Team:Leeds/Results
From 2013.igem.org
Results from the lab so far. Please note, many of the images have been uploaded at high resolution, and are best viewed from their file page. Access this by clicking the image. We may later introduce a gallery of the images to maximise the screen usage, but this is dependent upon Coffee-Induced coding by the Code Monkey.
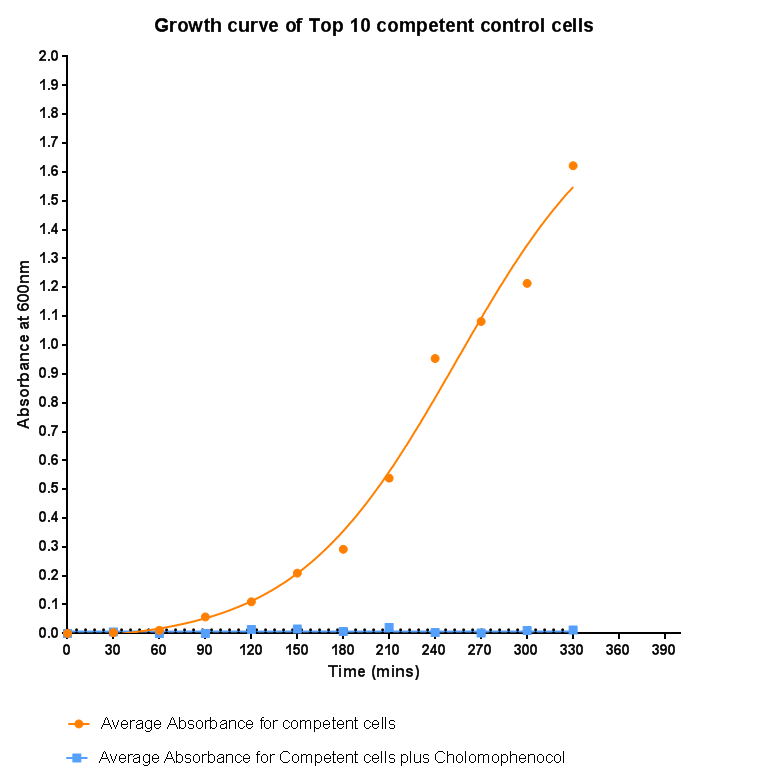
Bacterial growth curvesWe are monitoring the growth of our bacteria containing our devices to see if the genes we inserted have any effect on bacterial growth. Control Bacterial Growth CurveIn this experiment we used our chosen expression cells and monitored their growth on SB media both in the presence of chloramphenicol and without chloramphenicol. As the bacteria do not contain our plasmid that codes for chloramphenicol resistance, we didn't expect them to grow when it was present. The graph produced from the absorbance measurements taken over time, shows the growth of the bacteria. It has a clear lag and log phase when the bacteria are grown in SB media only. But as expected, the graph shows that the bacteria do not grow in the presence of chloramphenicol. This graph will be used as a control and other growth curves of bacteria containing our device will be compared to it in order to determine whether the genes we inserted cause an effect on cell growth.
Characterisation of Device 1
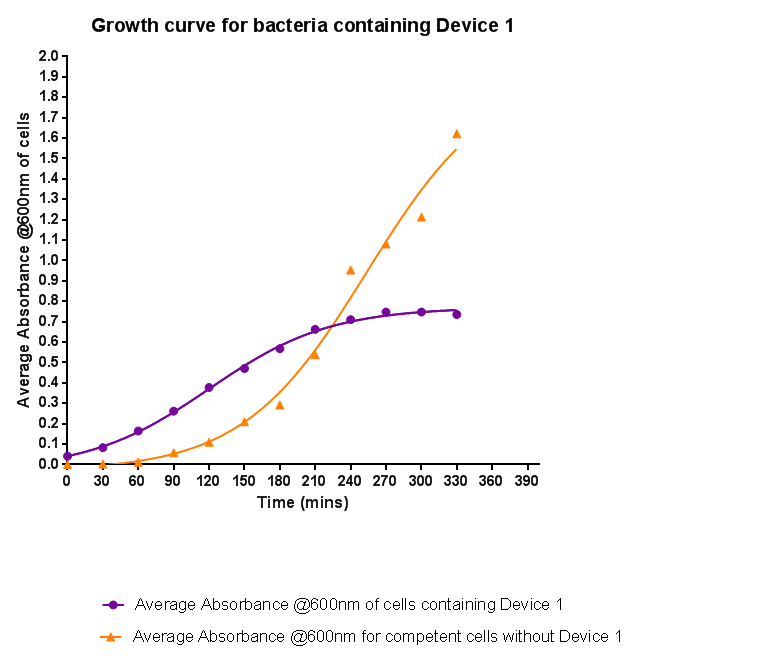
Growth Curve
Bacteria were grown which contained Device 1; their absorbance was monitored over time and then the rate of growth compared to bacteria that do not contain Device 1. The graph clearly shows that inserting Device 1 into cells considerably slows their growth compared with the growth of cells without Device 1. However the graph still shows a clear lag and log phase for both growths, the difference being that bacteria containing Device 1 don't grow to as high optical density in log phase as the bacteria without Device 1.
Restriction Digests
Plate Imaging
Membrane Stress - Hydrophobic Stress
Membrane Stress - Changes in pH
Membrane Stress- Atomic Force Microscopy
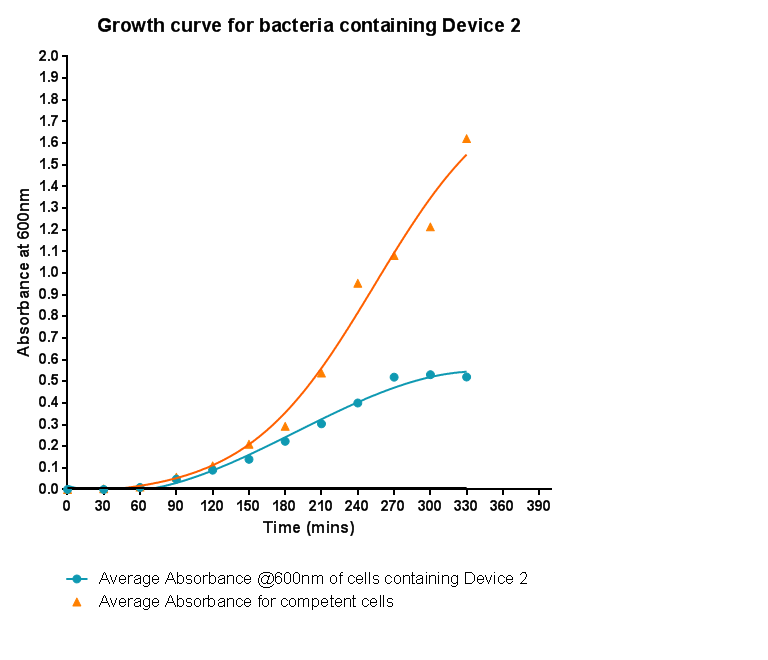
Characterisation of Device 2Growth CurveHere the growth of cells containing device 2 was monitored via absorbance and compared to the growth of cells that do not contain device 2. As was seen when device 1 was inserted into the cells, the growth is slowed down considerably. Although the graph shows a log and a lag phase for the cells containing Device 2, the absorbance reached in log phase is very low compared to cells without Device 2; it is even lower than the absorbance reached for cells containing Device 1.
Restriction Digests
Plate Imaging
Binding assay-washing beads
Binding assay-Confocal Microscopy
Characterisation of Device 3
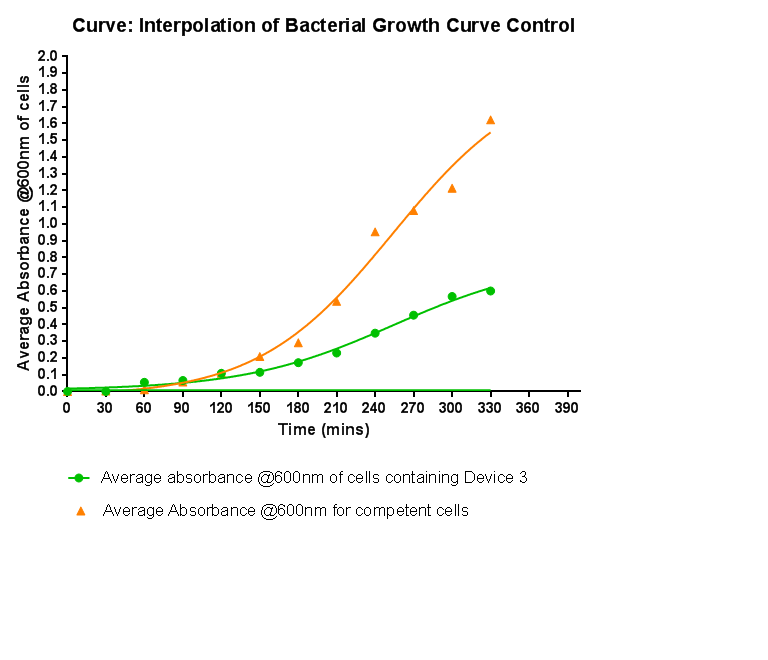
Growth CurveThis graph shows the results of monitoring the growth of cells with contain Device 3. As with cells containing Device 1 and Device 2, the growth of the cells is considerably slowed down when compared with cells that do not contain any device at all. A slight change in the gradient of the graph can be seen showing the transition of the cell growth from lag to log phase, so the cells are still growing normally, but the insertion of Device 3 is restricting the growth of the cells.
Restriction Digests
Plate Imaging
Binding Assay-Beads cause fluorescence?
| |||||||
 |
| ||||||

| |||||||

| |||||||
 "
"