mM</td> | <td>0.01 mM</td> | ||
<td>15 mM</td> | <td>15 mM</td> | ||
| - | <td>~ 1 minute</td> | + | <td><b>~ 1 minute</b></td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
<td bgcolor="#F5CCE0">5-Bromo-6-Chloro-3-indolyl N-acetyl-β-D-glucosaminide (Magenta GluNAc) | <td bgcolor="#F5CCE0">5-Bromo-6-Chloro-3-indolyl N-acetyl-β-D-glucosaminide (Magenta GluNAc) | ||
</td> | </td> | ||
| - | |||
<td>Magenta,<br>565 nm</td> | <td>Magenta,<br>565 nm</td> | ||
<td>0.1 M in DMSO</td> | <td>0.1 M in DMSO</td> | ||
<td>1 mM, supplemented with BSA</td> | <td>1 mM, supplemented with BSA</td> | ||
<td>50 mM, with BSA added</td> | <td>50 mM, with BSA added</td> | ||
| - | <td>~ 15 minutes</td> | + | <td><b>~ 15 minutes</b></td> |
</tr> | </tr> | ||
<tr> | <tr> | ||
<td bgcolor="#F0FAFF">5-Bromo-4-Chloro-3-indolyl N-acetyl-β-D-glucosaminide (X-GluNAc) | <td bgcolor="#F0FAFF">5-Bromo-4-Chloro-3-indolyl N-acetyl-β-D-glucosaminide (X-GluNAc) | ||
</td> | </td> | ||
| - | |||
<td>Blue,<br>615 nm</td> | <td>Blue,<br>615 nm</td> | ||
<td>0.1 M in DMSO</td> | <td>0.1 M in DMSO</td> | ||
<td>1 mM, supplemented with BSA</td> | <td>1 mM, supplemented with BSA</td> | ||
<td>50 mM, with BSA added</td> | <td>50 mM, with BSA added</td> | ||
| - | <td>~ 15 minutes</td> | + | <td><b>~ 15 minutes</b></td> |
</tr> | </tr> | ||
</table> | </table> | ||
| Line 131: | Line 141: | ||
<br clear="all"> | <br clear="all"> | ||
<br><br> | <br><br> | ||
| - | < | + | <html><a id="aes" class="frog"></a></html> |
<h1>Acetyl esterase (Aes)</h1> | <h1>Acetyl esterase (Aes)</h1> | ||
| - | + | ||
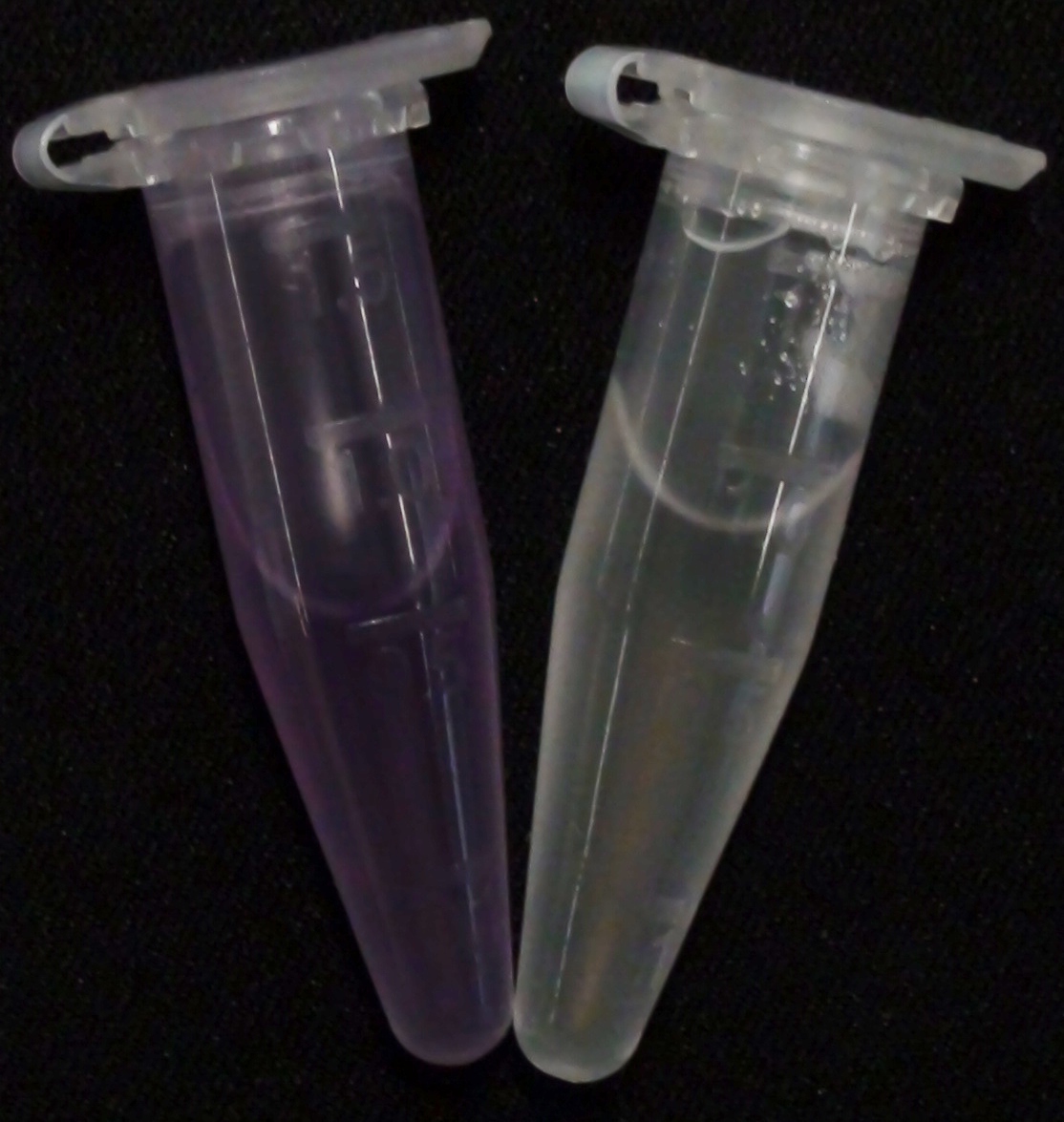
| - | [[File:Aes MagentaButyrate.JPG|thumb|right|200px| <b>Figure 2.</b> Liquid | + | [[File:Aes MagentaButyrate.JPG|thumb|right|200px| <b>Figure 2.</b> Liquid cultures of the Δ''aes''Δ''gusA''Δ''nagZ'' strain overexpressing Aes. Left: magenta-butyrate added; right: no substrate added.]] |
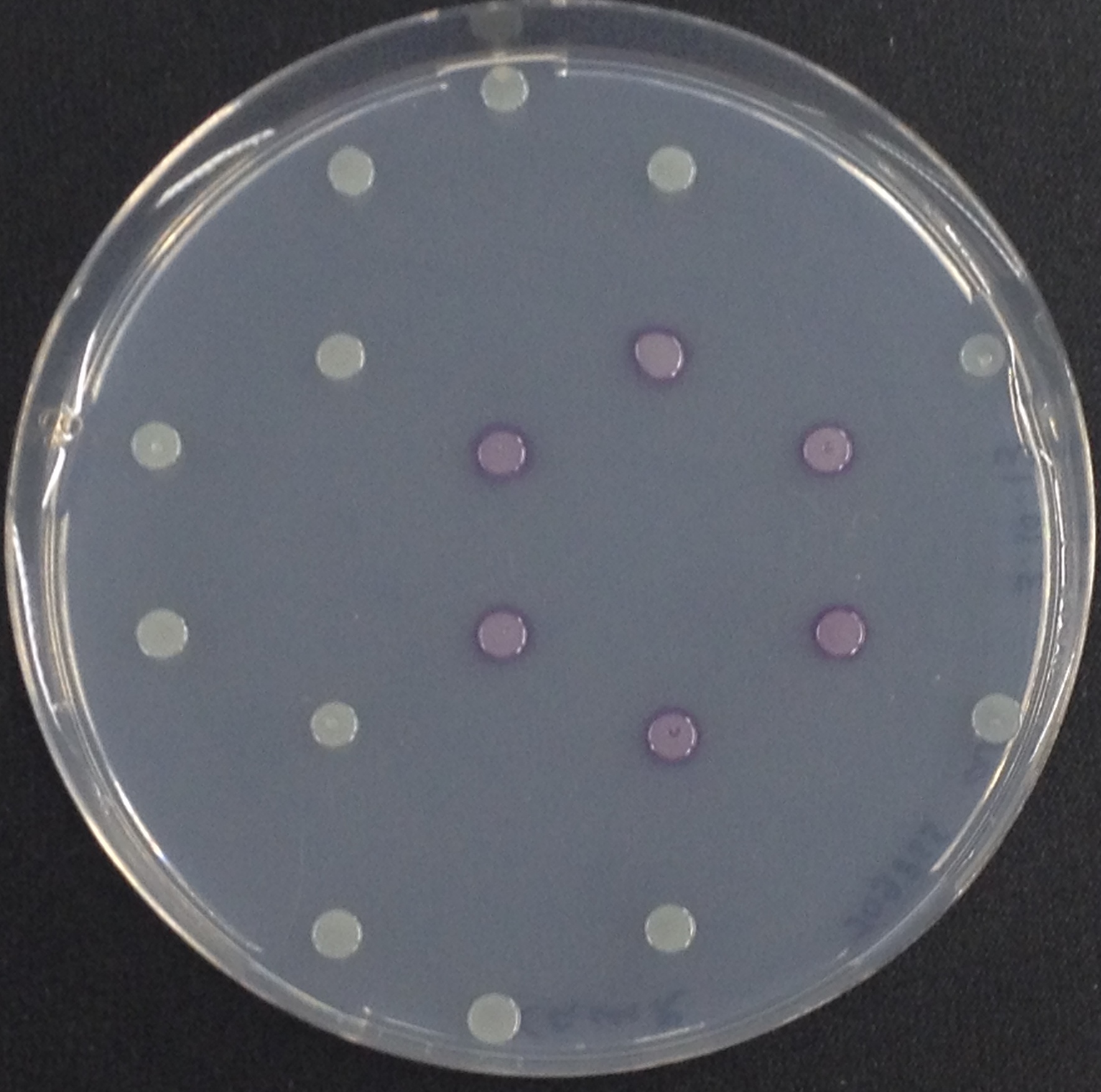
| - | [[File:Aes and Magenta Butyrate.jpeg|thumb|right|200px| <b>Figure 3 | + | [[File:Aes and Magenta Butyrate.jpeg|thumb|right|200px| <b>Figure 3.</b> Colonies of the triple knockout ''Escherichia coli'' strain overexpressing the Aes. Colonies produce magenta color after addition of magenta butyrate.]] |
| - | [[File:MagentaCaprylate.png|thumb|right|200px| <b> Figure | + | [[File:AesBackgroundTest.png|thumb|left|285px| <b>Figure 5. Background activity.</b> Images taken 5 minutes, 6 hours and 12 hours after addition of various concentrations of magenta butyrate onto colonies of the triple knockout strain overexpressing (left) and not overexpressing (right) Aes.]] |
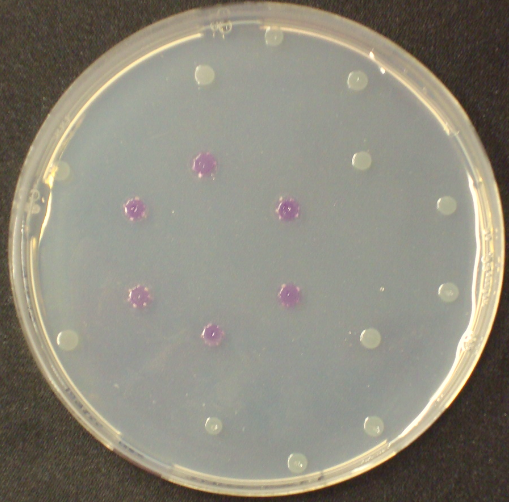
| - | <p align="justify"><b>Chromogenic assay</b><br>To assess color development after reaction of the enzyme with the chromogenic substrate, a liquid culture of our triple knockout <i>Escherichia coli</i> strain overexpressing Aes was grown until an OD<sub>600</sub> of 0.4 - 0.6 was reached before addition of 5-Bromo-6-Chloro-3-indolyl butyrate (magenta butyrate, dissolved in acetone) to a final concentration of 100 µM (Figure 2). To study the color development in the actual Colisweeper game setup, colonies were plated by pipetting 1.5 µl of the triple knockout <i>Escherichia coli</i> liquid culture (OD<sub>600</sub> of 0.4 - 0.6) on an M9 agar plate and incubated overnight. Addition of 1.5 µl of 20 mM magenta butyrate onto each colony results in color generation visible after two to three minutes at room temperature (Figure 3 | + | [[File:MagentaCaprylate.png|thumb|right|200px| <b> Figure 4.</b> Colonies of the triple knockout ''Escherichia coli'' strain overexpressing the hydrolase Aes. Upon addition of magenta caprylate, a magenta color is produced after approximately 30 minutes incubation at 37°C.]] |
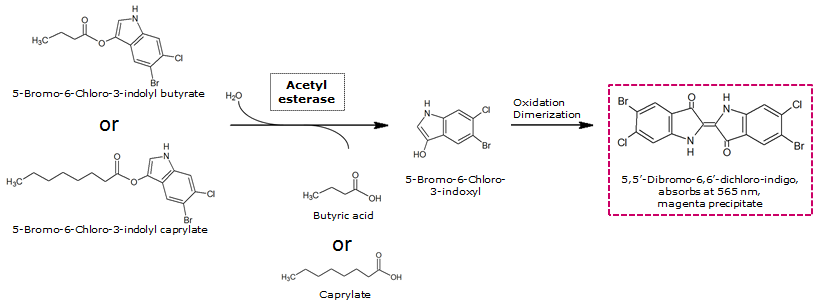
| - | Unfortunately, substrate tests in our triple knockout strain without Aes overexpression have shown that when using the butyrate as a substrate, there is background activity observable, meaning that enzymes other than the Aes can catalyze hydrolysis of the butyrate substrate. Therefore, 5-Bromo-6-Chloro-3-indolyl caprylate (magenta caprylate) was chosen as an alternative substrate, which has shown to be more specifically cleaved by the Aes in our knockout strain. However, when using magenta caprylate as the substrate, color on colonies developed only after at least half an hour of incubation at 37°C, or overnight at room temperature (Figure | + | [[File:AesSubstrates.png|thumb|left|740px| <b>Figure 6.</b> Aes catalyzed hydrolysis reaction of magenta butyrate or magenta caprylate.]] |
| - | Because the player of Colisweeper is not expected to incubate plates after each move, we made an assay comparing time differences of the color development in colonies overexpressing and not overexpressing Aes, using different concentrations of the substrates as well. Results have shown that with the butyrate substrate (20 mM in DMSO) and at room temperature (RT), colonies overexpressing Aes already developed color within the first few minutes, whereas colonies not overexpressing Aes did not show color until six hours after addition of the substrate. Even at twelve hours after adding the substrate, only a faint color was observed in the colonies not overexpressing Aes. Therefore, we continued using magenta butyrate for further tests and finally for the game, due to speed and intensity of color development compared to magenta caprylate. | + | <p align="justify"><b>Chromogenic assay</b><br>To assess color development after reaction of the enzyme with the chromogenic substrate, a liquid culture of our triple knockout <i>Escherichia coli</i> strain overexpressing Aes ([http://parts.igem.org/Part:BBa_K1216002 BBa_K1216002]) was grown until an OD<sub>600</sub> of 0.4 - 0.6 was reached before addition of 5-Bromo-6-Chloro-3-indolyl butyrate (magenta butyrate, dissolved in acetone), a magenta color yielding substrate for Aes (Figure 6), to a final concentration of 100 µM (Figure 2). To study the color development in the actual Colisweeper game setup, colonies were plated by pipetting 1.5 µl of the triple knockout <i>Escherichia coli</i> liquid culture (OD<sub>600</sub> of 0.4 - 0.6) on an M9 agar plate and incubated overnight. Addition of 1.5 µl of 20 mM magenta butyrate onto each colony results in color generation visible after two to three minutes at room temperature (Figure 3).<br> |
| - | </p> | + | Unfortunately, substrate tests in our triple knockout strain without Aes overexpression have shown that when using the butyrate as a substrate, there is background activity observable, meaning that enzymes other than the Aes can catalyze hydrolysis of the butyrate substrate. Therefore, 5-Bromo-6-Chloro-3-indolyl caprylate (magenta caprylate) was chosen as an alternative substrate, which has shown to be more specifically cleaved by the Aes in our knockout strain. However, when using magenta caprylate as the substrate, color on colonies developed only after at least half an hour of incubation at 37°C, or overnight at room temperature (Figure 4).</p> |
| - | + | <p align="justify">Because the player of Colisweeper is not expected to incubate plates after each move, we made an assay comparing time differences of the color development in colonies of the triple knockout ''Escherichia coli'' strain overexpressing and not overexpressing Aes, using different concentrations of the substrates as well (Figure 5). Results have shown that with the butyrate substrate (20 mM in DMSO) and at room temperature (RT), colonies overexpressing Aes already developed color within the first few minutes, whereas colonies not overexpressing Aes did not show color until six hours after addition of the substrate. Even at twelve hours after adding the substrate, only a faint color was observed in the colonies not overexpressing Aes. Therefore, we continued using magenta butyrate for further tests and finally for the game, due to speed and intensity of color development compared to magenta caprylate.</p> | |
<br clear="all"/> | <br clear="all"/> | ||
<b>Enzyme kinetics</b> | <b>Enzyme kinetics</b> | ||
| - | [[File:Aes_fluorescent.png|thumb|right|200px|<b>Figure | + | [[File:Aes_fluorescent.png|thumb|right|200px|<b>Figure 7.</b> Cell lysate of the triple knockout ''Escherichia coli'' strain overexpressing Aes after reaction with 4-MU-butyrate.]] |
| - | <p align="justify">For the kinetics assay, the ''Escherichia coli'' lysate overexpressing Aes was tested with the substrate 4-MU-butyrate which after cleavage gives a fluorescent product. The picture on the right (Figure | + | <p align="justify">For the kinetics assay, the ''Escherichia coli'' lysate overexpressing Aes was tested with the substrate 4-MU-butyrate which after cleavage gives a fluorescent product. The picture on the right (Figure 7) was taken with a common single lens reflex camera mounted on a dark hood at λ<sub>Ex</sub> 365 nm.</p> |
| - | [[File:AES_reaction.png|thumb|center|400px|<b>Figure | + | [[File:AES_reaction.png|thumb|center|400px|<b>Figure 8. Enzymatic reaction of Aes with 4-MU-butyrate.</b>]]<br> |
<p align="justify">To characterize the enzyme, we conducted Michaelis Menten kinetics to obtain the K<sub>m</sub> value. ''Escherichia coli'' cells were harvested and lysed, and the cell free extract (CFX) was then collected for the fluorometric assay. The properly diluted CFX was measured on a 96 well plate in triplicates per substrate concentration. The plate reader took measurements at λ<sub>Ex</sub> 365 nm and λ<sub>Em</sub> 445 nm and determined a K<sub>m</sub> value of 31.5 ± 12.5 μM. The obtained data was then evaluated and fitted to Michaelis-Menten-Kinetics with SigmaPlot™:</p> | <p align="justify">To characterize the enzyme, we conducted Michaelis Menten kinetics to obtain the K<sub>m</sub> value. ''Escherichia coli'' cells were harvested and lysed, and the cell free extract (CFX) was then collected for the fluorometric assay. The properly diluted CFX was measured on a 96 well plate in triplicates per substrate concentration. The plate reader took measurements at λ<sub>Ex</sub> 365 nm and λ<sub>Em</sub> 445 nm and determined a K<sub>m</sub> value of 31.5 ± 12.5 μM. The obtained data was then evaluated and fitted to Michaelis-Menten-Kinetics with SigmaPlot™:</p> | ||
| - | [[File:Hydrolase AES.png|thumb|center|400px|<b>Figure | + | [[File:Hydrolase AES.png|thumb|center|400px|<b>Figure 9.</b> Michaelis-Menten-Kinetics of cell lysate from <i>Escherichia coli</i> overexpressing Aes with the chromogenic substrate: plots velocity versus substrate concentration (10 μL, 30 μL, 100 μL, 200 μL, 400 μL) in 20 mM Tris buffer of pH 8. A kinetic value for K<sub>m</sub> obtained by fitting the raw data to standard the Michaelis Menten equation; K<sub>m</sub> = 31.5 ± 12.5 μM. All assays were carried out in triplicates, results are presented as means.]] |
| - | + | ||
<br clear="all"/> | <br clear="all"/> | ||
| - | + | <html><a id="phoA" class="frog"></a></html> | |
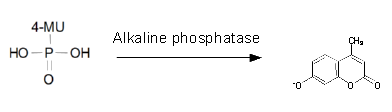
<h1>Alkaline phosphatase (PhoA)</h1> | <h1>Alkaline phosphatase (PhoA)</h1> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | <b> | + | [[File:PhoA_PhoA_his_compared_colored.png|thumb|right|200px|<b>Figure 10.</b> Liquid culture of the triple knockout ''Escherichia coli'' strain overexpressing PhoA or PhoA-His respectively after reacting with pNPP. The negative control tube contains liquid culture without pNPP.]] |
| - | [[File:Phoahis.png|left|400px|thumb|<b>Figure | + | <p align ="justify"><b>Chromogenic assay</b><br>To test the functionality of PhoA ([http://parts.igem.org/Part:BBa_K1216001 BBa_K1216001]) and our improved biobrick PhoA-His ([http://parts.igem.org/Part:BBa_K1216001 BBa_K1216005]), a liquid culture of the triple knockout ''Escherichia coli'' strain overexpressing ''phoA'' or ''phoA-His'' was grown until an OD<sub>600</sub> of 0.4 - 0.6 was reached before addition of ''p''-nitrophenyl phosphate (pNPP, dissolved in DEA) to a final concentration of 5 mM. This substrate gives rise to yellow color after hydrolysis by PhoA (Figure 10). In the Colisweeper game, substrates are pipetted onto colonies. Figure 10 shows color development on colonies five minutes after addition of 1.5 µl of a 0.5 M pNPP solution. The ''phoA'' gene is not a knockout in our host strain, resulting in the expression of additional chromosomal PhoA in all our cells. In our setup this is no problem, because the yellow color will be overlayed by either blue (mine), salmon and violett (mines nearby) if the cell is not a safe cell. <br> |
| - | <p align="justify">We introduced a HIS tag to the PhoA enzyme. The HIS tag enables to purify the enzyme and therby e.g. making a full Michalis Menten studies which can give exact values for the model. Adding of a HIS tag can lead to an unfunctional enzyme, so we first tested the functionality of both PhoA and PhoA HIS tag (see Figure | + | However, problems with the blue color-yielding substrate for NagZ (expressed in mine colonies) have led to the idea to exchange the colors used for PhoA and NagZ. 5-Bromo-4-Chloro-3-indolyl phosphate (BCIP), which is commonly used for detection of alkaline phosphatases in biochemistry, would have been the blue color-producing alternative substrate for PhoA.</p> |
| + | [[File:PhoAandblueliquid.JPG|thumb|right|200px|<b>Figure 11.</b> Liquid culture of the triple knockout ''Escherichia coli'' strain overexpressing PhoA after reacting with BCIP. The negative control tube contains liquid culture without BCIP.]] | ||
| + | [[File:PhoA and pNPP.jpeg|thumb|right|200px| <b>Figure 12.</b> Colonies of the triple knockout ''Escherichia coli'' strain overexpressing the ''Citrobacter'' PhoA. Colonies produce yellow color within 1 minute at RT after addition of pNPP.]] | ||
| + | [[File:Phoahis.png|left|400px|thumb|<b>Figure 15. PhoA-HIS-tag characterization by western blotting.</b> The red stripe indicates the anti-anti-HIS antibody carrying the red fluorescent dye. The white circles indicate the PhoA-HIS protein in the SDS-PAGE gel as well as on the western blot membrane. The molecular weight of this protein is 53kDa.]] | ||
| + | [[File:DSC00096.JPG|thumb|right|200px| <b>Figure 13.</b> Colonies of the triple knockout ''Escherichia coli'' overexpressing the ''Citrobacter'' PhoA. Colonies produce blue color after addition of BCIP.]] | ||
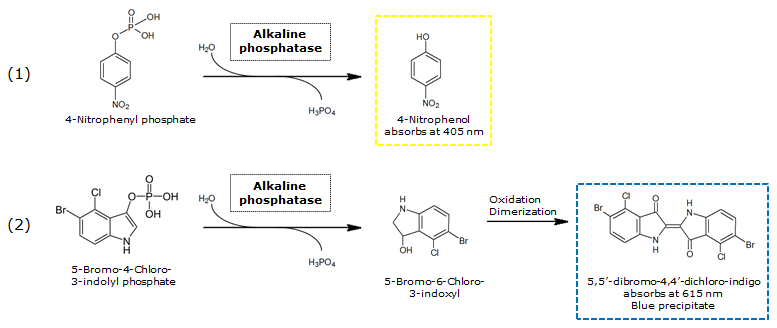
| + | [[File:PhoASubstrates.png|thumb|680px|center|<b>Figure 14.</b> Reaction of PhoA catalyzed hydrolysis of pNPP (1) and BCIP (2).]] | ||
| + | <br><b>Characterization of PhoA-HIS tagged protein</b> | ||
| + | <p align="justify">We introduced a HIS tag to the PhoA enzyme. The HIS tag enables to purify the enzyme and therby e.g. making a full Michalis Menten studies which can give exact values for the model. Adding of a HIS tag can lead to an unfunctional enzyme, so we first tested the functionality of both PhoA and PhoA HIS tag (see Figure 10) with our chromogenic substrate. In a second step, we confirmed the presents of the HIS tag by Western Blot: The PhoA and PhoA with HIS-tag were overexpressed in the ''Escherichia coli'' BL21 DE3 strain, induced by 5 μM IPTG (iso-propy-β-D-1-thiogalactopyranoside) and finally harvested in order to obtain the cell lysate by lysozyme lysis. This cell lysate of both cultures were analyzed next to each other in two SDS-PAGE gels, one for comassie staining (blue gel in the picture) and one for western blotting (black picture) with Anti-6X His tag® antibody from mouse and a second reporter goat anti mouse antibody with a IRDye 680RD. In the picture we can distinguish the PhoA His (53 kDa) on the western blot as well as on the SDS-PAGE gel (see white circles).</p> | ||
<br clear="all"/> | <br clear="all"/> | ||
| - | + | <b>Enzyme kinetics</b> | |
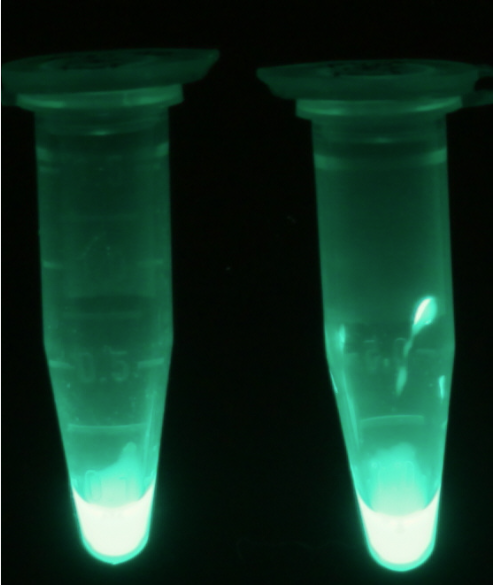
| - | [[File:PhoA_fluorescent.png|thumb|right|200px|<b>Figure | + | [[File:PhoA_fluorescent.png|thumb|right|200px|<b>Figure 16.</b> Cell lysate from <i>Escherichia coli</i> overexpressing PhoA-HIS (left) and PhoA (right) after reaction with 4-MU-phosphate.]] |
| - | <p align="justify">For the kinetics assay, we tested both the ''Citrobacter'' ''phoA'' and ''phoA''-HIS encoded proteins with the fluorescent substrate 4-MU-phosphate. The image in Figure | + | <p align="justify">For the kinetics assay, we tested both the ''Citrobacter'' ''phoA'' and ''phoA''-HIS encoded proteins with the fluorescent substrate 4-MU-phosphate. The image in Figure 16 was taken with a common single lens reflex camera mounted on a dark hood at λ<sub>Ex</sub> 365 nm.</p> |
| - | [[File:phoa_fluorescent_reaction.png|frame|center|<b>Figure | + | [[File:phoa_fluorescent_reaction.png|frame|center|<b>Figure 17. Enzymatic reaction of PhoA with 4-MU-phosphate.</b>]] |
<p align="justify">To characterize the enzymes, we conducted fluorometric assays to obtain K<sub>m</sub> values. ''Escherichia coli'' were harvested and lysed, and the cell free extract (CFX) was then collected for the fluorometric assay. The properly diluted CFX was measured on a 96 well plate in triplicates per substrate concentration. The plate reader took measurements at λ<sub>Ex</sub> 365 nm and λ<sub>Em</sub> 445 nm, determining a K<sub>m</sub> values of 105.9 ± 5.3 μM for PhoA and 72.0 ± 7.3 μM for Pho-HIS.<br> | <p align="justify">To characterize the enzymes, we conducted fluorometric assays to obtain K<sub>m</sub> values. ''Escherichia coli'' were harvested and lysed, and the cell free extract (CFX) was then collected for the fluorometric assay. The properly diluted CFX was measured on a 96 well plate in triplicates per substrate concentration. The plate reader took measurements at λ<sub>Ex</sub> 365 nm and λ<sub>Em</sub> 445 nm, determining a K<sub>m</sub> values of 105.9 ± 5.3 μM for PhoA and 72.0 ± 7.3 μM for Pho-HIS.<br> | ||
The obtained data was evaluated and finally fitted to Michaelis-Menten-Kinetics with SigmaPlot™:</p> | The obtained data was evaluated and finally fitted to Michaelis-Menten-Kinetics with SigmaPlot™:</p> | ||
| - | [[File:PhoA and phoAHIS kinetics.png|frame|center|500px|<b>Figure | + | [[File:PhoA and phoAHIS kinetics.png|frame|center|500px|<b>Figure 18.</b> Michaelis-Menten-Kinetics of cell lysate from <i>Escherichia coli</i> overexpressing the ''Citrobacter'' PhoA or PhoA-HIS: plots velocity versus substrate concentration (2.5 μL, 5 μL, 10 μL, 30 μL, 60 μL, 120 μL, 240 μL) in 20 mM Tris buffer of pH 8. A kinetic value for K<sub>m</sub> obtained by fitting the raw data to standard the Michaelis Menten equation; K<sub>m</sub> (PhoA) = 105.9 ± 5.3 μM and K<sub>m</sub> (PhoA-HIS) = 72.0 ± 7.3 μM. All assays were carried out in triplicates, results are presented as means.]] |
| - | + | ||
| - | + | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
<br clear="all"/> | <br clear="all"/> | ||
| + | <html><a id="lacZ" class="frog"></a></html> | ||
| + | <h1>β-Galactosidase (LacZ)</h1> | ||
| + | [[File:LacZ_GreenGal_XGal.JPG|thumb|right|200px| <b>Figure 19.</b> Liquid cultures of the triple knockout <i>Escherichia coli</i> strain; the natively expressed LacZ reacts with the added substrates Green-Gal (left) and X-Gal (middle). The negative control (right) contains liquid culture without substrates added.]] | ||
| + | <p align="justify"><b>Chromogenic assay</b><br>The β-Galactosidase is natively expressed in the triple knockout ''Escherichia coli'' strain used in Colisweeper. Its commonly used chromogenic substrate is 5-Bromo-4-Chloro-3-indolyl-β-D-Galactopyranoside (X-Gal), which yields a blue-colored precipitate (Figure 19, middle). In Colisweeper, this protein catalyzes hydrolysis of the flagging substrate, which is N-Methyl-3-indolyl-β-D-Galactopyranoside (Green-β-D-Gal) and produces a green color upon cleavage of the chromophore (Figure 19, left tube). For the color assay in liquid culuture, we used ''Escherichia coli'' cultures grown to an OD<sub>600</sub> of 0.4 - 0.6, then added the substrates to a final concentration of 1 mM. On colonies, drops of 50 mM were applied, and development of color showed approximately 10 - 15 minutes after addition of the substrates.</p><br> | ||
| + | [[File:LacZsubstrates.png|thumb|750px|left|<b>Figure 20.</b> Hydrolysis of Green-β-D-Gal (1) or X-Gal (2), catalyzed by LacZ.]] | ||
| + | [[File:LacZ and GreenGal.jpeg|thumb|right|200px| <b>Figure 21.</b> Colonies of our triple knockout strain ''Escherichia coli'' in the hexagonal grid setup. Colonies natively produce LacZ, and addition of X-Gal gives rise to green color.]] | ||
| + | <br clear="all"/> | ||
| + | <html><a id="gusA" class="frog"></a></html> | ||
<h1>β-Glucuronidase (GusA)</h1> | <h1>β-Glucuronidase (GusA)</h1> | ||
| - | + | ||
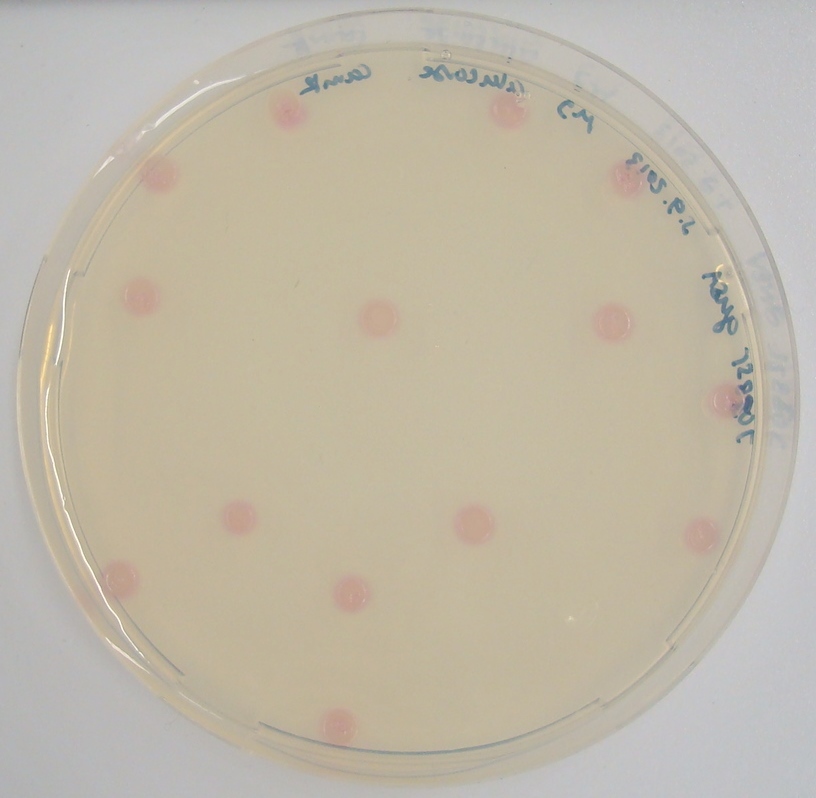
| - | [[File:GusA SalmonGlc.JPG|thumb|right|200px|<b>Figure | + | [[File:GusA SalmonGlc.JPG|thumb|right|200px|<b>Figure 22.</b> Liquid culture of the triple knockout ''Escherichia coli'' strain overexpressing GusA; after reaction with Salmon-Gluc. The negative control (right) is a liquid culture without the substrate added.]] |
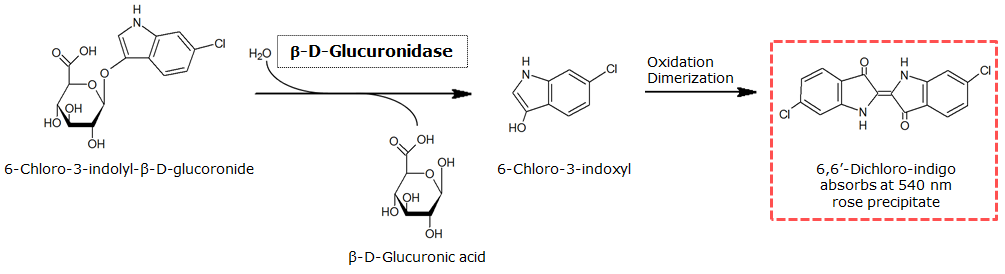
| - | <p align="justify"><b>Chromogenic assay</b><br>To characterize the reaction of GusA with its substrate, a liquid culture of | + | [[File:GusAgrid substrate.JPG|thumb|right|200px|<b>Figure 23.</b> Colonies of the triple knockout ''Escherichia coli'' strain overexpressing the ''Bacillus subtilis'' GusA. Colonies produce salmon color after adding substrate.]] |
| - | [[File:GusAReaction.png|thumb| | + | <p align="justify"><b>Chromogenic assay</b><br>In the Colisweeper game, GusA is expressed in colonies which are surrounded by one mine. To characterize the reaction of GusA with its substrate, a liquid culture of the triple knockout ''Escherichia coli'' strain overexpressing GusA ([http://parts.igem.org/Part:BBa_K1216000 BBa_K1216000]) was grown until an OD<sub>600</sub> of 0.4 - 0.6 was reached before addition of 6-Chloro-3-indolyl-β-glucuronide (Salmon-Gluc), a substrate for GusA which produces a rose/salmon color after cleavage (Figure 22). The final concentration of the substrate in liquid culture was 1 mM and the conversion of this substrate was done by incubation at 37°C. On colonies, 1.5 μl drops of the diluted substrate solution were added, resulting in production of salmon color after approximately 5 minutes at RT.</p><br> |
| + | [[File:GusAReaction.png|thumb|center|800px|<b>Figure 24.</b> Enzymatic reaction of GusA with the chromogenic substrate Salmon-Gluc.]]<br clear="all"/> | ||
<b>Enzyme kinetics</b> | <b>Enzyme kinetics</b> | ||
| - | [[File:GusA_fluorescent.png|thumb|right|200px|<b>Figure | + | [[File:GusA_fluorescent.png|thumb|right|200px|<b>Figure 25.</b> Cell lysate from <i>Escherichia coli</i> overexpressing GusA, after reaction with 4-MU-β-D-Glucuronide.]] |
| - | <p align="justify">For the kinetics assay, we tested both ''gusA'' and ''gusA''-HIS encoded proteins with the fluorescent substrate 4-MU-β-D-Glucuronide. The picture on the right (Figure | + | <p align="justify">For the kinetics assay, we tested both ''gusA'' and ''gusA''-HIS encoded proteins with the fluorescent substrate 4-MU-β-D-Glucuronide. The picture on the right (Figure 25) was taken with a common single lens reflex camera mounted on a dark hood at λ<sub>Ex</sub> 365 nm.</p> |
| - | [[File:gusa_fluorescent_reaction.png|frame|center|<b>Figure | + | [[File:gusa_fluorescent_reaction.png|frame|center|<b>Figure 26.</b> Enzymatic reaction of GusA with the fluorogenic substrate 4-MU-β-D-Glucuronide.]] |
<p align="justify">For the enzyme kinetics of GusA, we conducted fluorometric assays to obtain the K<sub>m</sub> value. ''Escherichia coli'' overexpressing GusA were harvested and lysed, and the cell free extract (CFX) was then collected for the fluorometric assay. The properly diluted CFX was measured on a 96 well plate in triplicates per substrate concentration. A plate reader took measurements at λ<sub>Ex</sub> 365 nm and λ<sub>Em</sub> 445 nm, and determined a K<sub>m</sub> value of 141.1 ± 5.3 μM for GusA, and 201.9 ± 51.1 μM for GusA-HIS. | <p align="justify">For the enzyme kinetics of GusA, we conducted fluorometric assays to obtain the K<sub>m</sub> value. ''Escherichia coli'' overexpressing GusA were harvested and lysed, and the cell free extract (CFX) was then collected for the fluorometric assay. The properly diluted CFX was measured on a 96 well plate in triplicates per substrate concentration. A plate reader took measurements at λ<sub>Ex</sub> 365 nm and λ<sub>Em</sub> 445 nm, and determined a K<sub>m</sub> value of 141.1 ± 5.3 μM for GusA, and 201.9 ± 51.1 μM for GusA-HIS. | ||
The obtained data was evaluated and finally fitted to Michaelis-Menten-Kinetics with SigmaPlot™:</p> | The obtained data was evaluated and finally fitted to Michaelis-Menten-Kinetics with SigmaPlot™:</p> | ||
| - | [[File:GusA and gusAHIS kinetics.png|frame|center|<b>Figure | + | [[File:GusA and gusAHIS kinetics.png|frame|center|<b>Figure 27. Michaelis-Menten-Kinetics of <i>Escherichia coli</i> overexpressing GusA and GusA-HIS:</b> plots velocity versus substrate concentration (8 μL, 16 μL, 32 μL, 65 μL, 130 μL, 260 μL, 520 μL)) in 20 mM Tris buffer of pH 8. A kinetic value for K<sub>m</sub> obtained by fitting the raw data to standard the Michaelis Menten equation; K<sub>m</sub> (GusA) = 141.1 ± 5.3 μM and K<sub>m</sub> (GusA-HIS) = 201.9 ± 51.1 μM. All assays were carried out in triplicates, results are presented as means.]] |
| - | + | ||
| - | + | ||
| + | <br clear="all"/> | ||
| + | <html><a id="nagZ" class="frog"></a></html> | ||
<h1>β-N-Acetylglucosaminidase (NagZ)</h1> | <h1>β-N-Acetylglucosaminidase (NagZ)</h1> | ||
| - | + | ||
| - | [[File:NagZSubstrateLiquid.JPG|thumb|right|200px|<b>Figure | + | [[File:NagZSubstrateLiquid.JPG|thumb|right|200px|<b>Figure 28.</b> Liquid culture of the triple knockout <i>Escherichia coli</i> strain overexpressing NagZ; after reaction with pNP-GluNAc (yellow), Magenta-GluNAc (magenta) and X-GluNAc (blue). The negative control is a liquid culture in M9 medium without any substrate added.]] |
| - | <p align="justify"><b>Chromogenic assay</b><br>NagZ was proposed to be constitutively expressed in the mine colonies, processing its blue color-yielding substrate 5-Bromo-4-Chloro-3-indolyl-β-N-acetylglucosaminide (X-GluNAc). However, we could not initially manage to get color from the reaction with this enzyme-substrate pair. Using p-nitrophenyl-β-N-acetylglucosaminide (pNP-GluNAc) at an end concentration of 0.01 mM in an ''Escherichia coli'' liquid culture with an OD<sub>600</sub> of 0.4 - 0.6, development of yellow color could be observed (Figure | + | <p align="justify"><b>Chromogenic assay</b><br>NagZ ([http://parts.igem.org/Part:BBa_K1216003 BBa_K1216003]) was proposed to be constitutively expressed in the mine colonies, processing its blue color-yielding substrate 5-Bromo-4-Chloro-3-indolyl-β-N-acetylglucosaminide (X-GluNAc). However, we could not initially manage to get color from the reaction with this enzyme-substrate pair. Using p-nitrophenyl-β-N-acetylglucosaminide (pNP-GluNAc) at an end concentration of 0.01 mM in an ''Escherichia coli'' liquid culture with an OD<sub>600</sub> of 0.4 - 0.6, development of yellow color could be observed (Figure 28), confirming that NagZ is expressed in our mine cells.<br> |
| - | According to Orenga et al. ([https://2013.igem.org/Team:ETH_Zurich/Experiments_3#hydrolase_reference 1]), the substrate incorporating the indoxyl chromophore is less preferentially hydrolyzed by bacterial NagZ in comparison to yeast NagZ, and more preferentially hydrolyzes the alizarine-based substrate they applied. To increase hydrolysis of the indoxyl-based substrate, we added the substrate to a liquid culture supplemented with BSA for stabilization of the enzyme ([https://2013.igem.org/Team:ETH_Zurich/Experiments_3#hydrolase_reference 2]), which enhanced hydrolysis by NagZ dramatically. The end concentration in liquid culture of both X-GluNAc and 5-Bromo-6-Chloro-3-indolyl-β-N-acetylglucosaminide (Magenta-GluNAc), a magenta color-yielding substrate for NagZ, were at 1 mM (Figure | + | According to Orenga et al. ([https://2013.igem.org/Team:ETH_Zurich/Experiments_3#hydrolase_reference 1]), the substrate incorporating the indoxyl chromophore is less preferentially hydrolyzed by bacterial NagZ in comparison to yeast NagZ, and more preferentially hydrolyzes the alizarine-based substrate they applied. To increase hydrolysis of the indoxyl-based substrate, we added the substrate to a liquid culture supplemented with BSA for stabilization of the enzyme ([https://2013.igem.org/Team:ETH_Zurich/Experiments_3#hydrolase_reference 2]), which enhanced hydrolysis by NagZ dramatically. The end concentration in liquid culture of both X-GluNAc and 5-Bromo-6-Chloro-3-indolyl-β-N-acetylglucosaminide (Magenta-GluNAc), a magenta color-yielding substrate for NagZ, were at 1 mM (Figure 28).</p> |
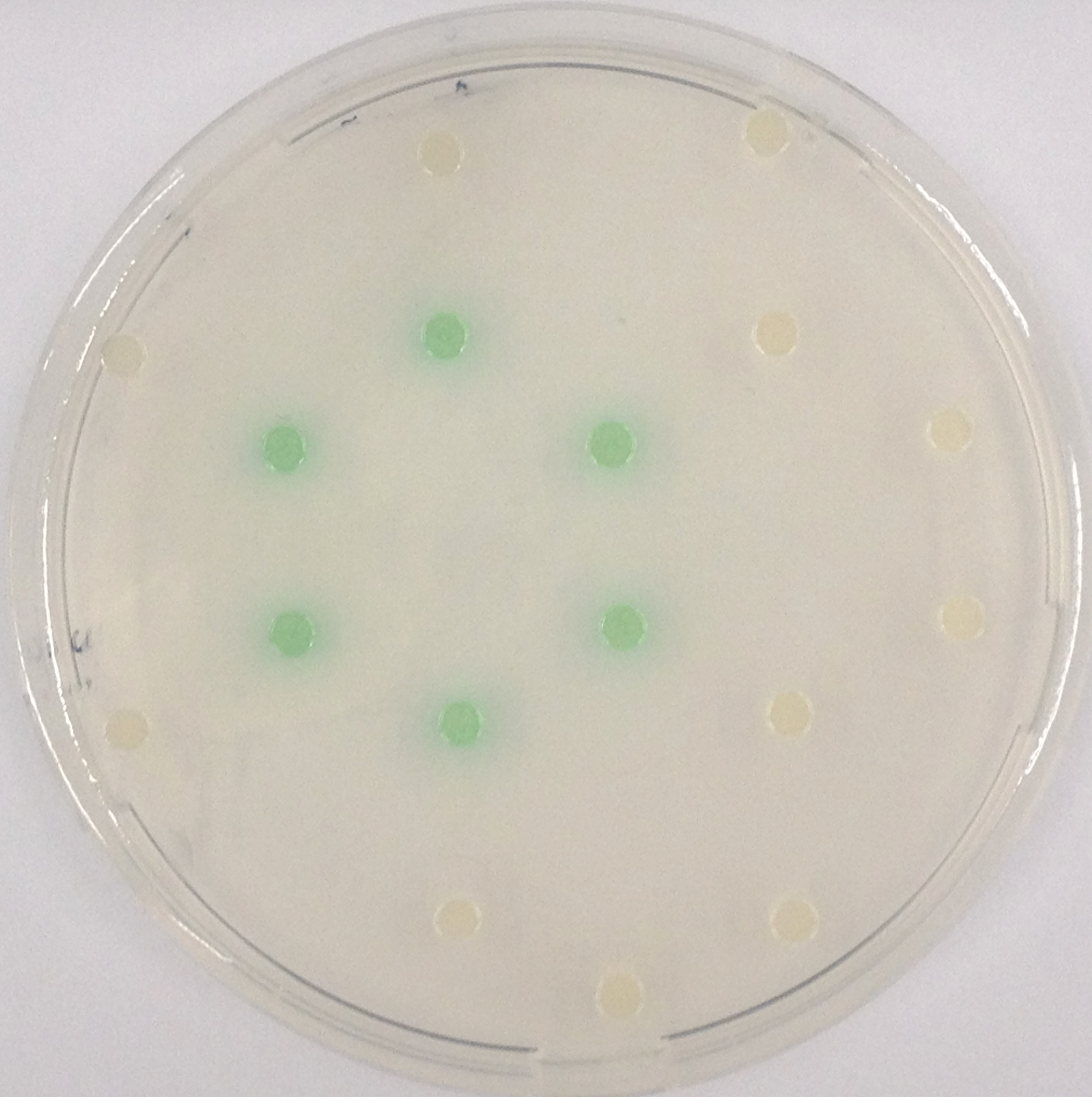
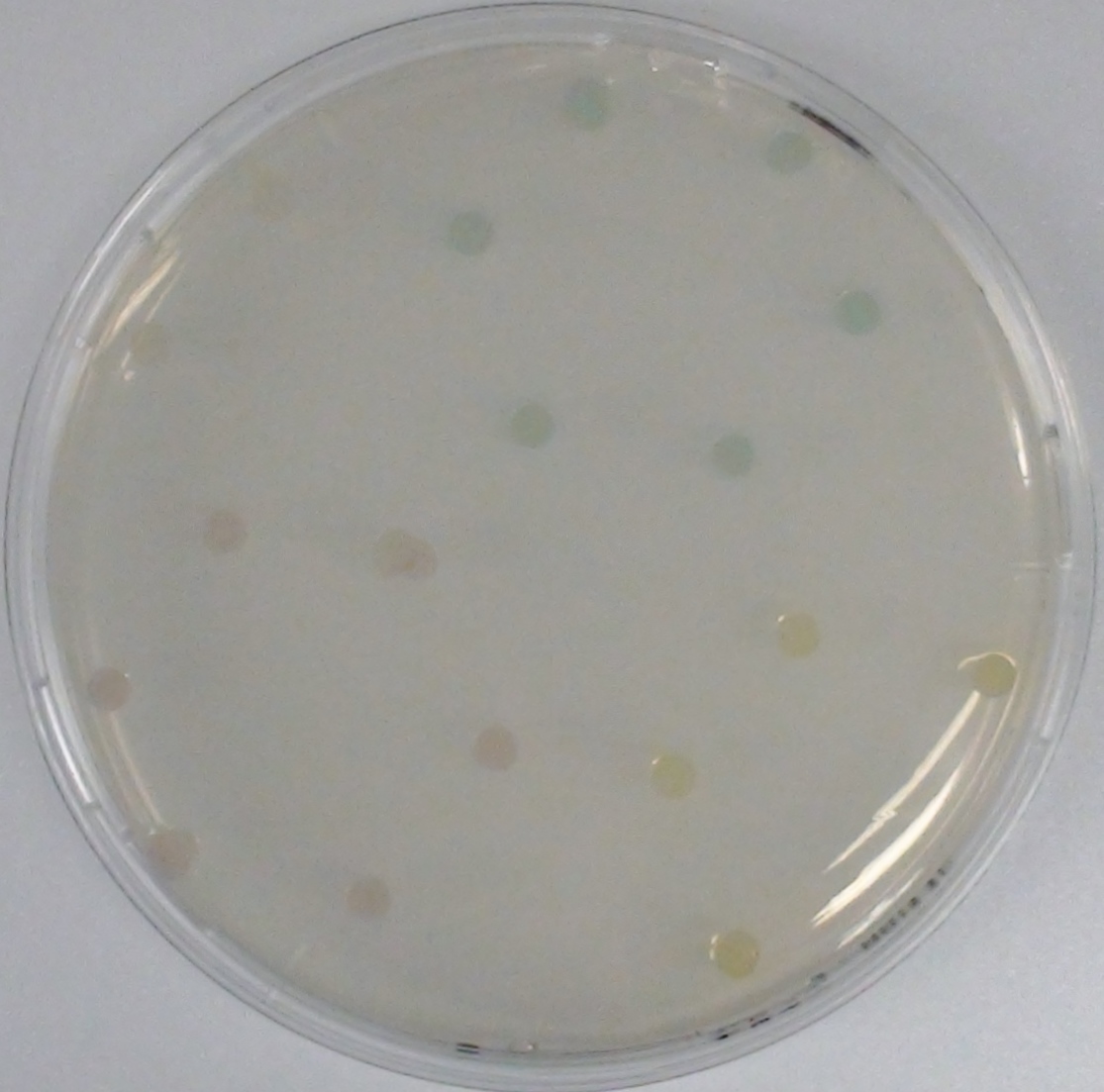
| - | [[File:NagZcoloniesthreesubstrates.JPG|thumb|right|200px|<b>Figure | + | [[File:NagZcoloniesthreesubstrates.JPG|thumb|right|200px|<b>Figure 29.</b> Colonies of the triple knockout <i>Escherichia coli</i> strain overexpressing NagZ; after addition of the NagZ substrates pNP-GluNAc (yellow colonies), Magenta-GluNAc (pink colonies) and X-GluNAc (blue colonies). This image was taken only 15 minutes after the addition of substrates.]] |
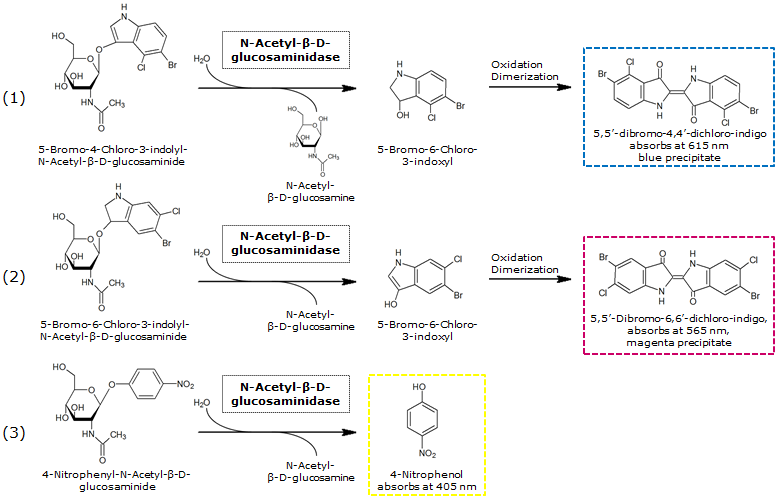
| - | [[File:NagZandSubstrates.png|thumb|left|700px| <b>Figure | + | [[File:NagZandSubstrates.png|thumb|left|700px| <b>Figure 30.</b> β-N-Acetylglucosaminidase catalyzed hydrolysis of the chromogenic substrates X-β-N-acetylglucosaminide (1), Magenta-GluNAc (2) and pNP-GluNAc (3).]]<br> |
| - | + | ||
<br clear="all"/> | <br clear="all"/> | ||
| + | <html><div id="orhtogonal" class=frog"></div></html> | ||
<h1>Crosstalk</h1> | <h1>Crosstalk</h1> | ||
<p align="justify">To ensure specificity of the enzyme-substrate pairs used in Colisweeper, a crosstalk test was done to make sure that all overexpressed enzymes specifically cleave their assigned substrate.</p> | <p align="justify">To ensure specificity of the enzyme-substrate pairs used in Colisweeper, a crosstalk test was done to make sure that all overexpressed enzymes specifically cleave their assigned substrate.</p> | ||
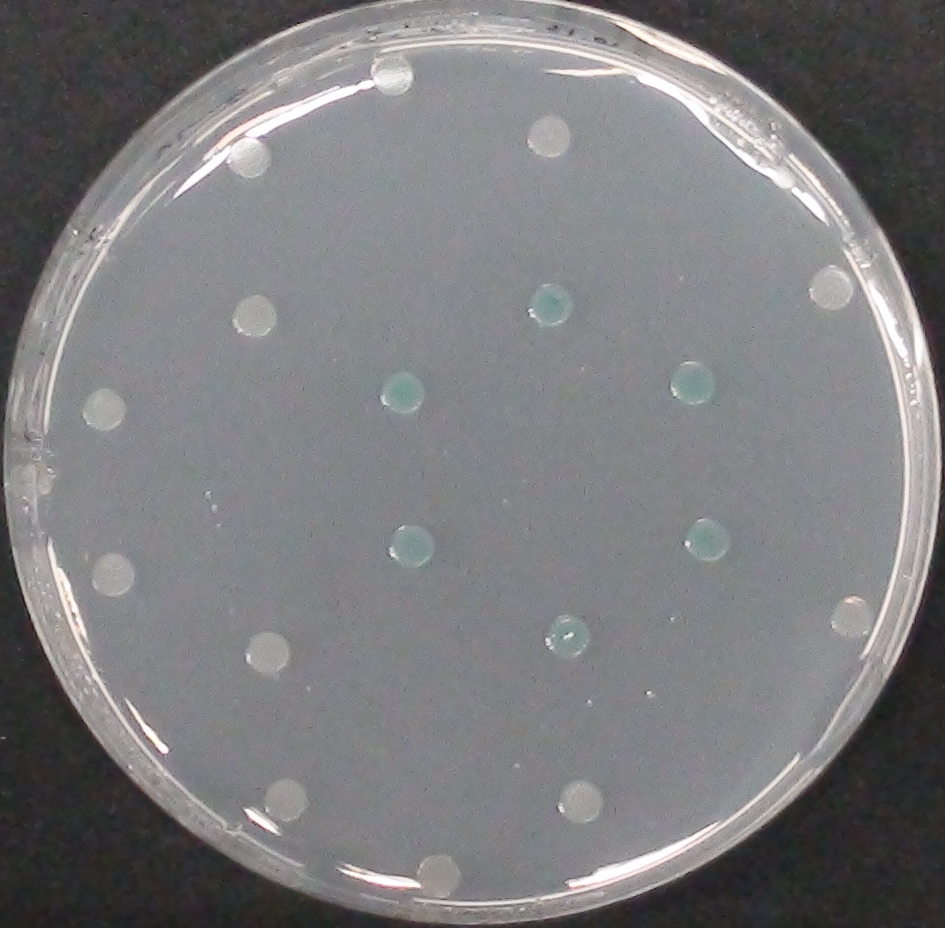
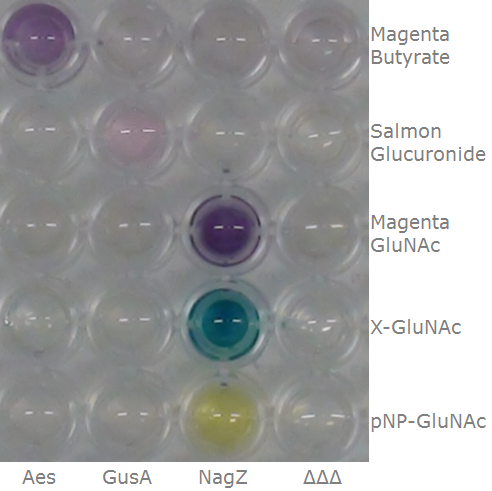
| - | [[File:CrosstalkSubstrates.png|thumb| | + | [[File:CrosstalkSubstrates.png|thumb|left|450px| <b>Figure 31.</b> Liquid cultures of the Δ''aes''Δ''gusA''Δ''nagZ'' <i>Escherichia coli</i> strain overexpressing Aes, GusA, NagZ or none in a 96-well plate, with substrates indicated on the left added horizontally.]] |
| - | <p align="justify">This crosstalk test was done in a 96-well plate, each well containing 200 μl from liquid cultures of our Δ''aes''Δ''gusA''Δ''nagZ'' <i>Escherichia coli</i> strain overexpressing either Aes, GusA, NagZ or none, each distributed among the column-wells of the plate. Horizontally, the chromogenic substrates were pipetted to the liquid cultures in the same order as their corresponding hydrolase. If specificity of the chosen enzyme-substrates pairs were given, we would expect an output as shown in the figure below (Figure | + | <p align="justify">This crosstalk test was done in a 96-well plate, each well containing 200 μl from liquid cultures of our Δ''aes''Δ''gusA''Δ''nagZ'' <i>Escherichia coli</i> strain overexpressing either Aes, GusA, NagZ or none, each distributed among the column-wells of the plate. Horizontally, the chromogenic substrates were pipetted to the liquid cultures in the same order as their corresponding hydrolase: magenta-butyrate for Aes, Salmon-Gluc for GusA, and magenta-, X- and pNP-GluNAc for NagZ. If specificity of the chosen enzyme-substrates pairs were given, we would expect an output as shown in the figure below (Figure 32). As Figure 31 shows, the overexpressed hydrolases cleave only the substrates they were expected to. |
</p> | </p> | ||
| - | [[File:CrosstalkExpected.png|frame| | + | [[File:CrosstalkExpected.png|frame|left| <b>Figure 32. Expected outcome.</b> Added substrates should be specifically cleaved by their hydrolases.]] |
<br clear="all"/> | <br clear="all"/> | ||
| + | <html><div id="overlay" class=frog"></div></html> | ||
<h1>Color overlay</h1> | <h1>Color overlay</h1> | ||
| - | [[File: | + | [[File:ColoroverlayWhite.png|thumb|left|450px| <b>Figure 33.</b> Mixture of liquid cultures of the Δ''aes''Δ''gusA''Δ''nagZ'' <i>Escherichia coli</i> strain overexpressing hydrolases after addition of their substrates: PhoA with pNPP in (1), PhoA with pNPP and GusA with Salmon-Gluc in (2), PhoA with pNPP, GusA with Salmon-Gluc and Aes with Magenta butyrate in (3), and PhoA with pNPP and NagZ with X-GluNAc in (4).]] |
<p align="justify">To play Colisweeper, colors for each colony identity have to be clearly distinguishable by eye. Because we are using high-pass filters to differentially process certain AHL levels, we had to make sure that a mix of cleaved product would show distinguishable colors.</p> | <p align="justify">To play Colisweeper, colors for each colony identity have to be clearly distinguishable by eye. Because we are using high-pass filters to differentially process certain AHL levels, we had to make sure that a mix of cleaved product would show distinguishable colors.</p> | ||
| + | <p align="justify">For this purpose, we mixed liquid cultures of our Δ''aes''Δ''gusA''Δ''nagZ'' triple knockout strain which overexpress PhoA, GusA, Aes or NagZ, and added the corresponding substrates to the cultures. Figure 33 shows the resulting colors after hydrolysis of the substrates: PhoA with pNPP in (1), PhoA with pNPP and GusA with Salmon-Gluc in (2), PhoA with pNPP, GusA with Salmon-Gluc and Aes with Magenta butyrate in (3), and PhoA with pNPP and NagZ with X-GluNAc in (4). These colors are clearly distinguishable from one another and therefore can be used together as reporters of a system incorporating high-pass filters.</p> | ||
<br clear="all"/> | <br clear="all"/> | ||
Latest revision as of 03:30, 29 October 2013
Contents |
Enzyme-substrate reactions
To generate visible output by adding substrates to colonies in Colisweeper, we made use of orthogonal enzyme-substrate reactions. A set of chromogenic substrates was chosen to produce different colors depending on the abundant hydrolases and thereby to uncover the identity of each colony to the player.
The set of enzyme-substrate pairs chosen for the Colisweeper project, and characterizations of their reactions, are described below. Information on other possible substrates that can be used for the enzymes of the Colisweeper reporter system can be found in the reporter system section.
To characterize the hydrolases used in Colisweeper, we conducted substrate tests in liquid cultures as well as on colonies, enzyme kinetics and detection of HIS-tagged protein by Western Blot. Additionally, crosstalk assays and color overlay tests (multiple enzyme-substrate reactions together) have been performed with the triple knockout ΔaesΔgusAΔnagZ for this project. A listing of chromogenic and fluorogenic substrates used can be found in the overview table below.
Overview of enzyme-substrate reactions
| Hydrolase | Substrate | Absorption λmax or Excitation/Emission | Stock solution | Liquid culture: final concentration | Colonies: 1.5 μl of substrate solution |
Response time on E. coli colonies, at RT |
|---|---|---|---|---|---|---|
| LacZ | 5-Bromo-4-Chloro-3-indoxyl-β-D-galactopyranoside (X-Gal) | Blue, 615 nm |
0.5 M in DMSO | 1 mM | 50 mM | ~ 10 minutes |
| N-Methyl-3-indolyl-β-D-galactopyranoside (Green-Gal) | Green, 665 nm |
0.1 M in DMSO | 1 mM | 50 mM | ~ 15 minutes | |
| GusA | 6-Chloro-3-indolyl-β-D-glucuronide (Salmon-Glc) | Salmon, 540 nm |
0.3 M in DMSO | 1.5 mM | 0.1 M | ~ 5 minutes |
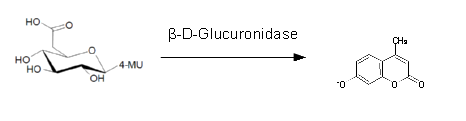
| 4-MU-β-D-Glucuronide | Blue (fluorescent), 372 nm (λEx), 445 nm (λEm) |
50 mM in DMSO | 100 μM | - | ~ 5 minutes | |
| PhoA | 4-Nitrophenoyl-phosphate (pNPP) | Yellow, 405 nm |
0.5 M in DEA | 50 mM | 0.5 M | ~ 1 minute |
| 5-Bromo-4-Chloro-3-indolyl phosphate (BCIP) | Blue, 615 nm |
0.1 M in H2O | 1 mM | 50 mM | ~ 30 minutes | |
| 4-MU-phosphate | Blue (fluorescent), 372 nm (λEx), 445 nm (λEm) |
50 mM in DMSO | 50 μM | - | ~ 5 minutes | |
| Aes | 5-Bromo-6-Chloro-3-indoxyl butyrate (Magenta butyrate) | Magenta, 565 nm |
0.5 M in Acetone | 0.1 mM | 20 mM | ~ 2 minutes |
| 5-Bromo-6-Chloro-3-indoxyl caprylate (Magenta caprylate) | Magenta, 565 nm |
0.2 M in Acetone | 1 mM | 0.2 M | overnight | |
| 4-MU-butyrate | Blue (fluorescent), 372 nm (λEx), 445 nm (λEm) |
50 mM in DMSO | 150 μM | - | ~ 5 minutes | |
| NagZ | 4-Nitrophenyl- N-acetyl-β-D-glucosaminide (pNP-GluNAc) | Yellow, 405 nm |
15 mM in H2O | 0.01 mM | 15 mM | ~ 1 minute |
| 5-Bromo-6-Chloro-3-indolyl N-acetyl-β-D-glucosaminide (Magenta GluNAc) | Magenta, 565 nm |
0.1 M in DMSO | 1 mM, supplemented with BSA | 50 mM, with BSA added | ~ 15 minutes | |
| 5-Bromo-4-Chloro-3-indolyl N-acetyl-β-D-glucosaminide (X-GluNAc) | Blue, 615 nm |
0.1 M in DMSO | 1 mM, supplemented with BSA | 50 mM, with BSA added | ~ 15 minutes |
Acetyl esterase (Aes)
Chromogenic assay
To assess color development after reaction of the enzyme with the chromogenic substrate, a liquid culture of our triple knockout Escherichia coli strain overexpressing Aes ([http://parts.igem.org/Part:BBa_K1216002 BBa_K1216002]) was grown until an OD600 of 0.4 - 0.6 was reached before addition of 5-Bromo-6-Chloro-3-indolyl butyrate (magenta butyrate, dissolved in acetone), a magenta color yielding substrate for Aes (Figure 6), to a final concentration of 100 µM (Figure 2). To study the color development in the actual Colisweeper game setup, colonies were plated by pipetting 1.5 µl of the triple knockout Escherichia coli liquid culture (OD600 of 0.4 - 0.6) on an M9 agar plate and incubated overnight. Addition of 1.5 µl of 20 mM magenta butyrate onto each colony results in color generation visible after two to three minutes at room temperature (Figure 3).
Unfortunately, substrate tests in our triple knockout strain without Aes overexpression have shown that when using the butyrate as a substrate, there is background activity observable, meaning that enzymes other than the Aes can catalyze hydrolysis of the butyrate substrate. Therefore, 5-Bromo-6-Chloro-3-indolyl caprylate (magenta caprylate) was chosen as an alternative substrate, which has shown to be more specifically cleaved by the Aes in our knockout strain. However, when using magenta caprylate as the substrate, color on colonies developed only after at least half an hour of incubation at 37°C, or overnight at room temperature (Figure 4).
Because the player of Colisweeper is not expected to incubate plates after each move, we made an assay comparing time differences of the color development in colonies of the triple knockout Escherichia coli strain overexpressing and not overexpressing Aes, using different concentrations of the substrates as well (Figure 5). Results have shown that with the butyrate substrate (20 mM in DMSO) and at room temperature (RT), colonies overexpressing Aes already developed color within the first few minutes, whereas colonies not overexpressing Aes did not show color until six hours after addition of the substrate. Even at twelve hours after adding the substrate, only a faint color was observed in the colonies not overexpressing Aes. Therefore, we continued using magenta butyrate for further tests and finally for the game, due to speed and intensity of color development compared to magenta caprylate.
Enzyme kinetics
For the kinetics assay, the Escherichia coli lysate overexpressing Aes was tested with the substrate 4-MU-butyrate which after cleavage gives a fluorescent product. The picture on the right (Figure 7) was taken with a common single lens reflex camera mounted on a dark hood at λEx 365 nm.
To characterize the enzyme, we conducted Michaelis Menten kinetics to obtain the Km value. Escherichia coli cells were harvested and lysed, and the cell free extract (CFX) was then collected for the fluorometric assay. The properly diluted CFX was measured on a 96 well plate in triplicates per substrate concentration. The plate reader took measurements at λEx 365 nm and λEm 445 nm and determined a Km value of 31.5 ± 12.5 μM. The obtained data was then evaluated and fitted to Michaelis-Menten-Kinetics with SigmaPlot™:
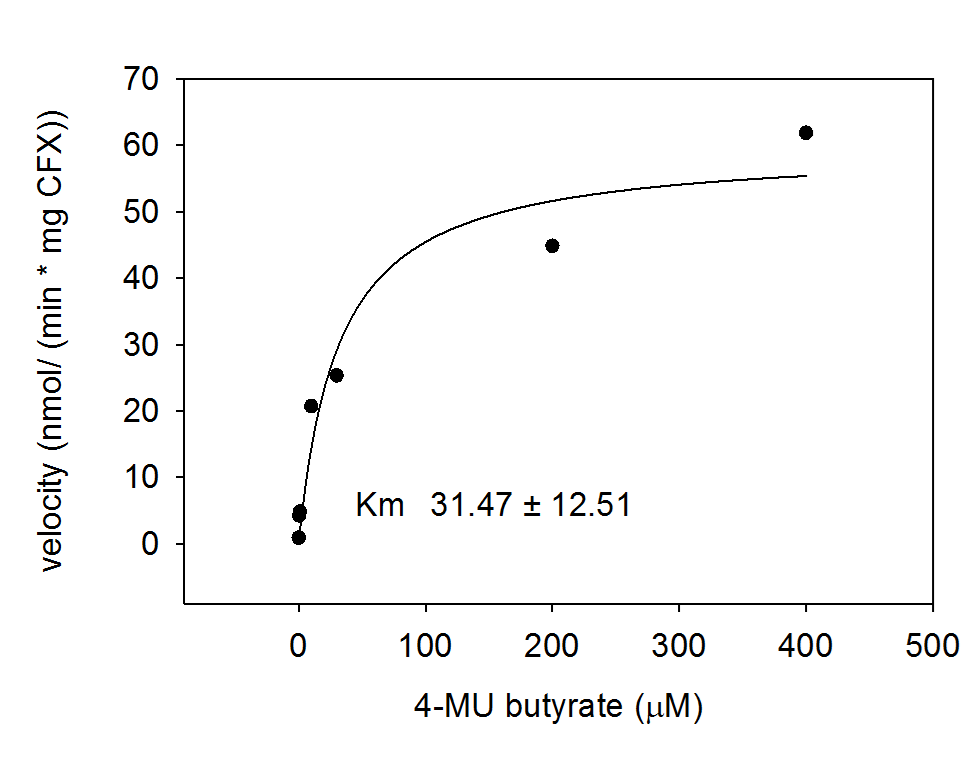
Alkaline phosphatase (PhoA)
Chromogenic assay
To test the functionality of PhoA ([http://parts.igem.org/Part:BBa_K1216001 BBa_K1216001]) and our improved biobrick PhoA-His ([http://parts.igem.org/Part:BBa_K1216001 BBa_K1216005]), a liquid culture of the triple knockout Escherichia coli strain overexpressing phoA or phoA-His was grown until an OD600 of 0.4 - 0.6 was reached before addition of p-nitrophenyl phosphate (pNPP, dissolved in DEA) to a final concentration of 5 mM. This substrate gives rise to yellow color after hydrolysis by PhoA (Figure 10). In the Colisweeper game, substrates are pipetted onto colonies. Figure 10 shows color development on colonies five minutes after addition of 1.5 µl of a 0.5 M pNPP solution. The phoA gene is not a knockout in our host strain, resulting in the expression of additional chromosomal PhoA in all our cells. In our setup this is no problem, because the yellow color will be overlayed by either blue (mine), salmon and violett (mines nearby) if the cell is not a safe cell.
However, problems with the blue color-yielding substrate for NagZ (expressed in mine colonies) have led to the idea to exchange the colors used for PhoA and NagZ. 5-Bromo-4-Chloro-3-indolyl phosphate (BCIP), which is commonly used for detection of alkaline phosphatases in biochemistry, would have been the blue color-producing alternative substrate for PhoA.
Characterization of PhoA-HIS tagged protein
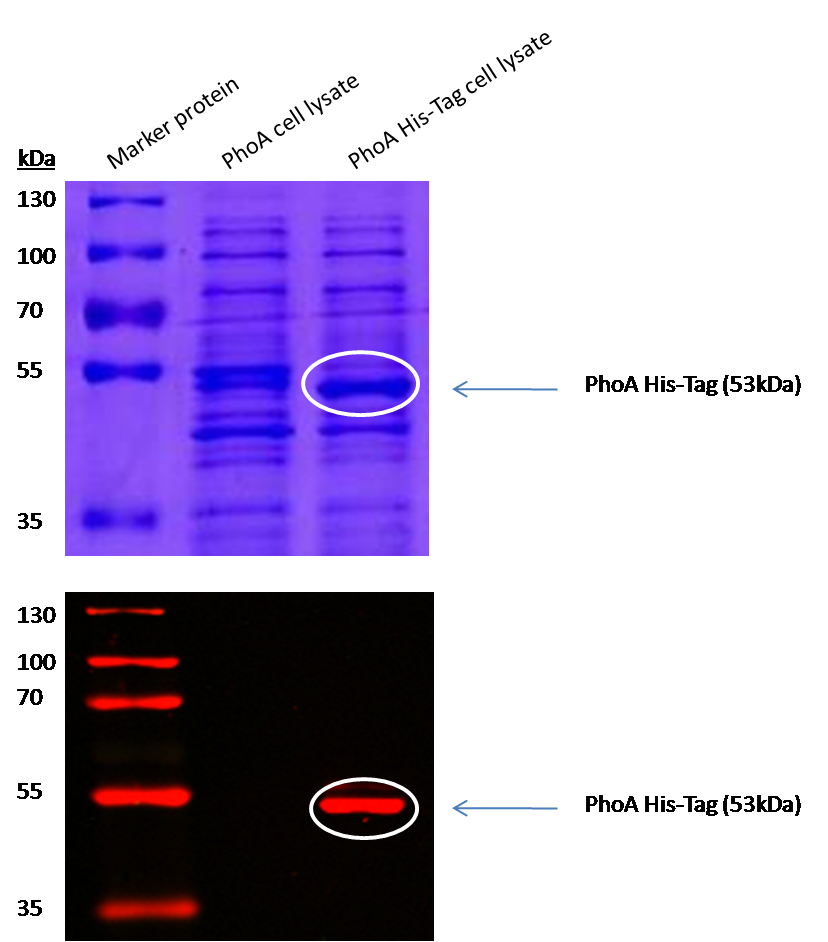
We introduced a HIS tag to the PhoA enzyme. The HIS tag enables to purify the enzyme and therby e.g. making a full Michalis Menten studies which can give exact values for the model. Adding of a HIS tag can lead to an unfunctional enzyme, so we first tested the functionality of both PhoA and PhoA HIS tag (see Figure 10) with our chromogenic substrate. In a second step, we confirmed the presents of the HIS tag by Western Blot: The PhoA and PhoA with HIS-tag were overexpressed in the Escherichia coli BL21 DE3 strain, induced by 5 μM IPTG (iso-propy-β-D-1-thiogalactopyranoside) and finally harvested in order to obtain the cell lysate by lysozyme lysis. This cell lysate of both cultures were analyzed next to each other in two SDS-PAGE gels, one for comassie staining (blue gel in the picture) and one for western blotting (black picture) with Anti-6X His tag® antibody from mouse and a second reporter goat anti mouse antibody with a IRDye 680RD. In the picture we can distinguish the PhoA His (53 kDa) on the western blot as well as on the SDS-PAGE gel (see white circles).
Enzyme kinetics
For the kinetics assay, we tested both the Citrobacter phoA and phoA-HIS encoded proteins with the fluorescent substrate 4-MU-phosphate. The image in Figure 16 was taken with a common single lens reflex camera mounted on a dark hood at λEx 365 nm.
To characterize the enzymes, we conducted fluorometric assays to obtain Km values. Escherichia coli were harvested and lysed, and the cell free extract (CFX) was then collected for the fluorometric assay. The properly diluted CFX was measured on a 96 well plate in triplicates per substrate concentration. The plate reader took measurements at λEx 365 nm and λEm 445 nm, determining a Km values of 105.9 ± 5.3 μM for PhoA and 72.0 ± 7.3 μM for Pho-HIS.
The obtained data was evaluated and finally fitted to Michaelis-Menten-Kinetics with SigmaPlot™:
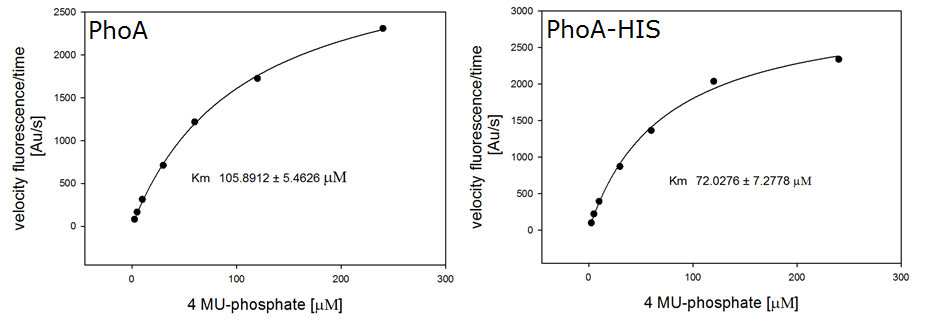
β-Galactosidase (LacZ)
Chromogenic assay
The β-Galactosidase is natively expressed in the triple knockout Escherichia coli strain used in Colisweeper. Its commonly used chromogenic substrate is 5-Bromo-4-Chloro-3-indolyl-β-D-Galactopyranoside (X-Gal), which yields a blue-colored precipitate (Figure 19, middle). In Colisweeper, this protein catalyzes hydrolysis of the flagging substrate, which is N-Methyl-3-indolyl-β-D-Galactopyranoside (Green-β-D-Gal) and produces a green color upon cleavage of the chromophore (Figure 19, left tube). For the color assay in liquid culuture, we used Escherichia coli cultures grown to an OD600 of 0.4 - 0.6, then added the substrates to a final concentration of 1 mM. On colonies, drops of 50 mM were applied, and development of color showed approximately 10 - 15 minutes after addition of the substrates.
β-Glucuronidase (GusA)
Chromogenic assay
In the Colisweeper game, GusA is expressed in colonies which are surrounded by one mine. To characterize the reaction of GusA with its substrate, a liquid culture of the triple knockout Escherichia coli strain overexpressing GusA ([http://parts.igem.org/Part:BBa_K1216000 BBa_K1216000]) was grown until an OD600 of 0.4 - 0.6 was reached before addition of 6-Chloro-3-indolyl-β-glucuronide (Salmon-Gluc), a substrate for GusA which produces a rose/salmon color after cleavage (Figure 22). The final concentration of the substrate in liquid culture was 1 mM and the conversion of this substrate was done by incubation at 37°C. On colonies, 1.5 μl drops of the diluted substrate solution were added, resulting in production of salmon color after approximately 5 minutes at RT.
Enzyme kinetics
For the kinetics assay, we tested both gusA and gusA-HIS encoded proteins with the fluorescent substrate 4-MU-β-D-Glucuronide. The picture on the right (Figure 25) was taken with a common single lens reflex camera mounted on a dark hood at λEx 365 nm.
For the enzyme kinetics of GusA, we conducted fluorometric assays to obtain the Km value. Escherichia coli overexpressing GusA were harvested and lysed, and the cell free extract (CFX) was then collected for the fluorometric assay. The properly diluted CFX was measured on a 96 well plate in triplicates per substrate concentration. A plate reader took measurements at λEx 365 nm and λEm 445 nm, and determined a Km value of 141.1 ± 5.3 μM for GusA, and 201.9 ± 51.1 μM for GusA-HIS. The obtained data was evaluated and finally fitted to Michaelis-Menten-Kinetics with SigmaPlot™:
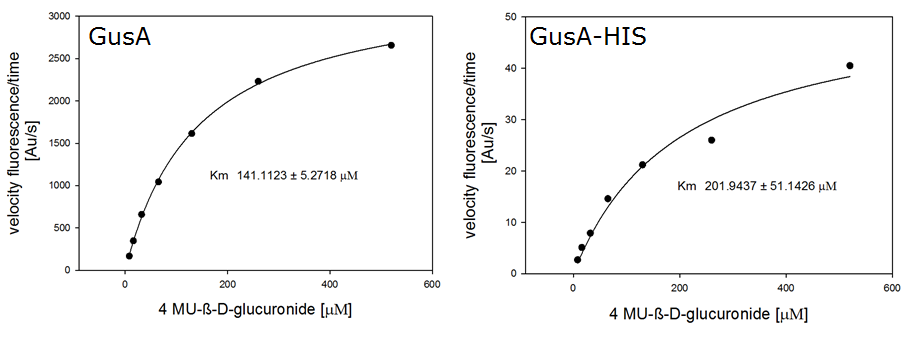
β-N-Acetylglucosaminidase (NagZ)
Chromogenic assay
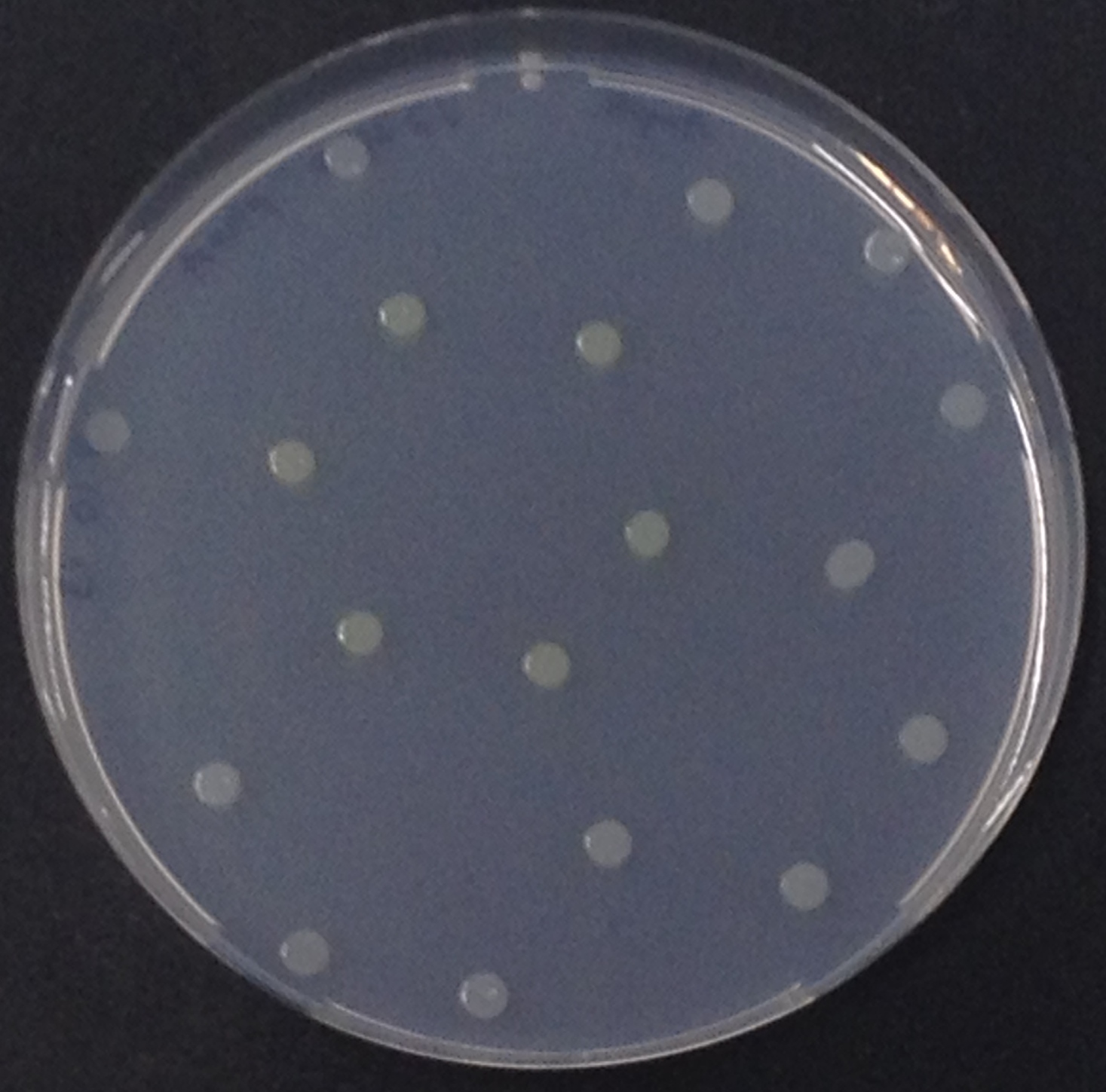
NagZ ([http://parts.igem.org/Part:BBa_K1216003 BBa_K1216003]) was proposed to be constitutively expressed in the mine colonies, processing its blue color-yielding substrate 5-Bromo-4-Chloro-3-indolyl-β-N-acetylglucosaminide (X-GluNAc). However, we could not initially manage to get color from the reaction with this enzyme-substrate pair. Using p-nitrophenyl-β-N-acetylglucosaminide (pNP-GluNAc) at an end concentration of 0.01 mM in an Escherichia coli liquid culture with an OD600 of 0.4 - 0.6, development of yellow color could be observed (Figure 28), confirming that NagZ is expressed in our mine cells.
According to Orenga et al. (1), the substrate incorporating the indoxyl chromophore is less preferentially hydrolyzed by bacterial NagZ in comparison to yeast NagZ, and more preferentially hydrolyzes the alizarine-based substrate they applied. To increase hydrolysis of the indoxyl-based substrate, we added the substrate to a liquid culture supplemented with BSA for stabilization of the enzyme (2), which enhanced hydrolysis by NagZ dramatically. The end concentration in liquid culture of both X-GluNAc and 5-Bromo-6-Chloro-3-indolyl-β-N-acetylglucosaminide (Magenta-GluNAc), a magenta color-yielding substrate for NagZ, were at 1 mM (Figure 28).
Crosstalk
To ensure specificity of the enzyme-substrate pairs used in Colisweeper, a crosstalk test was done to make sure that all overexpressed enzymes specifically cleave their assigned substrate.
This crosstalk test was done in a 96-well plate, each well containing 200 μl from liquid cultures of our ΔaesΔgusAΔnagZ Escherichia coli strain overexpressing either Aes, GusA, NagZ or none, each distributed among the column-wells of the plate. Horizontally, the chromogenic substrates were pipetted to the liquid cultures in the same order as their corresponding hydrolase: magenta-butyrate for Aes, Salmon-Gluc for GusA, and magenta-, X- and pNP-GluNAc for NagZ. If specificity of the chosen enzyme-substrates pairs were given, we would expect an output as shown in the figure below (Figure 32). As Figure 31 shows, the overexpressed hydrolases cleave only the substrates they were expected to.
Color overlay

To play Colisweeper, colors for each colony identity have to be clearly distinguishable by eye. Because we are using high-pass filters to differentially process certain AHL levels, we had to make sure that a mix of cleaved product would show distinguishable colors.
For this purpose, we mixed liquid cultures of our ΔaesΔgusAΔnagZ triple knockout strain which overexpress PhoA, GusA, Aes or NagZ, and added the corresponding substrates to the cultures. Figure 33 shows the resulting colors after hydrolysis of the substrates: PhoA with pNPP in (1), PhoA with pNPP and GusA with Salmon-Gluc in (2), PhoA with pNPP, GusA with Salmon-Gluc and Aes with Magenta butyrate in (3), and PhoA with pNPP and NagZ with X-GluNAc in (4). These colors are clearly distinguishable from one another and therefore can be used together as reporters of a system incorporating high-pass filters.
References
(1) Orenga S, Roger-Dalbert C, James A, Perry J, US Patent 20090017481 (2009).
(2) Magnelli PE, Bielik A, Guthrie E, Methods Mol. Biol., 801, 189-211 (2011).
Absorption: Biosynth
 "
"