Team:TU-Delft/Sensing
From 2013.igem.org
Jfkooijman (Talk | contribs) |
|||
| (36 intermediate revisions not shown) | |||
| Line 6: | Line 6: | ||
| - | <div style="margin-left:30px;margin-right:30px; width:900px;float:left;"> | + | <div style="margin-left:30px;margin-right:30px;margin-top:10px; width:900px;float:left;"> |
<h2 align="center">Sensing Device</h2> | <h2 align="center">Sensing Device</h2> | ||
<html> | <html> | ||
<center> | <center> | ||
| - | <a href="https://static.igem.org/mediawiki/2013/ | + | <a href="https://static.igem.org/mediawiki/2013/f/fa/PBADRecGFP.jpg" target="_blank"><img src="https://static.igem.org/mediawiki/2013/f/fa/PBADRecGFP.jpg"width="500px" height="354px"/></a> |
</center> | </center> | ||
<center> | <center> | ||
| Line 18: | Line 18: | ||
<div style="margin-left:30px;margin-right:30px; width:900px;float:left;"> | <div style="margin-left:30px;margin-right:30px; width:900px;float:left;"> | ||
<p align="justify"> | <p align="justify"> | ||
| - | In bacteria quorum sensing is an important mechanism for monitoring the state of the population. <i>S. aureus</i> uses its quorum sensing system to regulate a large part of its virulence genes, as having a large population is beneficial for <i>S. aureus</i> during infection. In this project we | + | In bacteria quorum sensing is an important mechanism for monitoring the state of the population. <i>S. aureus</i> uses its quorum sensing system to regulate a large part of its virulence genes, as having a large population is beneficial for <i>S. aureus</i> during infection <a href="https://2013.igem.org/Team:TU-Delft/Sensing#references">[1]</a>. In this project we hijack the native <i>S. aureus</i> system and implement that in the antimicrobial peptide expression host <i>E.coli</i>. |
</p> | </p> | ||
<p align="justify"> | <p align="justify"> | ||
| Line 24: | Line 24: | ||
</p> | </p> | ||
<p align="justify"> | <p align="justify"> | ||
| - | The system is composed out of the transmembrane receptor protein AgrC and the cytoplasmic protein AgrA. AgrA is phosphorylated by AgrC upon induction with auto-inducing peptide (AIP), after which it will act as a transcription factor positively acting on the promoter P2. AIP is made from the precursor peptide AgrD, which is circularized and secreted by AgrB (Figure 1). | + | The system is composed out of the transmembrane receptor protein AgrC and the cytoplasmic protein AgrA. AgrA is phosphorylated by AgrC upon induction with auto-inducing peptide (AIP), after which it will act as a transcription factor positively acting on the promoter P2. AIP is made from the precursor peptide AgrD, which is circularized and secreted by AgrB (Figure 1). In the final construct used in Peptidor, pP2 will not be in front of GFP, but in front of the SUMO-peptides as explained <a href="https://2013.igem.org/Team:TU-Delft/Peptides" target="blank">here</a>. |
</p> | </p> | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
<br> | <br> | ||
<h4>AIP Sensing Experiment: </h4> | <h4>AIP Sensing Experiment: </h4> | ||
<p align="justify"> | <p align="justify"> | ||
| - | The construct <a href="http://parts.igem.org/Part:BBa_K1022100" style="text-decoration: none"" target="_blank">BBa_K1022100</a> | + | The construct <a href="http://parts.igem.org/Part:BBa_K1022100" style="text-decoration: none"" target="_blank">BBa_K1022100</a> ,pBAD AIP Receiver GFP, (fig. 2) is the test construct for the first module of the project in order to sense the auto-inducing peptides (AIPs) produced by <i>S. aureus</i>. |
</p> | </p> | ||
<h4>Diagram of the Part</h4> | <h4>Diagram of the Part</h4> | ||
| - | In this sensor test-construct AgrC and AgrA are expressed, after which AgrC can act as a receptor for AIP. After binding AIP, AgrC will phosphorylate AgrA which then starts acting as a postitive transcription factor for pP2 <a href="https://2013.igem.org/Team:TU-Delft/Sensing#references">[ | + | In this sensor test-construct AgrC and AgrA are expressed, after which AgrC can act as a receptor for AIP. After binding AIP, AgrC will phosphorylate AgrA which then starts acting as a postitive transcription factor for pP2 <a href="https://2013.igem.org/Team:TU-Delft/Sensing#references">[2]</a>. Therefore, upon induction by AIP, the construct will give a GFP signal. This is tested using the following experiment. |
| + | |||
| + | <center> | ||
| + | <img src="https://static.igem.org/mediawiki/2013/3/38/Part100.jpg"> | ||
| + | <p>Figure 2: Part:BBa_K1022100 </p> | ||
| + | </center> | ||
| + | |||
| + | |||
<br><br> | <br><br> | ||
<h4>Experimental Set Up</h4> | <h4>Experimental Set Up</h4> | ||
<p align="justify"> | <p align="justify"> | ||
| - | Cells are induced with 0.1 % Arabinose (to induce pBAD) and | + | Cells are induced with 0.1 % Arabinose (to induce pBAD) and AIP, and with only AIP. AIP Induction is done at O.D 0.5 with 10 µM AIP. 3 hours after induction fluorescence-activated cell sorting (FACS) is used to check for GFP signal. The controls are BL21(DE3) cells, AIP receiver GFP containing BL21(DE3)without any induction and a strain constitutively expressing GFP. The AIP used to induce the cells was synthesized by Eurogentec. |
</p> | </p> | ||
| + | |||
| + | <a href="https://2013.igem.org/Team:TU-Delft/Protocol_13#protocol_13" target="_blank"><i>Full protocol</i></a> | ||
<p align="justify"> | <p align="justify"> | ||
| - | The cells induced with arabinose all died, which could be seen by the fact the signal given by the FACS, | + | The cells induced with arabinose all died, which could be seen by the fact the signal given by the FACS. FACS gives a fluorescent signal for every cell passing the reader, live cells give a higher signal than cell debris. From the fact the arabinose induced cells showed a lower signal than the BL21(DE3) control we concluded these cells were dead. This cell lysis could be due the fact AgrA and/or AgrC is toxic in high concentrations. The detection of AIP by <i>E. coli</i> without induction of the Agr genes is explainable due the fact pBAD is a bit leaky, making low levels of transcription possible. Apparently only a small amount of activated AgrA is required to give a measurable level of GFP. |
</p> | </p> | ||
<p align="justify"> | <p align="justify"> | ||
| - | Earlier work on the detection of AIPs by Gram-negatives has been done, but with unclear or non-convincing results <a href="https://2013.igem.org/Team:TU-Delft/Sensing#references">[ | + | Earlier work on the detection of AIPs by Gram-negatives has been done, but with unclear or non-convincing results <a href="https://2013.igem.org/Team:TU-Delft/Sensing#references">[3,4]</a>. Often the outer-membrane of E. coli was seen as an obstacle for the implementation of auto-inducing peptide sensing systems in Gram negatives. In the AIP sensing experiments preformed by the iGEM Cambridge 2007 team a FepA construct was used as this is known to make the outer membrane more permeable<a href="https://2013.igem.org/Team:TU-Delft/Sensing#references">[8]</a>. Although, after intensive literature research we found a molecule very similar to the AIP molecule used in these experiments, that is known to pass the outer membrane. The molecule bacitracin [5] has been shown to interfere with cell wall synthesis in Gram negative species, therefore being able to pass the outer membrane as cell wall synthesis occurs in the periplasm. Comparison of the molecular structures of bacitracin and AIP regarding size, molecular weight, polarity and charge (fig. 3) gave us an indication AIP should be possible to pass through the outer membrane of <i>E. coli</i> without any modifications done on the outer membrane. |
</p> | </p> | ||
| + | |||
| + | <center> | ||
| + | <img src="https://static.igem.org/mediawiki/2013/1/13/BacitracinAip.png"> | ||
| + | <p >Figure 3: Molecule structures of bacitracin and autoinducing peptides </p> | ||
| + | </center> | ||
| + | |||
<h4>Results</h4> | <h4>Results</h4> | ||
<p align="justify"> | <p align="justify"> | ||
| - | Using Cyflogic 1.2.1 software <a href="https://2013.igem.org/Team:TU-Delft/Sensing#references">[ | + | Using Cyflogic 1.2.1 software <a href="https://2013.igem.org/Team:TU-Delft/Sensing#references">[6]</a>, FACS data was analysed, showing a clear induction of the AIP receiver GFP construct by AIP. The difference between the blue and the red line (218) indicates the difference in fluorescent signal of uninduced and induced cells. Untransformed Bl21(DE3) and constitutively expressed GFP were used as respectively negative and positive control (fig. 4). The significant difference between the uninduced and the induced samples is determined with a P-value >10<sup>-4</sup> as described in the next paragraph. |
</p> | </p> | ||
| Line 65: | Line 72: | ||
<img src="https://static.igem.org/mediawiki/2013/f/f0/FinalSensing.jpg"> | <img src="https://static.igem.org/mediawiki/2013/f/f0/FinalSensing.jpg"> | ||
<p align="justify"> | <p align="justify"> | ||
| - | + | Figure 4: Fluorescent signal measured with the FACS. The white line refers to the control BL21(DE3) cells. In blue the uninduced pBAD AIP receiver GFP is shown. The red line represents the AIP induced receiver cells. The green line shows cells constitutively expressing GFP. The fluorescence intensity scale is logarithmic and the total number of cells counted per sample is 10,000. | |
| + | </p> | ||
| + | </center> | ||
| + | <br> | ||
| + | <h4>Significance test</h4> | ||
| + | <p align="justify"> | ||
| + | To test if the difference between the uninduced and induced result is significant a statistic test is performed as in <a href="https://2013.igem.org/Team:TU-Delft/Sensing#references">[7]</a>. Here it is assumed that both results are normal distributions, as the number of datasets is large (central limit theorem) <a href="https://2013.igem.org/Team:TU-Delft/Sensing#references">[7]</a>. Then the critical value equation becomes a simple one: | ||
| + | </p> | ||
| + | <center> | ||
| + | <img src="https://static.igem.org/mediawiki/2013/a/a2/Eq_stat.jpg" width="250"> | ||
| + | <p align="justify"> | ||
| + | Based on the induced and uninduced data as in Table 1, this t-value is calculated as 52.7. This is a very high value and corresponds to a one-sided p value of less than 10<sup>-4</sup>. This means that there is a significant difference between the induced and uninduced curves. | ||
</p> | </p> | ||
</center> | </center> | ||
| + | |||
| + | |||
| + | <style type="text/css"> | ||
| + | .CSSTableGenerator { | ||
| + | margin:0px;padding:0px; | ||
| + | width:100%; | ||
| + | box-shadow: 10px 10px 5px #888888; | ||
| + | border:1px solid #000000; | ||
| + | |||
| + | -moz-border-radius-bottomleft:0px; | ||
| + | -webkit-border-bottom-left-radius:0px; | ||
| + | border-bottom-left-radius:0px; | ||
| + | |||
| + | -moz-border-radius-bottomright:0px; | ||
| + | -webkit-border-bottom-right-radius:0px; | ||
| + | border-bottom-right-radius:0px; | ||
| + | |||
| + | -moz-border-radius-topright:0px; | ||
| + | -webkit-border-top-right-radius:0px; | ||
| + | border-top-right-radius:0px; | ||
| + | |||
| + | -moz-border-radius-topleft:0px; | ||
| + | -webkit-border-top-left-radius:0px; | ||
| + | border-top-left-radius:0px; | ||
| + | }.CSSTableGenerator table{ | ||
| + | width:100%; | ||
| + | height:100%; | ||
| + | margin:0px;padding:0px; | ||
| + | }.CSSTableGenerator tr:last-child td:last-child { | ||
| + | -moz-border-radius-bottomright:0px; | ||
| + | -webkit-border-bottom-right-radius:0px; | ||
| + | border-bottom-right-radius:0px; | ||
| + | } | ||
| + | .CSSTableGenerator table tr:first-child td:first-child { | ||
| + | -moz-border-radius-topleft:0px; | ||
| + | -webkit-border-top-left-radius:0px; | ||
| + | border-top-left-radius:0px; | ||
| + | } | ||
| + | .CSSTableGenerator table tr:first-child td:last-child { | ||
| + | -moz-border-radius-topright:0px; | ||
| + | -webkit-border-top-right-radius:0px; | ||
| + | border-top-right-radius:0px; | ||
| + | }.CSSTableGenerator tr:last-child td:first-child{ | ||
| + | -moz-border-radius-bottomleft:0px; | ||
| + | -webkit-border-bottom-left-radius:0px; | ||
| + | border-bottom-left-radius:0px; | ||
| + | }.CSSTableGenerator tr:hover td{ | ||
| + | |||
| + | } | ||
| + | .CSSTableGenerator tr:nth-child(odd){ background-color:#ffffff; } | ||
| + | .CSSTableGenerator tr:nth-child(even) { background-color:#ffffff; }.CSSTableGenerator td{ | ||
| + | vertical-align:middle; | ||
| + | |||
| + | |||
| + | border:1px solid #000000; | ||
| + | border-width:0px 1px 1px 0px; | ||
| + | text-align:left; | ||
| + | padding:7px; | ||
| + | font-size:10px; | ||
| + | font-family:Arial; | ||
| + | font-weight:normal; | ||
| + | color:#000000; | ||
| + | }.CSSTableGenerator tr:last-child td{ | ||
| + | border-width:0px 1px 0px 0px; | ||
| + | }.CSSTableGenerator tr td:last-child{ | ||
| + | border-width:0px 0px 1px 0px; | ||
| + | }.CSSTableGenerator tr:last-child td:last-child{ | ||
| + | border-width:0px 0px 0px 0px; | ||
| + | } | ||
| + | .CSSTableGenerator tr:first-child td{ | ||
| + | background:-o-linear-gradient(bottom, #007f7f 5%, #007f7f 100%); background:-webkit-gradient( linear, left top, left bottom, color-stop(0.05, #007f7f), color-stop(1, #007f7f) ); | ||
| + | background:-moz-linear-gradient( center top, #007f7f 5%, #007f7f 100% ); | ||
| + | filter:progid:DXImageTransform.Microsoft.gradient(startColorstr="#007f7f", endColorstr="#007f7f"); background: -o-linear-gradient(top,#007f7f,007f7f); | ||
| + | |||
| + | background-color:#007f7f; | ||
| + | border:0px solid #000000; | ||
| + | text-align:center; | ||
| + | border-width:0px 0px 1px 1px; | ||
| + | font-size:14px; | ||
| + | font-family:Arial; | ||
| + | font-weight:bold; | ||
| + | color:#ffffff; | ||
| + | } | ||
| + | .CSSTableGenerator tr:first-child:hover td{ | ||
| + | background:-o-linear-gradient(bottom, #007f7f 5%, #007f7f 100%); background:-webkit-gradient( linear, left top, left bottom, color-stop(0.05, #007f7f), color-stop(1, #007f7f) ); | ||
| + | background:-moz-linear-gradient( center top, #007f7f 5%, #007f7f 100% ); | ||
| + | filter:progid:DXImageTransform.Microsoft.gradient(startColorstr="#007f7f", endColorstr="#007f7f"); background: -o-linear-gradient(top,#007f7f,007f7f); | ||
| + | |||
| + | background-color:#007f7f; | ||
| + | } | ||
| + | .CSSTableGenerator tr:first-child td:first-child{ | ||
| + | border-width:0px 0px 1px 0px; | ||
| + | } | ||
| + | .CSSTableGenerator tr:first-child td:last-child{ | ||
| + | border-width:0px 0px 1px 1px; | ||
| + | } | ||
| + | </style> | ||
| + | |||
| + | |||
| + | <html> | ||
| + | <center>Table 1: Data of the induced and non-induced FACS </center> | ||
| + | <div class="CSSTableGenerator" > | ||
| + | <table > | ||
| + | <tr> | ||
| + | <td> | ||
| + | Dataset | ||
| + | </td> | ||
| + | <td >Mean (μ)</td> | ||
| + | <td>Standard deviation (σ)</td> | ||
| + | <td>Number of samples (N)</td> | ||
| + | |||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td> | ||
| + | Non-induced | ||
| + | </td> | ||
| + | <td > | ||
| + | 465 | ||
| + | </td> | ||
| + | <td> | ||
| + | 242.6 | ||
| + | </td> | ||
| + | <td>10.000</td> | ||
| + | |||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> | ||
| + | Induced | ||
| + | </td> | ||
| + | <td > | ||
| + | 683 | ||
| + | </td> | ||
| + | <td> | ||
| + | 334.5 | ||
| + | </td> | ||
| + | <td>10.000</td> | ||
| + | |||
| + | </tr> | ||
| + | |||
| + | </table> | ||
| + | </div> | ||
| + | <br> | ||
| + | <h4>Discussion</h4> | ||
| + | <p align="justify"> | ||
| + | Taking the results described above into account, we are able to sense the presence of a Gram positive species with the use of <i>E. coli</i> transformed with our AIP receiver GFP construct. To our knowledge, we are the first to show the interspecies communication between Gram negative and Gram positive bacterial species. In this case the sensing of the AIP of <i>S. aureus</i> by <i>E. coli</i>. For the final application described in the introduction (link) page though, the level of activation of the pP2 promoter by AgrA might be too low. In order to make the sensing system able to activate a peptide production system capable of producing a concentration high enough to kill or inhibit the growth of S. aureus, an amplification circuit could be used. | ||
| + | </p> | ||
<a name="references"></a> | <a name="references"></a> | ||
<h2 align="center">References</h2> | <h2 align="center">References</h2> | ||
<ol> | <ol> | ||
| + | <li>Textbook of bacteriology,available on:<a href="http://textbookofbacteriology.net/staph_2.html" target="blank"> www.textbookofbacteriology</a></li> | ||
<li>Philip Mdowell, Zina Affas, et al.,Structure, activity and evolution of the group I thiolactone peptide quorum-sensing system of Staphylococcus aureus, Molecular Microbiology, Volume 41, Issue 2, pages 503–512, Jul 2001.</li> | <li>Philip Mdowell, Zina Affas, et al.,Structure, activity and evolution of the group I thiolactone peptide quorum-sensing system of Staphylococcus aureus, Molecular Microbiology, Volume 41, Issue 2, pages 503–512, Jul 2001.</li> | ||
<li><a href="https://2007.igem.org/wiki/index.php/Cambridge" target="blank">https://2007.igem.org/wiki/index.php/Cambridge</a></li> | <li><a href="https://2007.igem.org/wiki/index.php/Cambridge" target="blank">https://2007.igem.org/wiki/index.php/Cambridge</a></li> | ||
<li><a href="https://2012.igem.org/Team:HIT-Harbin" target="blank">https://2012.igem.org/Team:HIT-Harbin</a></li> | <li><a href="https://2012.igem.org/Team:HIT-Harbin" target="blank">https://2012.igem.org/Team:HIT-Harbin</a></li> | ||
<li>T.J. Pollock, L. Thorne, Mechanism of bacitracin resistance in gram-negative bacteria that synthesize exopolysaccharides, J Bacteriol ; 176(20): 6229–6237, Oct 1994 </li> | <li>T.J. Pollock, L. Thorne, Mechanism of bacitracin resistance in gram-negative bacteria that synthesize exopolysaccharides, J Bacteriol ; 176(20): 6229–6237, Oct 1994 </li> | ||
| - | <li><a href="http://www.cyflogic.com/" target="blank">http://www.cyflogic.com/</a> | + | <li>Cyflogic 1.2.1., available on:<a href="http://www.cyflogic.com/" target="blank">http://www.cyflogic.com/</a>. |
</li> | </li> | ||
| - | <li>F.M. Dekker, C. Kraaikaamp et al., ‘A Modern Introduction to Probability and Statistics’, January 2005 | + | <li>F.M. Dekker, C. Kraaikaamp et al., ‘A Modern Introduction to Probability and Statistics’, Springer,January 2005.</li> |
| + | <li><a href="https://2007.igem.org/wiki/index.php/Cambridge/Signalling_project#FepA_permeability_device" target="blank">www.2007.igem.org/wiki/cambridge</a></li> | ||
</ol> | </ol> | ||
Latest revision as of 14:18, 4 October 2013

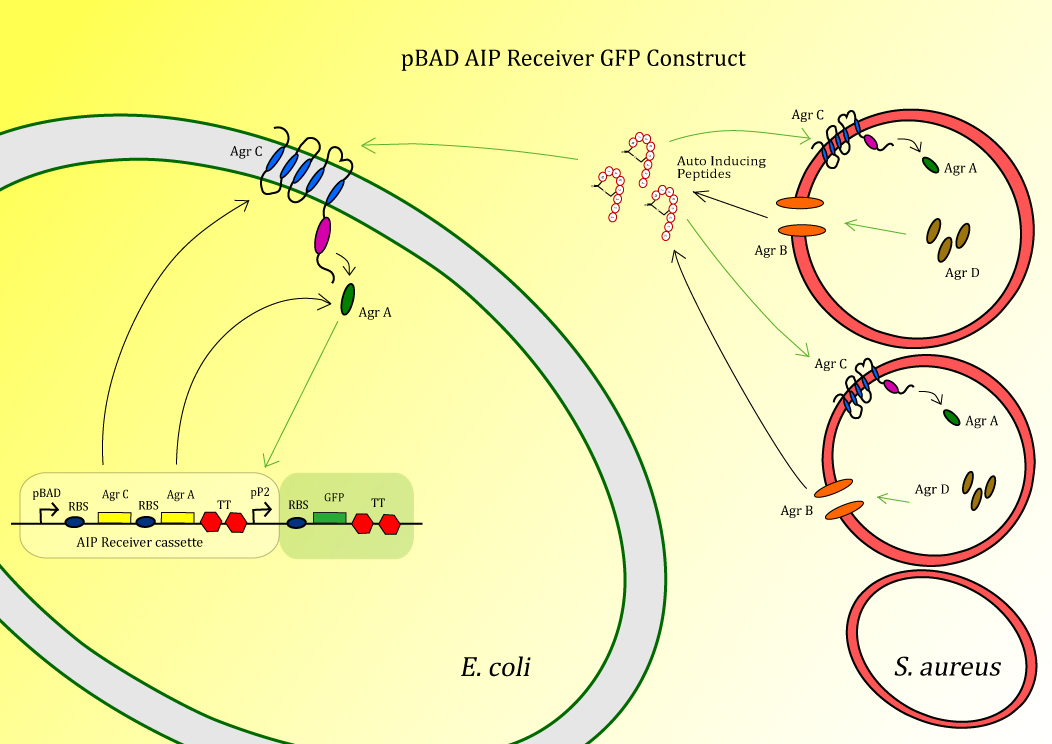
In bacteria quorum sensing is an important mechanism for monitoring the state of the population. S. aureus uses its quorum sensing system to regulate a large part of its virulence genes, as having a large population is beneficial for S. aureus during infection [1]. In this project we hijack the native S. aureus system and implement that in the antimicrobial peptide expression host E.coli.
We used the quorum sensing system of S. aureus in our peptidor in order to make sure the antimicrobial peptides will only be produced when S. aureus is present. This way there is less chance of other microbes becoming resistant against the peptides and therefore indirectly, through horizontal gene transfer, preventing S. aureus becoming resistant.
The system is composed out of the transmembrane receptor protein AgrC and the cytoplasmic protein AgrA. AgrA is phosphorylated by AgrC upon induction with auto-inducing peptide (AIP), after which it will act as a transcription factor positively acting on the promoter P2. AIP is made from the precursor peptide AgrD, which is circularized and secreted by AgrB (Figure 1). In the final construct used in Peptidor, pP2 will not be in front of GFP, but in front of the SUMO-peptides as explained here.
AIP Sensing Experiment:
The construct BBa_K1022100 ,pBAD AIP Receiver GFP, (fig. 2) is the test construct for the first module of the project in order to sense the auto-inducing peptides (AIPs) produced by S. aureus.
Diagram of the Part
In this sensor test-construct AgrC and AgrA are expressed, after which AgrC can act as a receptor for AIP. After binding AIP, AgrC will phosphorylate AgrA which then starts acting as a postitive transcription factor for pP2 [2]. Therefore, upon induction by AIP, the construct will give a GFP signal. This is tested using the following experiment.
Figure 2: Part:BBa_K1022100
Experimental Set Up
Cells are induced with 0.1 % Arabinose (to induce pBAD) and AIP, and with only AIP. AIP Induction is done at O.D 0.5 with 10 µM AIP. 3 hours after induction fluorescence-activated cell sorting (FACS) is used to check for GFP signal. The controls are BL21(DE3) cells, AIP receiver GFP containing BL21(DE3)without any induction and a strain constitutively expressing GFP. The AIP used to induce the cells was synthesized by Eurogentec.
Full protocolThe cells induced with arabinose all died, which could be seen by the fact the signal given by the FACS. FACS gives a fluorescent signal for every cell passing the reader, live cells give a higher signal than cell debris. From the fact the arabinose induced cells showed a lower signal than the BL21(DE3) control we concluded these cells were dead. This cell lysis could be due the fact AgrA and/or AgrC is toxic in high concentrations. The detection of AIP by E. coli without induction of the Agr genes is explainable due the fact pBAD is a bit leaky, making low levels of transcription possible. Apparently only a small amount of activated AgrA is required to give a measurable level of GFP.
Earlier work on the detection of AIPs by Gram-negatives has been done, but with unclear or non-convincing results [3,4]. Often the outer-membrane of E. coli was seen as an obstacle for the implementation of auto-inducing peptide sensing systems in Gram negatives. In the AIP sensing experiments preformed by the iGEM Cambridge 2007 team a FepA construct was used as this is known to make the outer membrane more permeable[8]. Although, after intensive literature research we found a molecule very similar to the AIP molecule used in these experiments, that is known to pass the outer membrane. The molecule bacitracin [5] has been shown to interfere with cell wall synthesis in Gram negative species, therefore being able to pass the outer membrane as cell wall synthesis occurs in the periplasm. Comparison of the molecular structures of bacitracin and AIP regarding size, molecular weight, polarity and charge (fig. 3) gave us an indication AIP should be possible to pass through the outer membrane of E. coli without any modifications done on the outer membrane.

Figure 3: Molecule structures of bacitracin and autoinducing peptides
Results
Using Cyflogic 1.2.1 software [6], FACS data was analysed, showing a clear induction of the AIP receiver GFP construct by AIP. The difference between the blue and the red line (218) indicates the difference in fluorescent signal of uninduced and induced cells. Untransformed Bl21(DE3) and constitutively expressed GFP were used as respectively negative and positive control (fig. 4). The significant difference between the uninduced and the induced samples is determined with a P-value >10-4 as described in the next paragraph.

Figure 4: Fluorescent signal measured with the FACS. The white line refers to the control BL21(DE3) cells. In blue the uninduced pBAD AIP receiver GFP is shown. The red line represents the AIP induced receiver cells. The green line shows cells constitutively expressing GFP. The fluorescence intensity scale is logarithmic and the total number of cells counted per sample is 10,000.
Significance test
To test if the difference between the uninduced and induced result is significant a statistic test is performed as in [7]. Here it is assumed that both results are normal distributions, as the number of datasets is large (central limit theorem) [7]. Then the critical value equation becomes a simple one:

Based on the induced and uninduced data as in Table 1, this t-value is calculated as 52.7. This is a very high value and corresponds to a one-sided p value of less than 10-4. This means that there is a significant difference between the induced and uninduced curves.
| Dataset | Mean (μ) | Standard deviation (σ) | Number of samples (N) |
| Non-induced | 465 | 242.6 | 10.000 |
| Induced | 683 | 334.5 | 10.000 |
Discussion
Taking the results described above into account, we are able to sense the presence of a Gram positive species with the use of E. coli transformed with our AIP receiver GFP construct. To our knowledge, we are the first to show the interspecies communication between Gram negative and Gram positive bacterial species. In this case the sensing of the AIP of S. aureus by E. coli. For the final application described in the introduction (link) page though, the level of activation of the pP2 promoter by AgrA might be too low. In order to make the sensing system able to activate a peptide production system capable of producing a concentration high enough to kill or inhibit the growth of S. aureus, an amplification circuit could be used.
References
- Textbook of bacteriology,available on: www.textbookofbacteriology
- Philip Mdowell, Zina Affas, et al.,Structure, activity and evolution of the group I thiolactone peptide quorum-sensing system of Staphylococcus aureus, Molecular Microbiology, Volume 41, Issue 2, pages 503–512, Jul 2001.
- https://2007.igem.org/wiki/index.php/Cambridge
- https://2012.igem.org/Team:HIT-Harbin
- T.J. Pollock, L. Thorne, Mechanism of bacitracin resistance in gram-negative bacteria that synthesize exopolysaccharides, J Bacteriol ; 176(20): 6229–6237, Oct 1994
- Cyflogic 1.2.1., available on:http://www.cyflogic.com/.
- F.M. Dekker, C. Kraaikaamp et al., ‘A Modern Introduction to Probability and Statistics’, Springer,January 2005.
- www.2007.igem.org/wiki/cambridge
 "
"