Team:Evry/Modelmeta1
From 2013.igem.org
Sensor Model
Introduction
In order to determine our Iron Coli's enterobactin production rate, we first have to know how much time our bacteria will take to sense the ambient iron. So, this first part of the Enterobactin production model focuses on the synthetic sensing system our team implemented in the bacteria.
Observations
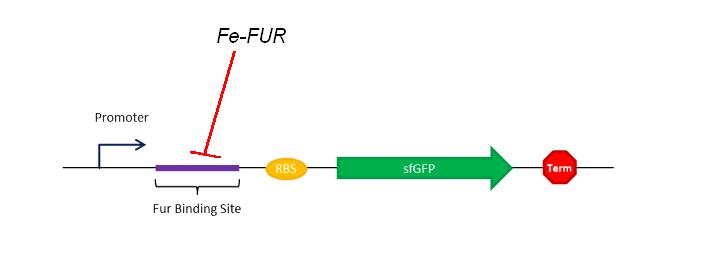
As shown on the Figure 1, our sensing system relies on a FUR Binding Site (FBS). In order to easily model the sensing delay and analyse its results, the first construction is composed of a GFP placed right after the FBS.
Goals
Our goal in this part of the model is to create a generic FBS-related sensing model so that:
- We can determine the iron-sensing delay of our bacteria
- The model can can be reused by other iron related projects
Materials and methods

From Iron to FBS:
The iron-FUR complex is simply formed that way:

We reduced this equation to:

Which is not annoying, since we just have to divide our [FeFur] by two to get the real complex concentration.
We can easily write down both the formation (v) and the dissociation (v') speeds:

We chose to model the iron input in the bacteria using a linear function of the external iron concentration Ferext, the factor p being the cell-wall permeability for iron.
The FUR on the other hand is produced by the bacteria. Its evolution can also be considered as linear, using a mean production rate of Fur0.


In this model, we only track the free Fe-FUR complex and not those attached to a FUR Binding Site. FBS is the number of inhibited Fur Binding Sites.

GFP expression:
To simulate the inhibition phenomenon, we chose to use our Logistic function under its differential form. Since it is the Fe-FUR that represses it, the LacI can be expressed as a logistic fuction of the Fe-FUR:

Ki1 is a non-dimensional parameter that repesents the inhibition power, and Kf is the fixation rate of the Fe-FUR on the FBS. Finally, Npla1 is the number of pasmids containing the GFP.
GFP Production:
The [mRNA] and [GFP] equations are alike. The prodction rates are Kr for the mRNA and Kp for the GFP. Since FBS represents the number of inhibited Fur Binding Sites, we have to substract it from Npla1.
Both variables also have a negative degadation term:

Tuning the model
The only parameter we couldn't find in the litterature is Ki1. Nevertheless, thanks to our construction and the experimental results we obtained with a plate reader, we can tune this parameter to obtain a realistic sensing model!
The Figure 2 is a set of experimental results showing the sensing efficiency for different iron concentrations. We can notice a substantial behaviour change between 10-4M and 10-5M of iron : the activity of our bacteria leaps at 10-5M.
Therefore, we ran simulations with different Ki1 values to try to determine the most fitting Ki1.
The Figure 3 shows that our model can lead to 3 different leaps, depending on the Ki1 value. The leap we are interested in is the 10-4M -> 10-5M leap.
We thus zoomed in, and performed the simulations with a more adapted range of Ki1.
The Figure 4 shows a more precise graph. It is with those simulation values that we calculated the most realistic Ki1, by maximizing the leap it creates.
Finally, we set Ki1 at 6.3*10-5.
Results
Conclusion
Models and scripts
References:
 "
"