Team:INSA Toulouse/contenu/project/biological construction/output
From 2013.igem.org
| (3 intermediate revisions not shown) | |||
| Line 80: | Line 80: | ||
<p class="texte">The output needed to be a signal that can be easily seen without any complicated device or apparatus, something visual like the color of the organism bearing the full adder. | <p class="texte">The output needed to be a signal that can be easily seen without any complicated device or apparatus, something visual like the color of the organism bearing the full adder. | ||
| - | <br> <br> | + | |
| + | |||
| + | |||
| + | |||
| + | |||
| + | <br> <br>We decided to use the RFP (Red Fluorescent Protein) for the following reasons: | ||
<div class="list"> | <div class="list"> | ||
<ul class="circlearrow"> | <ul class="circlearrow"> | ||
| - | <li>RFP is a pigment that can be measured even with low expression levels. The RFP protein was | + | <li>RFP is a pigment that can be measured even with low expression levels. The RFP protein was engineered from the classical GFP to have a different color and a strong fluorescence quantum yield. The fluorescence detection can be performed with UV or visible light. |
</li> | </li> | ||
| - | </div> | + | </div> |
<div class="list"> | <div class="list"> | ||
<ul class="circlearrow"> | <ul class="circlearrow"> | ||
| - | <li>The RFP expression is not really toxic and the pigment | + | <li>The RFP expression is not really toxic and the pigment can be easily accumulated in the cell. Even with low expression, accumulation could still lead to visible phenotype. |
</li> | </li> | ||
</div> | </div> | ||
<div class="list"> | <div class="list"> | ||
<ul class="circlearrow"> | <ul class="circlearrow"> | ||
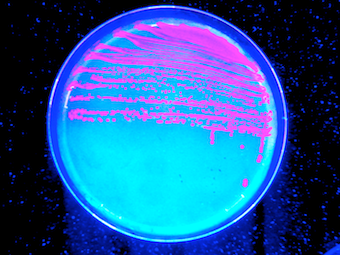
| - | <li>We love the color of RFP expressing cells on a petri dish under UV light ;) | + | <li>We love the color of RFP expressing cells on a petri dish under UV light ;-) |
</li> | </li> | ||
</div> | </div> | ||
| Line 101: | Line 106: | ||
<img src="https://static.igem.org/mediawiki/2013/2/29/RFP-UV-340px.png" class="imgcontent" /> | <img src="https://static.igem.org/mediawiki/2013/2/29/RFP-UV-340px.png" class="imgcontent" /> | ||
| + | <p class="texteleft"> <i>General principle</i> | ||
| + | <br>The gene encoding the RFP protein was directly used as the reporter system. It was placed directly under the control of the different promoters belonging to the logic gates. | ||
| + | </p> | ||
| + | |||
| + | <p class="texteright"> <i>Output module characterization </i> | ||
| + | <br>As simple as it seems. Colonies are red or white. Visual inspection of the clones is the first criteria. When time was really limiting, we also used the UV light and a camera to distinguish between expressing and non expressing bacteria. For quantitative characterization, we used a fluorimeter. | ||
| + | </p> | ||
Latest revision as of 16:24, 4 October 2013
Biological Modules: Output
The output needed to be a signal that can be easily seen without any complicated device or apparatus, something visual like the color of the organism bearing the full adder.
We decided to use the RFP (Red Fluorescent Protein) for the following reasons:
- RFP is a pigment that can be measured even with low expression levels. The RFP protein was engineered from the classical GFP to have a different color and a strong fluorescence quantum yield. The fluorescence detection can be performed with UV or visible light.
- The RFP expression is not really toxic and the pigment can be easily accumulated in the cell. Even with low expression, accumulation could still lead to visible phenotype.
- We love the color of RFP expressing cells on a petri dish under UV light ;-)
General principle
The gene encoding the RFP protein was directly used as the reporter system. It was placed directly under the control of the different promoters belonging to the logic gates.
Output module characterization
As simple as it seems. Colonies are red or white. Visual inspection of the clones is the first criteria. When time was really limiting, we also used the UV light and a camera to distinguish between expressing and non expressing bacteria. For quantitative characterization, we used a fluorimeter.
 "
"

